Molecular profiles and urinary biomarkers of upper tract urothelial carcinomas associated with aristolochic acid exposure
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Funding information: Croatian Science Foundation, Grant/Award Number: 04/38; Ministry of Science and Technology, Croatia, Grant/Award Number: 108-0000000-0329; National Institute of Environmental Health Sciences, Grant/Award Number: P01 ES004068; Fogarty International Center, Grant/Award Number: R03 TW007042; National Cancer Institute, Grant/Award Number: P30 CA016087; Laufer Family Foundation
Abstract
Recurrent upper tract urothelial carcinomas (UTUCs) arise in the context of nephropathy linked to exposure to the herbal carcinogen aristolochic acid (AA). Here we delineated the molecular programs underlying UTUC tumorigenesis in patients from endemic aristolochic acid nephropathy (AAN) regions in Southern Europe. We applied an integrative multiomics analysis of UTUCs, corresponding unaffected tissues and of patient urines. Quantitative microRNA (miRNA) and messenger ribonucleic acid (mRNA) expression profiling, immunohistochemical analysis by tissue microarrays and exome and transcriptome sequencing were performed in UTUC and nontumor tissues. Urinary miRNAs of cases undergoing surgery were profiled before and after tumor resection. Ribonucleic acid (RNA) and protein levels were analyzed using appropriate statistical tests and trend assessment. Dedicated bioinformatic tools were used for analysis of pathways, mutational signatures and result visualization. The results delineate UTUC-specific miRNA:mRNA networks comprising 89 miRNAs associated with 1,862 target mRNAs, involving deregulation of cell cycle, deoxyribonucleic acid (DNA) damage response, DNA repair, bladder cancer, oncogenes, tumor suppressors, chromatin structure regulators and developmental signaling pathways. Key UTUC-specific transcripts were confirmed at the protein level. Exome and transcriptome sequencing of UTUCs revealed AA-specific mutational signature SBS22, with 68% to 76% AA-specific, deleterious mutations propagated at the transcript level, a possible basis for neoantigen formation and immunotherapy targeting. We next identified a signature of UTUC-specific miRNAs consistently more abundant in the patients' urine prior to tumor resection, thereby defining biomarkers of tumor presence. The complex gene regulation programs of AAN-associated UTUC tumors involve regulatory miRNAs prospectively applicable to noninvasive urine-based screening of AAN patients for cancer presence and recurrence.
Abstract
What's new?
Ingestion of aristolochic acid (AA) via contaminated wheat-containing food products is a major cause of endemic neuropathy and urologic carcinogenesis in southeastern Europe. Here, using integrated multi-omics analysis, the authors identified molecular programs underlying upper tract urothelial tumors (UTUC) in patients in Southern Europe with past carcinogenic AA exposure. Analyses reveal associations between 89 miRNAs and 1,862 target mRNAs, with confirmation of UTUC-specific transcripts at protein level. AA-specific mutations in UTUC and deleterious mutations were uncovered at both gene and transcript levels. The findings suggest that tumors in the urinary tract can be monitored by urine miRNA signature.
Abbreviations
-
- AA
-
- aristolochic acid
-
- AAN
-
- aristolochic acid nephropathy
-
- AL-dA
-
- aristolactam-I-deoxyadenosine
-
- BAM
-
- binary alignment map
-
- bp
-
- base pair
-
- COSMIC
-
- Catalogue of Somatic Mutations in Cancer
-
- cRNA
-
- complementary RNA
-
- DAPPLE
-
- Disease Association Protein-Protein Link Evaluator
-
- dbSNP
-
- database of single nucleotide polymorphisms
-
- DNA
-
- deoxyribonucleic acid
-
- EN
-
- endemic nephropathy
-
- FDR
-
- false discovery rate
-
- GATK
-
- Genome Analysis Toolkit
-
- GO
-
- Gene Ontology
-
- GSEA
-
- gene set enrichment analysis
-
- HH
-
- hedgehog
-
- IARC
-
- International Agency for Research on Cancer
-
- IPA
-
- Ingenuity Pathway Analysis
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- MAF
-
- minor allele frequency
-
- miRNA
-
- microRNA
-
- mRNA
-
- messenger ribonucleic acid
-
- q-PCR
-
- quantitative polymerase chain reaction
-
- RCF
-
- relative centrifugal force
-
- RNA
-
- ribonucleic acid
-
- RNA-seq
-
- ribonucleic acid sequencing
-
- SAM
-
- sequence alignment map
-
- SBS
-
- single base substitution
-
- TLDA
-
- TaqMan Low Density Array
-
- TMA
-
- tissue microarray
-
- UTUC
-
- upper tract urothelial carcinoma
-
- WES
-
- whole exome sequencing
1 INTRODUCTION
Ingestion of Aristolochia herbs containing aristolochic acid (AA) leads to aristolochic acid nephropathy (AAN), marked by severe renal damage and cancer formation in the urinary tract, renal cortex, liver, bile duct and possibly other anatomical sites.1-4 Millions of people worldwide may develop AAN and associated cancers following exposure to AA (International Agency for Research on Cancer [IARC] Group I carcinogen5), particularly in Asia where Aristolochia is prescribed and used as traditional medicine2, 3, 6-8 and where the causal role for AA in urologic carcinogenesis had been supported by markedly reduced incidence rates of urologic cancers following a ban on the use of select AA-containing herbs.9 In the endemic nephropathy (EN) regions of Bosnia and Herzegovina, Bulgaria, Croatia, Romania and Serbia, AAN results from environmental contamination of wheat grains used for bread-baking, by the seeds of Aristolochia clematitis.10-12 As patients continue to be admitted for AAN-associated surgery decades after exposure, AAN is a serious public health problem in individuals with past exposure, including the farming families in the Balkan countries or the Belgian female patient cohort identified in the 1990s.13 AAN-associated upper tract urothelial carcinomas (UTUCs) develop years after exposure and often recur elsewhere in the urinary tract, including the urinary bladder.14, 15
Here we report a multiomics analysis of UTUCs from AAN patients from Croatia and Bosnia. We integrate global profiles of the deregulated messenger ribonucleic acid (mRNA) and microRNA (miRNA) transcriptomes, validate the key pathways at the protein level in the source tumors, investigate mutation spectra at both the deoxyribonucleic acid (DNA) and RNA levels and finally investigate urinary miRNAs as markers of urinary tract tumor presence. We describe broad UTUC-specific miRNA:mRNA networks, implicating massive deregulation of mutated oncogenes and tumor suppressor genes, effectors of cell-cell and cell-matrix remodeling, in regulators of chromatin dynamics, and inhibition of developmental morphogenic pathways and mesenchymal programs. We confirm the mutational signature of AA, present in both the DNA and RNA of UTUCs and identify a set of UTUC-specific miRNAs depleted in the patients' urine after surgical tumor resection. By generating a unique catalog of molecular alterations, we provide insights into complex candidate mechanisms of AA-associated UTUC carcinogenesis, and show suitability of urine miRNAs as noninvasive biomarkers of early recurrence of urothelial cancer in AAN patients.
2 MATERIALS AND METHODS
2.1 Patients and clinical data
Fifteen patients from the EN regions in Croatia and Bosnia and Herzegovina were hospitalized with urothelial cancer between 2009 and 2010. All patients had urothelial carcinoma, except for the patient B_AAN_27 (Tables S1 and S2) with sarcomatoid urothelial cancer. The patients underwent tumor surgery by radical nephroureterectomy or ureterectomy while two had also undergone cystectomy. The list of patients, their clinical data and information on samples used in molecular analyses is listed in Tables S1 and S2.
2.2 Nucleic acid isolation
DNA was extracted from fresh frozen samples of renal cortex, tumor and adjacent nontumor tissues and purified by standard phenol-chloroform extraction techniques. The quantity and purity of DNA were measured by NanoDrop 2000c (NanoDrop Technologies, Wilmington, DE).
Total RNA was isolated from fresh samples of tumor and adjacent normal tissue using miRNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions and stored in RNAlater. All samples were treated with DNase (Qiagen, Valencia, CA). RNA samples were stored at −80°C. The quantity and purity of RNA was measured using NanoDrop 2000c (NanoDrop Technologies, Wilmington, DE). Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) was used to determine the RNA integrity number which ranged between 8 and 10.
2.3 DNA adduct analysis and TP53 mutation analysis
The level of aristolactam-I-deoxyadenosine (AL-dA) DNA adducts in the renal cortex DNA (10-20 mg) was determined using 32P-postlabeling polyacrylamide gel electrophoresis as described previously.10 The UTUC TP53 mutation status was determined as previously described.16
2.4 Analysis of mRNA and miRNA abundance in tissues
Gene expression profiling was performed using the Human Genome U133 Plus 2.0 GeneChip (Affymetrix, Santa Clara, CA), measuring over 47,000 transcripts. The RNA was processed using 3′ in vitro transcription Express kit (Affymetrix, Santa Clara, CA), 500 ng of total purified RNA was transcribed into biotin-labeled complementary RNA (cRNA). Labeled cRNA probe was hybridized overnight to the microarrays. Following hybridization, arrays were washed, fluorescently tagged and scanned, using the Affymetrix GeneChip Scanner 3000 7G (Affymetrix, Santa Clara, CA). Usual quality measures and normalization for the Affymetrix GeneChip (3′/5′ ratios and trimmed mean normalization) were used. Raw data were imported into GeneSpring GX10 software (Agilent Technologies, Santa Clara, CA) and subjected to Robust Multichip Average (RMA) normalization. Differentially expressed genes were identified using the unpaired t test (P < .05 with Benjamini-Hochberg false discovery rate (FDR) multiple testing correction). Given the strict statistical parameters, the fold-change cutoff was set to at least 50% change in mRNA abundance.
MicroRNA profiling was performed by subjecting total RNA (500 ng) to high-capacity quantitative polymerase chain reaction (q-PCR) using a megaplex reverse-transcription primer set for ~754 human microRNAs (Sanger miRBase database v.14), analyzed by Applied Biosystems TaqMan Low Density Arrays (TLDA, TaqMan Array Human MicroRNA Panel A v2.1 and B v3.0) on the ABI 7900HT Sequence Detection System (Applied Biosystems, Waltham, MA), as described previously.17 The reactions were incubated in a 384-well plate at 50°C for 2 minutes and 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and at 60°C for 1 minute and were then held for 10 minutes at 72°C. The data were collected and processed using GeneSpring GX10 software (Agilent Technologies, Santa Clara, CA). The miRNA abundance was determined by the 2−Ct value formula which was followed by quantile normalization of all data. A union of unpaired t test (corrected P value < .05) and Pavlidis Template Matching (P < .01) yielded the differentially abundant miRNAs. Multiple-testing correction for t test was performed using Benjamini-Hochberg FDR. The fold-change cutoff was set at 1.5×, analogously to the differential mRNA abundance threshold.
2.5 Pathway analysis and bioinformatics
Gene Spring GX10 software (Agilent Technologies, Santa Clara, CA) was used for data management, analysis and visualization. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were used for annotation of specific genes and pathways. Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, Redwood CityCA, www.qiagen.com/ingenuity) based on the TargetScan high-confidence predictions and experimental validation was used to identify the mRNA targets of miRNAs (Table S5). The TargetScan algorithm in IPA searches for the presence of conserved 8-mer and 7-mer sites matching the seed region of a given microRNA, while the experimentally observed miRNA-mRNA relationships are based on the combined TarBase and miRecords database contents as well as the Ingenuity Knowledge Base containing information from published literature manually curated by the Ingenuity expert team. miRconnX was used to identify the top-ranking microRNA-mRNA relationships (http://mirconnx.csb.pitt.edu). NIH DAVID (Database for Annotation, Visualization and Integrated Discovery)18 and gene set enrichment analysis (GSEA), collections C2 and C519 were used for pathway analysis. Disease Association Protein-Protein Link Evaluator (DAPPLE) was used to determine and visualize significant physical connectivity among proteins encoded for by miRNA target genes, based on protein-protein interactions reported in public domain.20 Additional data visualization was done using CIRCOS version 0.6321 while MutSpec was used for mutation analysis22 and non-negative matrix factorization method for mutational signatures analysis using an R package.23, 24 Statistical analysis of the tissue microarray (TMA) data were performed using GraphPad Prism version 7.03 (GraphPad Software, La Jolla, CA).
2.6 Whole exome sequencing and RNA sequencing library preparation
Genomic DNAs were prepared for sequencing using the TruSeq LT DNA sample prep kit (Illumina, San Diego, CA). Genomic DNA was sheared using the Covaris S220 (Covaris, Woburn, MA) and the size distribution was verified using a high-sensitivity DNA assay on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Following end repair and 3′ adenylation of DNA fragments, a unique index adapter was ligated to each sample along with sequencing adapters. Fragment size selection was performing using a 2% agarose gel, and the excised bands were cleaned up using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). PCR amplification and subsequent Ampure XP (Beckman Coulter, Brea, CA) bead cleanup were performed as described in the TruSeq DNA prep protocol. The resulting library was verified for fragment size using the Bioanalyzer 2100, and quantified via q-PCR on the ViiA7 Real-Time PCR System (Applied Biosystems, Waltham, MA) using the KAPA Library Quantification kit (KAPA Biosystems, Wilmington, MA). The libraries were then enriched using the TruSeq Exome Enrichment kit (Illumina, San Diego, CA) prior to sequencing.
The quality and concentration of RNA were measured by NanoDrop 2000c (NanoDrop Technologies, Wilmington, DE). The starting amount was 3.5 μg of total RNA. Ribosomal RNA was removed using Ribo-Zero rRNA removal kits (Epicentre, Illumina, San Diego, CA). RNA-Seq libraries from depleted RNA were prepared using the ScriptSeq v2 RNASeq library prep kit (Epicentre, Illumina, San Diego, CA). In brief, after the addition of fragmentation buffer, the ribosomal ribonucleic acid-depleted sample is chemically fragmented. The fragmented RNA is reverse-transcribed using random-sequence primers containing a tagging sequence at 5′ ends. 3′ tagging is accomplished using the Terminal-Tagging Oligo. The di-tagged cDNA is purified by MinElute PCR Purification kit (Qiagen, Valencia, CA) and washed with EB buffer. cDNA is then amplified by limited-cycle PCR using PCR primer pairs that anneal to the tagging sequences of the di-tagged cDNA. Excess nucleotides and PCR primers are removed from the amplified double-stranded, adaptor-tagged cDNA, by Agencourt Ampure XP beads (Beckman Coulter, Brea, CA). The library products were then ready for sequencing analysis via Illumina HiSeq 2000 (Illumina, San Diego, CA).
2.7 RNA sequencing and data processing
RNA sequencing (RNA-seq) libraries were sequenced deeply (for number of generated reads, see Table S6), with either single-end for UTUC_20 or pair-end for UTUC_03 and UTUC_18 (at 51 bp read length) using the Illumina HiSeq 2000 sequencer.
All reads in FASTQ format were generated using Illumina CASAVA version 1.8 by trimming of 2 bp from both the 5′ end and 3′ end and yielding 47 bp sequence reads. All passed filtered and qualified reads were aligned to human genome version GRCh37 (hg19) using eland software of CASAVA version 1.8 with default settings. Binary alignment map (BAM) files and single base substitution (SBS) calls were generated by variant detection and counting software of CASAVA version 1.8. The bulk sequencing metrics for the RNA-seq are listed in the Table S6.
After removing duplicate reads, postprocessing steps have been applied to the raw SBS calls in order to obtain SBSs with confidence. SBSs with coverage less than 10× were removed. SBSs with <10% reads supporting for variant or >90% reads supporting for reference nucleotide per site were also filtered out. Last, we filtered out all SBSs that match the common database of single nucleotide polymorphisms (dbSNP) version 137 with minor allele frequency (MAF) ≥1%.
2.8 Whole exome sequencing, data processing and variant calling
Whole exome sequencing (WES) libraries were sequenced (51-bp single-read) using Illumina's HiSeq 2000. FASTQ reads were generated using Illumina CASAVA version 1.8 by trimming of 2 bp from both the 5′ end and 3′ end and yielding 47 bp sequence reads. All pass-filtered and qualified reads were aligned to human genome version GRCh37 (hg19) in sequence alignment map (SAM) format using BWA with default parameter settings. SAM tools were used to convert SAM files into BAM files. Mapped reads in the raw BAM files were then realigned locally, duplicates marked and base pair score recalibrated using Genome Analysis Toolkit (GATK) version 1.3 and Picard version 1.59/1.79. The UnifiedGenotyper variant caller (within GATK version 1.3) was used to call SBS. The bulk sequencing metrics for the WES analysis are listed in the Table S6.
After removing duplicate reads, several postprocessing steps have been applied to the raw SBS calls in order to obtain SBSs with confidence. SBSs with coverage less than 10× were filtered out. SBSs with <10% reads supporting for variant or >1% reads supporting for background control data per site were also removed. Finally, all SBSs that match the common dbSNP version 137 with MAF ≥1% were removed. Annotations of somatic SBS were performed by ANNOVAR and their functional impact was evaluated by the ANNOVAR-embedded SIFT and PolyPhen-2 modules.
2.9 TMA analysis
Immunohistochemistry on TMAs was performed following standard recommendations.25 TMAs were constructed from formalin-fixed paraffin-embedded blocks using three cores of tumor tissue and three cores of normal urothelium per sample and internal controls (skin, breast, muscle, kidney, thyroid, lymph node), all 1 mm in diameter. Once constructed, the blocks were sectioned using microtome. Slides were deparaffinized, then rehydrated before staining. For the antigen retrieval, slides were carried using Trilogy solution (Cell Marque, Rocklin, CA, ref 920P-04) or Citrate Buffer pH 6 (Vector Laboratories, Burlingame, CA, ref H3300) in a steamer (95°C) or by microwave, and maintained at this temperature for 10 minutes followed by a 20 minutes cool-down period at room temperature. Endogenous peroxidases were blocked with 3% H2O2 in methanol for 5 minutes. Slides were rinsed in distilled water, and immersed in wash buffer before immunostaining. Sections were processed by incubation with primary antibodies overnight at 4°C; incubation with the secondary antibody for 30 minutes at room temperature, signal amplification with ABS kit and visualization using 3,3′-diaminobenzidine solution for 15 minutes. Slides were subsequently counterstained with hematoxylin, dehydrated, mounted with permanent mounting medium, and coverslipped for visualization.
Antibodies that were used included (listed as the protein name, and in parentheses the antibody clone, type, company and dilution used): Vimentin (N/A, rabbit pAb, Epitomics, Burlingame, CA, 1:2000), Actin (C-4, mouse mAb, MP Biomedicals, Santa Ana, CA, 1:2000), ki67 (MM1, mouse mAb, Novocastra, Leica Biosystems, Wetzlar, Germany, 1:50), p53 (1C12, mouse mAb, Cell Signaling Technology, Danvers, MA, 1:10 000), p21 (Ab-1, mouse mAb, Calbiochem, San Diego, CA, 1:1000), MSH2 (Ab2, mouse mAb, Calbiochem, San Diego, CA, 1:500), IDH1 (H09, mouse mAb, Dianova, Hamburg, Germany, 1:500), PCNA (N/A, mouse mAb, Dako, Glostrup, Denmark, 1:1000), 3meH3K27 (K27, mouse mAb, Abcam, Cambridge, United Kingdom, 1:2000), EZH2 (AC22, mouse mAb, Cell Signaling Technology, Danvers, MA, 1:2000), SMAD3 (N/A, rabbit pAb, Abcam, Cambridge, United Kingdom, 1:2000), pSMAD3 (phospho S208) (N/A, rabbit pAb, Abcam, Cambridge, United Kingdom, 1:2000), S100A4 (N/A, rabbit pAb, Dako, Glostrup, Denmark, 1:2000), TWIST (2C1a, mouse mAb, Abcam, Cambridge, United Kingdom, 1:500), PTEN (A2B1, mouse mAb, Santa Cruz Biotechnology, Dallas, TX, 1:4000), GSK3-beta (N/A, rabbit pAb, Cell Signaling Technology, Danvers, MA, 1:1000), Phospho-GSK3B (Ser9, rabbit pAb, Cell Signaling Technology, Danvers, CA, 1:750), Cyclin D1 (SP4, mouse mAb, Neomarkers, Portsmouth, NH, prediluted), E-cadherin (NCH-38, mouse mAb, Dako, Glostrup, Denmark, 1:200), Brg1 (N-15, polygoat Ab, Santa Cruz Biotechnology, Dallas, TX, 1:2000), PAI (H135, rabbit pAb, Santa Cruz Biotechnology, Dallas, TX, 1:1000), BRCA1 (D-9, mouse mAb, Santa Cruz Biotechnology, Dallas, TX, 1:1000), CDC25A (F-6, mouse mAb, Santa Cruz Biotechnology, Dallas, TX, 1:1000), Nibrin (Ab-1, rabbit pAb, Oncogene Research Products, San Diego, CA, 1:1000), ATM (233, rabbit pAb, Serotech, Oxford, United Kingdom, 1:500).
Immunoreactivity of the stained slides was visualized by light microscopy and digitally by Leica SCN400 Scanner (Leica Biosystems, Wetzlar, Germany). The quantification of the results was done manually using intensity score ranging 0 to 3 and percentage of the stained cells on high magnification. The final score was calculated by multiplying intensity score and percentage on all three biological replicates. Nonparametric Mann-Whitney test was used to evaluate differences between the pathologic review scores of the tumor and normal tissues.
2.10 Profiling of urinary miRNAs
Spot urine samples of additionally involved five UTUC patients were collected prior and at 6 weeks and 3 months after the surgery; 15 mL of each sample was used for further analysis. After centrifugation for 10 minutes at 2,000 RCF, urine supernatant containing cell-free nucleic acids including exosomic miRNAs was kept in long-term preservative (NorgenBiotek, Thorold, ON, Canada). Supernatant was subjected to additional high-speed centrifugation (30 minutes at 10,000 RCF) as to fractionate exosomic miRNAs from cellular debris and was spiked by 10 pM of control miRNA (derived form Arabidopsis thaliana). The miRNA content from exosomes was isolated using Urine Exosome RNA Isolation Kit (NorgenBiotek, Thorold, ON, Canada). The miRNA analysis was performed using TaqMan Low Density Array (TLDA, TaqMan Array Human MicroRNA Panel A v2.1 and B v3.0) high-throughput q-PCR as described above for the UTUC and nontumor control tissues.
3 RESULTS AND DISCUSSION
3.1 miRNA:mRNA networks and target validation by TMAs
Integrated molecular profiling of UTUC tumors and unaffected tissues (Tables S1 and S2) identified consistent gene regulation aberrations. All patients had documented history of exposure to AA, with eight cases testing positive for the aristolactam-I-deoxyadenosine DNA adducts and/or A:T>T:A mutation(s) in the TP53 gene (Table S2). Differential analysis effectively segregated the UTUC and unaffected tissues (Figure 1A,B) involving 5,438 mRNAs (2,021 elevated, 3,417 reduced, see Table S3) and 138 miRNAs (74 elevated, 64 reduced, Table S4) significantly deregulated in UTUC and often comodulated when originating from discrete genomic clusters/families (Figure S1).
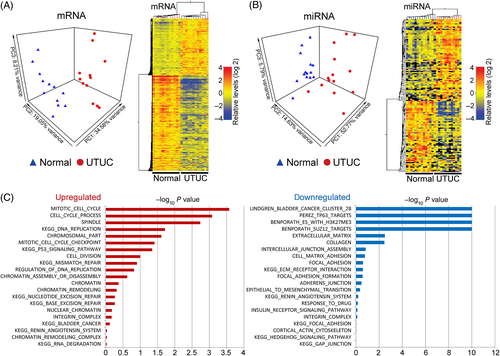
Increased UTUC mRNAs implicated cell cycle, DNA replication, TP53 signaling, DNA repair, chromatin remodeling and bladder cancer (Figure 1C). Downmodulated UTUC transcripts implicated bladder cancer, silencing by PRC2/3 via histone H3K27 methylation, cell-cell and cell-matrix interactions and epithelial-to-mesenchymal transition (Figure 1C). Collectively, the AAN UTUCs manifest with increased cell proliferation, DNA repair, chromatin remodeling/silencing and reduction of cell motility.
Highest abundance of UTUC-specific miRNAs were observed for the oncogenic miR-17-92 family, the oncogenic and stress response-associated miR-21, and the miR-182 and miR-183 cluster. We also observed prominent upregulation of the antimetastatic miR-200 family. Conversely, tumor suppressor miRNAs, including let-7c, the miR-143/miR-145 cluster and miR-23b, were downmodulated (Table S4). Thus, the UTUC-specific miRNAs indicate roles in tumor growth while limiting its invasive and metastatic capacity.
Networks of miRNAs inversely correlated with their predicted, experimentally validated mRNA targets implicated 25 and 14 miRNAs in high-score up- and downmodulated nodes, respectively (Figure S2A). Stringent TargetScan prediction identified a total of 45 upmodulated miRNAs associated with 1,159 downmodulated mRNAs, and 44 downmodulated miRNAs associated with 703 upmodulated mRNAs (Figure S2 and Table S5). The mutual interconnectivity of the miRNA:mRNA network components is summarized in Table S5. Pathway analysis of the UTUC upregulated miRNA targets identified roles in ATM, BRCA2 and CHEK2 networks and cell-cycle deregulation (Figure S2B). Among the prominent miRNA-repressed UTUC, programs were targets of EZH2, SUZ12 and methylated H3K27, consistent with the silencing role of PRC2/3, and also promigratory components of the transforming growth factor-β (TGF-β) and integrin signaling and extracellular matrix remodeling (Figure S2B).
TMA analysis confirmed the UTUC deregulation of cell cycle manifesting by elevated Ki67 (MKI67), PCNA and cyclin D1 (CCND1) proteins (Figure 2). The UTUC-specific DNA damage response and DNA repair manifested by elevated TP53 mRNA/protein (not shown), and by increased mRNA and proteins of the TP53 target p21 (CDNK1A) and BRCA1 (Figure 2). We observed UTUC-specific decrease of the tumor suppressor PTEN, increased levels of EZH2 and augmented UTUC epithelial phenotype (decreased vimentin (VIM), increased E-cadherin (CDH1)). DAPPLE analysis suggested extensive physical connectivity and orchestration among proteins corresponding to the derepressed UTUC genes collectively targeted by 44 reduced miRNAs (Figure S3).

Dysregulation of cancer genes involved 89 oncogenes and 20 tumor suppressor genes (Table S9). The upregulated oncogenes MYC, FGFR3, HRAS and KRAS and downregulated tumor suppressor PTEN (also depleted in UTUC as protein; Figure 2) and PTCH1 were likely coordinated by miRNAs (Figure S4A) with elevated MYC representing a central node.
The upstream miRNA regulators of the PRC2/3 silencing complex included let-7c, miR-1, miR-23b, miR-143, miR-145 and miR-150 (Figure S4B). Additional UTUC-enriched chromatin regulators included lysine demethylases (KDMs) of diverse specificities, IDH1 (implicated in widespread changes in histone and DNA methylation in cancer) and methyl-CpG binding domain protein MBD1. The miRNAs regulating these gene products (including tumor suppressors miR-143, miR-145, miR-23b and let-7c) were collectively depleted in UTUCs. Next, the downregulated epigenome regulators HDAC9 and DNA methyltransferase DNMT3A are targets of elevated miR-21 and the miR-17-92 and miR-200 families (Figure S4B).
The repression of promigratory targets of ZEB1 and ZEB2, inhibitors of E-cadherin (CDH1), correlated with elevated miR-200bc/429 and miR-200a/141 families (Figure S5). ZEB1 and ZEB2 were reduced in UTUC while E-cadherin was increased as mRNA and protein (Figures 2 and S5). TGF-β signaling, a positive regulator of both ZEB1 and ZEB2,26 was downregulated in UTUC, including the TGF-β receptors TGFBR1 and TGFBR2, and SMAD3 and phospho-SMAD3 proteins (basal and activated transducer of TGF-β signaling, respectively) (Figure S5).
UTUC-specific repression of WNT manifested as reduced components of the KEGG hsa04310:Wnt signaling pathway and GO:0016055:Wnt receptor signaling pathway categories, including the ligands WNT2B, WNT4, the frizzled members of the receptor complexes, signal transducer AXIN and the downstream target LEF1. Additionally, WIF1 (WNT inhibitory factor 1), DKK2 (Dickkopf WNT signaling pathway inhibitor 1) and SFRP (secreted frizzled-related protein 1) were also downregulated, consistent with observed, inversely modulated miRNAs targeting these Wnt components. Furthermore, TMA analysis identified both the glycogen synthase kinase 3 beta (GSK3β) protein and its phosphorylated form as downregulated in tumors (not shown). Similar to suppression of TGF-β, the downmodulation of the WNT, another effector of epithelial-to-mesenchymal transition, may further contribute to the nonmetastatic behavior of UTUCs.
We next observed UTUC-specific downmodulation of PTCH1 and GLI2, the canonical receptor/tumor suppressor protein and a signal transducer in the hedgehog (HH) signaling pathway, and further HH repression was identified by GSEA (Figure 1C). The deregulated miRNAs targeting PTCH1 and GLI2 are shown in Table S5. The downregulation of the HH pathway in UTUC suggests a reduction in its tumor suppressor function.
Affected bladder cancer programs involved deregulated cancer genes and upstream miRNA regulators (Figures 1C and S2B). The KEGG bladder cancer pathway hsa05219 was enriched with upregulated HRAS, CDKN1A, E2F3, CCND1, FGFR3, KRAS, VEGFA and CDH1. The networks with key downregulated miRNAs putatively targeting these genes are summarized in Figure S4 and in Table S5. We observed significant enrichment (48 of 141 [34%], P value 4.12E−32, FDR q-value 1.42E−30) of downmodulated UTUC genes previously observed as depleted in bladder cancer (GSEA set M1189627). We also identified deregulated bladder cancer-specific miRNAs: upregulated miR-103 and downmodulated miR-99a, miR-100, miR-125, miR-145 and miR-143 consistent with observations from papillary stage Ta bladder carcinoma studies.28 Additionally, miR-21 and miR-221 elevated in invasive bladder cancer (KEGG MicroRNAs in cancer) were upregulated in UTUCs.
In sum, our study identifies broad gene regulation programs in AA-associated UTUCs, together with a signature of 138 deregulated miRNAs including known cancer miRNAs.29 This data set partly overlapped with a smaller 40-miRNA set identified previously in AAN/EN associated UTUCs,30, 31 including the shared miR-205-5p, miR-1290 (also observed in our study as an urinary tumor marker) and miR-127-3p. We built mRNA:miRNA networks that generally characterized the phenotype of UTUC by revealing deregulation of cell cycle, DNA damage response and repair, chromatin regulation, bladder cancer, developmental signaling and a number of oncogenes and tumor suppressor genes. Importantly, the validation at the protein level using TMAs showed considerable concordance with the mRNA expression of key miRNA targets.
3.2 UTUC-specific DNA mutational signatures and RNA mutation spectra
WES was performed in three representative UTUC and matched nontumor renal cortex DNAs. A total of 2775 UTUC-specific somatic SBSs were identified (Figure 3A and Table S6) with varying SBS counts across samples (Figure 3A). However, the distribution of SBS classes remained consistent: 23% to 25% synonymous, 62% to 65% nonsynonymous, 7% to 10% introducing a stop codon and 5% to 8% affecting mRNA splicing (Figure 3B). Full description of UTUC-specific DNA mutations and their predicted impact on protein function is summarized in Table S7. The UTUC exome data showed a predominance of A:T>T:A transversions (56% of all mutation types) (Figure 3A and Table S7) with a prominent transcription-associated strand bias (2.7- to 3.5-fold enrichment on the nontranscribed DNA strand) (Figure 3A). The distributions of the A:T>T:A mutations into synonymous, nonsynonymous, stop-gain and affecting splicing matched that of all SBS and the impact of mutations was predicted as deleterious for most nonsynonymous A:T>T:As (Figure 3B). We identified distinct mutational signatures corresponding to the Catalogue of Somatic Mutations in Cancer (COSMIC) signature SBS22 (AA), signatures SBS2/13 (APOBEC) and signature SBS1 (age, clock-like, with a possible admixture of clock-like SBS5; Figure 3C), in keeping with previous reports.32, 33 The identified signature of AA exposure was nearly identical (cosine similarity of 0.97-0.99, with no remarkable peak differences) to the AA signatures determined previously by whole exome sequencing of UTUC tumors from East Asia and Southern Europe,32, 34, 35 including the consistent and significant transcription strand bias of A:T>T:A transversions accumulating prominently on the nontranscribed DNA strand (Figure S6).
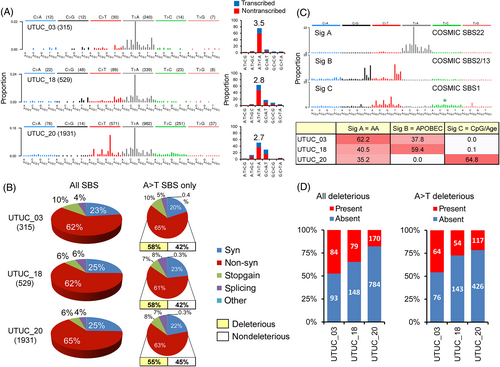
To examine the somatic alterations in the tumor RNA, deep whole transcriptome sequencing (RNA-seq) was performed on the UTUC_03, UTUC_18 and UTUC_20 samples used for WES. In the corresponding mRNA samples, we observed the respective counts of 143, 148 and 344 of the WES-identified SBS (Table S6). The SBS type distribution (34% synonymous, 63% nonsynonymous, 2% stop-gain, 1% affecting splicing) was consistent across all UTUC RNA samples (data not shown). The number of deleterious mutations detected in both DNA and mRNA was 79 to 170 (range across samples) for any mutation type, and 54 to 117 for A:T>T:As (Figure 3D). The mutations shared between DNA and RNA are listed in Table S7. Thus, a large proportion of deleterious DNA mutations in UTUCs are propagated at the mRNA level, and the AA-specific A:T>T:As are likely to have a considerable impact on normal protein function. Integrated exome and RNA-seq of a subset of UTUC thus revealed the presence of the AA mutational signature SBS2232, 34, 35 and extended the limited available data35 on the propagation of signature-like A:T>T:A transversions at the RNA level, by adding hundreds of mutated transcripts, including those deregulated in tumors. Despite the constrained scale, such observation is consistent with a recently published in silico prediction of an increased neoantigen load based on genome-scale DNA sequencing of AA signature-positive UTUCs of Chinese patients. The observed increased levels of infiltrating lymphocytes36 can help select the high mutation-load UTUCs linked to AA exposure for efficient targeting by immunotherapy.36-38 Additional studies are warranted to further evaluate the impact of AA-directed mutagenesis on the transcriptome and proteome functions, and the potential clinical utility.
3.3 Recurrently mutated cancer genes
Analysis of A:T>T:A counts as a function of chromosome size yielded Pearson product-moment correlation coefficients ranging between R = 0.85 and R = 0.89, recapitulating previous findings on the stochastic accumulation of A:T>T:A transversions in the genome.32 However, a set of 63 cancer genes were recurrently mutated (Figure S7 and Table S8). Eight of these (AHNAK, ATM, ARID1A, MKI67, MLL2, PLEC, SYNE1 and TP53) are established cancer driver genes39 previously reported as recurrently mutated in AAN UTUCs from Taiwan and from EN regions, although these previous studies focused only on mutations in the DNA.32, 34 Recurrently mutated ARID1A, ATM, FAT4, PTPRC, TP53 and UBR5 are listed in the COSMIC Cancer Gene Census.40 Interestingly, a number of recurrently hit genes (ATM, EPHA3, HMCN1, INPP5B, LYST, MLL2, RELN, TLR6 and USH2A) were mutated by multiple nonsynonymous A:T>T:A transversions in the same tumor. Finally, 35 driver mutations corresponding to 27 recurrently mutated genes (with a subset of 22 genes harboring 27 A:T>T:A transversions) were also detected at the level of mRNA (Figure S7 and in Table S8), a result adding a new dimension to the previous reports of recurrently mutated driver genes in AAN-associated UTUC.32, 34
3.4 Urinary miRNA as biomarkers of tumor presence
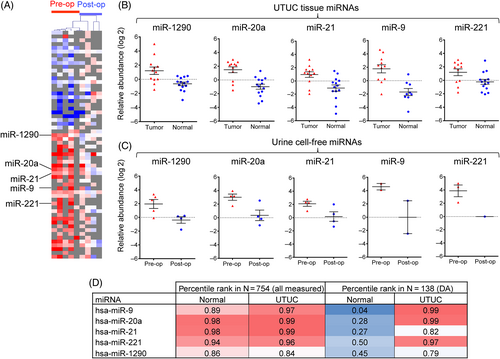
We next profiled cell-free miRNAs in the urine of five AAN UTUC patients (Table S10), to explore the utility of this biomaterial for determining the tumor presence in the urinary tract, and by extension for monitoring the tumor recurrence often associated with AAN cancers.15, 41, 42 MiRNA profiling of the presurgery and postsurgery urine supernatants revealed a signature of 60 differentially modulated miRNAs (35 high and 25 low in the presurgery urine samples) (Figure 4 and Table S11) of which five cancer-related miRNAs (miR-20a, miR-9, miR-21, miR-221 and miR-1290) were also enriched in primary UTUCs (Figure 1 and Table S4). This subset of urinary miRNAs exhibits high and stable relative abundance rank in both the normal (median percentile of 0.94) and in the tumor tissues (median percentile of 0.97), while it undergoes a major increase in its relative abundance rank in the context of the differentially abundant miRNAs in the UTUC compared to normal tissue (median percentile increase from 0.28 to 0.97; Figure 4D). We propose that due to this shift in abundance in tumors, this 5-miRNA signature in the urine could be UTUC-derived and more easily detectable at elevated levels prior to tumor resection. This is in contrast to other miRNAs that are more abundant in the presurgery urine but either not detected as present or as or differentially modulated in tissues. The signature may therefore serve as a noninvasive biomarker for clinical screening and monitoring of tumor presence and cancer recurrence, upon proper validation for robustness in further studies conducted in an extended set of patients. A number of studies performed on bladder cancer patients similarly implicated urine miR-21 and/or miR-221 as markers of tumor presence,43-45 and the promise of miRNAs as urinary tumor biomarkers have been extensively discussed.46-49 To our knowledge, our study is the first to report a specific urine miRNA signature in the context of recurrent upper tract urothelial cancers driven by exposure to AA, a highly potent environmental and iatrogenic carcinogen. As such, it complements recent studies demonstrating the presence of AA mutational signature as a readout of exposure in cell-free DNA in urine,36 or the sensitive detection of urothelial cancer-specific driver mutations by targeted resequencing.38, 50 Such noninvasive tests have to be further evaluated for the potentially best-suited application in clinical monitoring of AA-exposed individuals without cancer and of AA-associated cancer recurrence in the urinary tract.
4 CONCLUSIONS
While various modes of toxicities of AA have recently been extensively reviewed,51 the study presented here identifies specific and complex mRNA:miRNA networks of the AAN-associated UTUC, supported by adduct analysis, with components of key affected pathways further validated by high-throughput tissue microarray immunohistochemistry. Sequencing of patient exomes and transcriptomes confirmed a burden of somatic A:T>T:A transversions in UTUC and provided insights into deleteriously mutated expressed genes. Next, we identified a subset of UTUC-specific urinary miRNAs potentially applicable as noninvasive biomarkers in screening and monitoring the AAN patients for urothelial carcinogenesis. Previous studies focusing on singular-level omic analyses of AA-associated UTUCs have been recently systematically reviewed,52 yet the integration of various layers of molecular analyses in a single study has not yet been reported. We thus propose our work to be unique in design, scope and comprehensiveness, while offering new insights into the AA UTUC biology and a basis for the development of an important molecular cancer-screening tool.
ACKNOWLEDGMENTS
In memoriam of Dr Frederick Miller, to whom we are indebted for expert pathology evaluations. We thank Dr Matej Knezevic, Dr Branko Brdar, Dr Elisabetta Kuhn and Christine Carrera for expert assistance with sample collection and processing. Expert technical assistance provided by Andrea Fernandes, Gyongyi Mihalyne and Vincent Cahais is gratefully acknowledged. Funding for this study was provided by the U.S. NIH/National Institute of Environmental Health Sciences (grant P01 ES004068), the U.S. NIH Fogarty International Center (R03 TW007042), the Ministry of Science and Technology, Croatia (108-0000000-0329) and Croatian Science Foundation (Grant 04/38). Kathleen G. Dickman and Arthur P. Grollman gratefully acknowledge the financial support provided by Marsha and Henry Laufer (Laufer Family Foundation). The NYU Genome Technology Center received support from the U.S. NIH/National Cancer Institute (P30 CA016087).
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ETHICS STATEMENT
The study protocols included the patients' informed consent and were approved by the IARC Ethics Committee and the Institutional Review Boards of the participating institutions.
Open Research
DATA AVAILABILITY STATEMENT
The mRNA and miRNA profiling data were deposited to the NCBI Gene Expression Omnibus (GEO) public database (www.ncbi.nlm.nih.gov/gds) under the Series reference ID GSE166912. The WES and RNA-seq data generated in this study were deposited to the European Genome-Phenome Archive (EGA) under the Study ID EGAS00001005363. The list of all somatic SBSs identified by WES and their overlap with the RNA-seq profiling data is available in the Table S7.




