Genetic variants in SLC22A3 contribute to the susceptibility to colorectal cancer
Abstract
Previous a genome-wide association study (GWAS) of colorectal cancer in Japanese population has identified a risk region at the chromosome 6q26-q27 associated with colorectal cancer risk. However, the causal gene at this locus remained unclear. In our study, we enrolled a total of 14 candidate functional single nucleotide polymorphisms (SNPs) at 6q26-q27 (318 kb), and then genotyped them by TaqMan method in a Chinese population including 1,147 colorectal cancer cases and 1,203 controls. Among that, 5 SNPs were identified statistical association with colorectal cancer risk by logistic regression analysis. Of which, SNP rs420038 G > A in SLC22A3 was related to decreased risk of colorectal cancer (adjusted odds ratio (OR) = 0.79, 95% confidence interval (CI) = 0.67–0.94, p = 0.007), and also associated with lower expression of SLC22A3 (p = 0.040) using expression quantitative trait loci (eQTL) analysis. Moreover, by the luciferase assays, we found that compared to the G allele of rs420038, the A allele could suppress the activity of the promoter in SLC22A3. Furthermore, the expression of SLC22A3 was significantly higher in colorectal cancer tissues than that in paired normal tissues (p < 0.001). Meanwhile, the phenotypes of proliferation, migration, invasion, cell cycle and apoptosis of colorectal cancer cell were significantly affected by SLC22A3 in vitro. Our results revealed a novel susceptible locus, rs420038 in SLC22A3, which may be involved in colorectal cancer development and progression.
Abstract
What's new?
The identification of genes or loci associated with colorectal cancer (CRC) susceptibility can facilitate the discovery of molecular pathways underlying CRC development and progression. Here, the authors investigated a risk region at chromosome 6q26-q27, which previously was linked to CRC susceptibility in a Japanese population. Analyses of candidate functional single nucleotide polymorphisms (SNPs) at 6q26-q27 revealed a novel functional SNP, rs420038 G>A, in the SLC22A3 gene. While expression of SLC22A3 was elevated in CRC tissues, the novel SNP was associated with decreased CRC risk in a Chinese population. The A allele of rs420038 significantly suppressed SLC22A3 promoter activity.
Introduction
Colorectal cancer is one of the most common digestive system malignant tumors that endangers human health in the world. The incidence of colorectal cancer has obvious regional and ethnic differences and is significantly higher in the United States than in Asia.1 In China, colorectal cancer ranked the third highest in the incidence rate, while the mortality rate ranked fifth.2 The environmental risk factors for colorectal cancer include sedentary living, staying up all night, poor eating habits and so on.3-5 However, despite exposure to the same harmful environmental factors, the occurrence of colorectal cancer varies from person to person, indicating that the occurrence of colorectal cancer is a result of a combination of environmental and genetic factors. Previous studies have found that colorectal cancer patients with a family history of the disease could have twice the risk of those without a family history.6, 7 They also found that genetic factors accounted for 35% of the risk of developing sporadic colorectal cancer. Additionally, many case–control studies showed that genetic variations were closely related to colorectal cancer, among which single-nucleotide polymorphisms (SNPs) were one of the most common type.8, 9
Recently, multiple genome-wide association studies (GWASs) have reported several genes or loci associated with colorectal cancer susceptibility.10-14 For the Asian population, SNP rs7758229 located in the chromosome 6q26-q27 region was the first discovered to be associated with the risk of distant colorectal cancer in the Japanese population.15 However, in our previous validation study within the Chinese population, we found that rs7758229 showed no relationship with colorectal cancer risk.16 We attributed the possible causes of inconsistencies to differences in the genetic background and minor allele frequency (MAF) among races. Therefore, it is reasonable that different effects of SNPs on disease exist in different races. The effect of rs7758229 on the Japanese population found by GWASs might only be the chain embodiment of the adjacent pathogenic sites, and the real pathogenic sites might be located in other sites of the chromosome 6q26-q27 region.17-20 Therefore, we proposed that there could be genetic variation associated with susceptibility to colorectal cancer in this region.
Here, we conducted a comprehensive analysis on the chromosome 6q26-q27 region to further clarify the potential causal variants in this region related to colorectal cancer risk. our study will provide important clues to screen patients with high colorectal cancer risk, implementing individual treatment and assessing the prognosis of colorectal cancer patients in the future, as well as further elucidating the pathogenesis of colorectal cancer.
Methods
Patients or participants
All subjects of our study were from the Chinese Han individuals. A case–control study was performed with enrolling 1,147 colorectal cancer cases and 1,203 controls. All cases were diagnosed and confirmed by pathology at The First Affiliated Hospital and Nanjing First Hospital of Nanjing Medical University, without other tumor history. The controls matched age and sex to cases were randomly selected among the population for physical examination in the same geographical region without genetically related to the cases. Details of the study participants have been demonstrated previously.21 All subjects recruited for the study signed informed consent and provided 5 ml of peripheral blood. A consolidated face-to-face questionnaire survey was used for the case and control groups. All subjects who had smoked daily for over 1 year were considered smokers, and the remaining subjects were considered nonsmokers. Individuals who had consumed one or more glasses of alcohol weekly for at least 1 year were considered drinkers, and the remaining subjects were considered non-drinkers. The study protocol was performed in accordance with the Institutional Review Board of Nanjing Medical University.
Selection strategies of SNPs
Based on the genotyping data of CHB and JPT population from1000 Genomes Project Database (http://www.1000genomes.org), the strategies of selecting SNPs were as follows: (i) SNPs locating in the chromosome 6q26-q27 region (GRCh37/hg19, http://genome.ucsc.edu/); (ii) SNPs with MAF > 0.05, p value of Hardy–Weinberg Equilibrium >0.05 and call rate > 95%; (iii) tag SNPs using the Haploview 4.0 software according to linkage disequilibrium (LD) of r2 > 0.6; (iv) SNPs with high function scores referring to the online tools of HaploReg v4.1 (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php).
TaqMan genotyping
We derived genomic DNA from EDTA-venous blood by the Qiagen Blood Kit (Qiagen). The samples were genotyped by TaqMan assays with the ABI 7900HT Real-time PCR System (Applied Biosystems). All the primers and probes for the TaqMan assay are listed in Supporting Information Table S1. The reaction conditions of the assay were followed by manual. After the reaction was completed, the fluorescence was detected, and the amplification curve and genotyping data were derived by Sequence Detection System version 2.4 (SDS 2.4) software. Two samples of 1,203 controls failed to be genotyped due to the quality of them.
Luciferase activity
The 1,000-bp-containing rs420038 A or G alleles of the enhancer sequence and SLC22A3 promoter region were synthesized. Both of them were cloned into the pGL3-basic Vector (Promega) by the NheI and XhoI restriction sites and confirmed by DNA sequencing. In luciferase assays, HCT116 and SW620 cells were plated onto 24-well plates and transfected with plasmids above by Lipofectamine 2000 (Invitrogen). As an internal reference, 10 ng pRL-SV40, which contained the Renilla luciferase gene, was co-transfected with all plasmids. All of the transfections were performed in triplicate. After transfection for 24 h, the cells were collected for luciferase activity measurement using a Dual-Luciferase Reporter Assay System (Promega). The relative luciferase activity was compared by the two-sided t-test after normalized to Renilla luciferase.
Quantitative RT–PCR
Four cell lines of colorectal cancer (i.e., HT29, DLD1, HCT116 and SW620) were purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China) and maintained under standard conditions. Authenticity of any human cell lines used in our study has been proven by DNA profiling. Cell line authentication was conducted by the Genetic Testing Biotechnology Corporation (Suzhou, China) and the KeyCen BioTech (Nanjing, China) by short tandem repeat (STR) markers. The authentication results were compared to those of the American Type Culture Collections (ATCC) cell bank. No mycoplasma contamination was detected. Total RNA was isolated from cells and tissue samples by TRIzol (Invitrogen) and was quantified by ultraviolet spectrometry. The relative mRNA expression level of SLC22A3 and the internal reference genes were detected using the ABI 7900 Real-Time PCR system (Applied Biosystems). ACTINB was selected as endogenous control using geNorm39. The primer sets designed are shown in Supporting Information Table S1.
Western blotting
Western blot assays were performed as standard procedures. The primary antibodies used were monoclonal rabbit anti-SLC22A3 (1:1,000; ab151698; Abcam) and rabbit anti-b-actin (1:1,000; 13E5; Cell Signaling Technology). The secondary antibody was anti-rabbit HRP (1:1,000; BS13278, Bioworld Technology). The immune complexes were detected by enhanced chemiluminescence (Cell Signaling Technology).
Construction and transfection of overexpression and knockdown of SLC22A3
To overexpress and knock down of SLC22A3 in colorectal cancer cells, one SLC22A3 cDNA was cloned into pEGFP-C1 Vector (Clontech) by XhoI/BamHI restriction sites and confirmed by DNA sequencing. Two independent siRNAs were synthesized. The sequences of two siRNAs are shown in Supporting Information Table S1. The effect of SLC22A3 overexpression and knockdown was determined by quantitative RT–PCR and Western blotting. Lipofectamine 2000 transfection reagent (Invitrogen) was used in transfection. All the plasmid sequences were confirmed by sequencing.
Cell proliferation and cell death assays
Cell Counting Kit-8 (CCK-8; Dojindo) was performed at various time intervals by an Infinite M200 spectrophotometer (Tecan). The absorbance at an optical density of 450 nm represented cell proliferation. The cell cycle assay was performed using a FACS Calibur flow cytometer (Beckman Coulter) when cells were fixed with 75% ethyl alcohol and stained with propidium iodide. Flow cytometry was adopted to detect cell apoptosis when the cells were dealt with the Annexin V-FITC apoptosis detection kit (Invitrogen). All of our experiments were independently conducted three times at least, and the data were expressed as means ± standard deviation. Statistical comparisons were analyzed using two-sided t-test.
Statistical analysis
The association between the selected SNP and colorectal cancer risk was evaluated in the additive, dominant and recessive model after adjusted for age and sex by logistic regression. We assessed the correlations between each SNP genotype and the mRNA expression level through the analysis of linear model in expression quantitative trait loci (eQTL) analysis. The software of SAS 9.4 (SAS Institute) and Stata 10.0 (StataCorp LP) were both used.
Results
SNPs selection from chromosome 6q26-q27region
Supporting Information Table S2 summarized the characteristics of the subjects included in our study. The SNP selection process was shown in Figure 1. Shortly, a total of 6,371 SNPs were located in chromosome 6q26-q27 region, and 923 of them passed the standard quality control. Next, with additional LD of r2 > 0.6, 14 tag SNPs were screened for further analysis (Supporting Information Table S3).

Evaluation association of candidate SNPs with colorectal cancer risk
Next, we applied logistic regression to analyze the association between the selected SNPs and risk of colorectal cancer. As shown in Table 1, the distribution frequencies of the tag SNP genotypes of the control group were in accordance with HWE (p > 0.05), except for SNP rs2221750. Therefore, we removed it in the later evaluation. For rs420038, the GA/AA genotype was associated with a lower risk of colorectal cancer (adjusted OR = 0.79; 95% CI = 0.67–0.94; p = 0.007) than the GG genotype; for rs9456537, the TT/CT genotype was 19% higher than the CC genotype colorectal cancer (adjusted OR = 1.19; 95% CI = 1.01–1.40; p = 0.043); for rs2048328, compared to the GG genotype, the GA/AA genotype decreased the colorectal cancer risk (adjusted OR = 0.81; 95% CI = 0.69–0.96; p = 0.013); for rs3124784, compared to the GG genotype, the AA/GA genotype increased the risk of colorectal cancer (adjusted OR = 1.29; 95% CI = 1.07–1.56; p = 0.009); for rs7765803, the mutant genotype reduced the risk of colorectal cancer (adjusted OR = 0.77; 95% CI = 0.64–0.91; p = 0.003). Additionally, we did not find that the remaining 8 SNPs were associated with the colorectal cancer risk (p > 0.05).
| SNPs | MAF (1KG/case/control) | pHWE | MM/MN/NN | ORadditive (95% CI)a | pa | pFDR | ORdominant (95% CI)a | pa | pFDR | ORrecessive (95% CI)a | pa | pFDR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||||||
| rs668871 | 0.263/0.346/0.365 | 0.950 | 144/503/497 | 159/559/483 | 0.92 (0.82–1.03) | 0.165 | 0.230 | 0.88 (0.74–1.03) | 0.114 | 0.228 | 0.94 (0.74–1.20) | 0.618 | 0.808 |
| rs675162 | 0.335/0.384/0.408 | 0.591 | 161/558/428 | 195/590/416 | 0.90 (0.80–1.01) | 0.081 | 0.142 | 0.89 (0.75–1.05) | 0.177 | 0.275 | 0.84 (0.67–1.05) | 0.129 | 0.451 |
| rs420038 | 0.371/0.374/0.410 | 0.311 | 163/532/452 | 193/599/409 | 0.86 (0.76–0.97) | 0.011 | 0.037 | 0.79 (0.67–0.94) | 0.007 | 0.040 | 0.86 (0.69–1.08) | 0.196 | 0.458 |
| rs2221750 | 0.196/0.239/0.209 | 0.011 | 72/404/671 | 38/426/737 | 1.19 (1.04–1.37) | 0.013 | 0.037 | 1.13 (0.96–1.33) | 0.151 | 0.264 | 2.04 (1.36–3.05) | 0.001 | 0.007 |
| rs9456537 | 0.237/0.256/0.222 | 0.277 | 83/421/642 | 52/429/720 | 1.21 (1.06–1.39) | 0.005 | 0.037 | 1.19 (1.01–1.40) | 0.043 | 0.121 | 1.72 (1.21–2.46) | 0.003 | 0.020 |
| rs3123636 | 0.263/0.240/0.252 | 0.702 | 71/408/668 | 73/458/670 | 0.94 (0.82–1.07) | 0.350 | 0.408 | 0.90 (0.77–1.07) | 0.230 | 0.297 | 1.02 (0.73–1.43) | 0.922 | 0.922 |
| rs2661834 | 0.351/0.365/0.385 | 0.180 | 153/531/463 | 166/590/443 | 0.92 (0.81–1.03) | 0.154 | 0.230 | 0.87 (0.73–1.02) | 0.088 | 0.205 | 0.95 (0.75–1.21) | 0.693 | 0.808 |
| rs1810126 | 0.454/0.445/0.448 | 0.560 | 227/566/354 | 245/583/371 | 0.99 (0.88–1.11) | 0.856 | 0.856 | 1.01 (0.85–1.20) | 0.931 | 0.931 | 0.96 (0.78–1.17) | 0.674 | 0.808 |
| rs2048328 | 0.345/0.331/0.360 | 0.287 | 131/498/518 | 147/571/482 | 0.88 (0.78–0.98) | 0.031 | 0.072 | 0.81 (0.69–0.96) | 0.013 | 0.046 | 0.92 (0.71–1.18) | 0.496 | 0.771 |
| rs3124784 | 0.155/0.145/0.117 | 0.126 | 31/270/846 | 22/238/941 | 1.26 (1.07–1.49) | 0.007 | 0.037 | 1.29 (1.07–1.56) | 0.009 | 0.040 | 1.47 (0.85–2.57) | 0.171 | 0.458 |
| rs1801693 | 0.485/0.451/0.477 | 0.773 | 224/582/337 | 270/605/326 | 0.90 (0.80–1.01) | 0.079 | 0.142 | 0.90 (0.75–1.07) | 0.233 | 0.297 | 0.84 (0.69–1.03) | 0.090 | 0.418 |
| rs7765803 | 0.459/0.414/0.451 | 0.352 | 208/533/405 | 236/611/354 | 0.86 (0.77–0.97) | 0.012 | 0.037 | 0.77 (0.64–0.91) | 0.003 | 0.037 | 0.91 (0.74–1.11) | 0.349 | 0.638 |
| rs9347438 | 0.366/0.371/0.381 | 0.245 | 159/532/456 | 164/586/451 | 0.96 (0.85–1.08) | 0.512 | 0.551 | 0.92 (0.77–1.08) | 0.296 | 0.319 | 1.02 (0.81–1.29) | 0.856 | 0.922 |
| rs1800769 | 0.418/0.433/0.415 | 0.552 | 209/575/363 | 201/594/406 | 1.08 (0.96–1.21) | 0.214 | 0.272 | 1.10 (0.93–1.31) | 0.271 | 0.317 | 1.10 (0.89–1.37) | 0.365 | 0.638 |
- a Logistic regression analysis with adjustment for age and sex. MAF, minor allele frequency; 1KG, the 1,000 Genomes Projects(CHB and JPT); HWE, Hardy–Weinberg equilibrium; M, Mutant allele; N, Wild-type allele; MM/MN/NN, Additive model; (MM+ MN)/NN, Dominant model; MM/(NN + MN), Recessive model; OR, odds ratio; CI, confidence interval; FDR, false discovery rate.
In addition, we performed eQTL analysis using the genotyping and RNA sequencing data of colorectal cancer from The Cancer Genome Atlas (TCGA) to determine whether these risk SNPS (i.e., rs420038, rs9456537, rs2048328, rs3124784 and rs7765803) were correlated with the expression of their located genes. As shown in Supporting Information Figure S1, the eQTL analysis indicated that there was an inverse association of rs420038 with SLC22A3 expression in normal tissues (p = 0.040, Supporting Information Fig. S1a) but not in colorectal cancer tissues (p = 0.843, Supporting Information Fig. S1a), and also rs7765803 with LPA expression in normal tissues (p = 0.005, Supporting Information Fig. S1e) but not in tumor tissues (p = 0.241, Supporting Information Fig. S1e). Furthermore, we found that SLC22A3 was widely expressed in various organs (Supporting Information Fig. S2a), while LPA was specifically expressed in the liver (Supporting Information Fig. S2b). Considering the feasibility, we therefore focused on rs420038 in SLC22A3 for further studies.
Stratified analysis of the association of rs420038 with colorectal cancer risk
We further conducted a stratified analysis according to the basic characteristics such as age, sex, tumor location, smoking status, drinking status, tumor family history, pathological type, TNM staging and other clinical features. As shown in Supporting Information Tables S4 and S5, individuals with the AA/GA genotype had a lower risk of colorectal cancer than those with the GG genotype when they were at an age equal to or less than 60 years (adjusted OR = 0.64, 95% CI = 0.50–0.81, p < 0.001), male (adjusted OR = 0.69, 95% CI = 0.56–0.87, p = 0.001), non-smokers (adjusted OR = 0.77, 95% CI = 0.62–0.95, p = 0.013), drinkers (adjusted OR = 0.68, 95% CI = 0.48–0.96, p = 0.028), no family history (adjusted OR = 0.81, 95% CI = 0.67–0.98, p = 0.026) and Well/ Medium differentiation (adjusted OR = 0.79, 95% CI = 0.67–0.95, p = 0.010).
Potential regulatory effect of rs420038 on SLC22A3
We further explored the potential biological molecular mechanisms of rs420038 regulating the transcription of SLC22A3. The HaploReg v4.1 functional prediction showed that rs420038 was located in the region of H3K4me1, H3K4me3, H3K27ac and other histone modifications in digestive tract tumors (Supporting Information Table S6), suggesting that rs420038 could affect the expression of SLC22A3 by influencing its promoter activity.
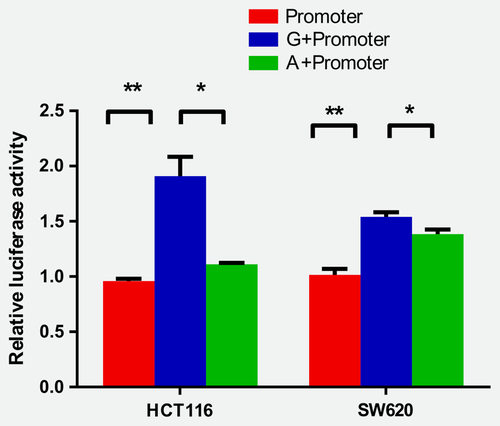
To validate whether different alleles of rs420038 changed the promoter activity of SLC22A3, we performed luciferase report assays. As shown in the map of the PGL3-Basic-Plasmid (Supporting Information Fig. S3), the sequence containing G or A allele of rs420038 was cloned upstream of promoter-luciferase reporter vector of SLC22A3 (Supporting Information Table S7). We then measured the luciferase activity after transfection, and the results showed that the transcriptional activity of the colorectal cancer cells with the A allele was significantly lower than that of those with the G allele (HCT116: p = 0.010; SW620: p = 0.030, Fig. 2), suggesting that different alleles of rs420038 could alter the transcriptional activity of SLC22A3.
Functional exploration of SLC22A3 in colorectal cancer
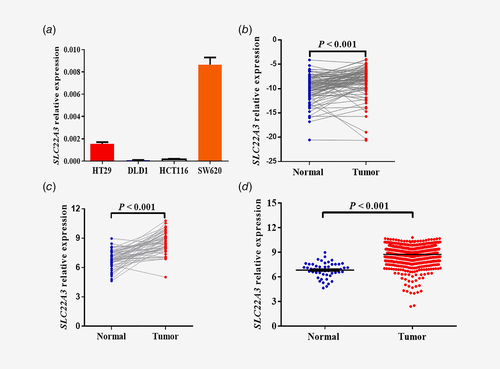
In order to choose proper cell line for the next experiment, four colorectal cancer cells were detected for mRNA expression levels of SLC22A3 (Fig. 3 a). Next, the SLC22A3 mRNA expression levels were detected in 94 pairs of colorectal cancer tissues and their adjacent normal tissues, and the results manifested that SLC22A3 was expressed more highly in tumor tissues than in adjacent normal tissues (p < 0.001, Fig. 3 b), a finding that was consistent with the data from the independent TCGA data comprising RNA-Seq of 50 paired colorectal tissues (p < 0.001, Fig. 3 c). Moreover, the data from the TCGA data consisting of 644 colorectal cancer tumor tissues and 51 normal tissues showed that SLC22A3 was highly expressed in tumor tissues (Fig. 3 d). Results were also demonstrated in the subgroups of the colon and rectum (Supporting Information Fig. S4). The TCGA data mentioned was up to June, 2016.
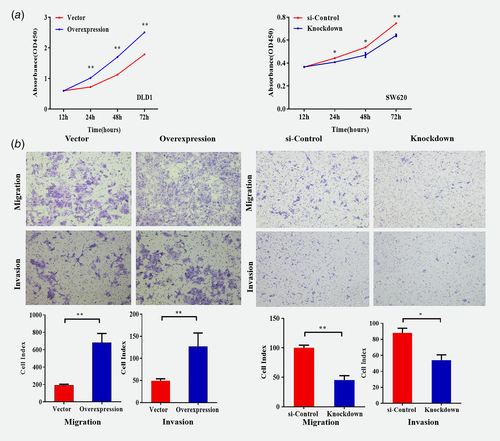
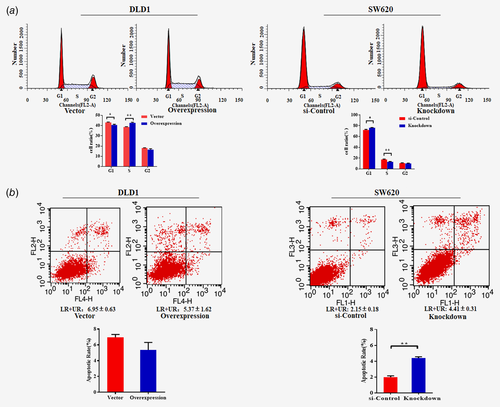
To explore the biological function of SLC22A3 in colorectal cancer, we transfected SLC22A3 overexpression and siRNA knockdown vectors into DLD1 and SW620 cells, respectively (Supporting Information Fig. S5). As shown in Figure 4 a, overexpression of SLC22A3 promoted proliferation (p < 0.05), whereas knockdown of SLC22A3 suppressed cellular growth (p < 0.05). Moreover, SLC22A3 overexpression significantly enhanced the migration and invasion ability of colorectal cancer, whereas knockdown of SLC22A3 exhibited the opposite results (Fig. 4 b). The further cell cycle and apoptosis were analyzed. With SLC22A3 knockdown, the progression of SW620 cells was arrested in the G1 phase compared to that in control cells, while the overexpression of SLC22A3 reduced the number of DLD1 cells in G1 phase compared to that in control cells (Fig. 5 a). Moreover, overexpression of SLC22A3 resulted in a decrease in DLD1 cell apoptosis compared to that in the control group, without a significant difference, but the finding was consistent with the trend that knockdown of SLC22A3 resulted in an increase in SW620 cell apoptosis compared to that in the control group (Fig. 5 b).
Discussion
SNP rs420038 in the chromosome 6q26-q27 region was first identified as a novel susceptibility locus for colorectal cancer risk in the Chinese population. compared to the GG genotype, the GA/AA genotype contributed to the reduction in colorectal cancer risk, especially in the subgroups of age equal to or less than 60 years, male gender, non-smoking, drinking and no family history of cancer.
Many studies have indicated that stress hormones were associated with cancer risk and progression,22, 23 and SLC22A3 was associated with the transportation and reuptake of norepinephrine.24-26 The group of youngers or men may suffer a more stressful life due to various kinds of pressure from family or the society, where norepinephrine would be more generated. In this condition, the inhibition of SLC22A3 in individuals with A allele would appear more obvious protective effect. However, as generally accepted, the mechanism of the oncogenesis, especially colorectal cancer, is a comprehensive combination of multiple risk factors including environmental conditions, dietary habits and genetic predispositions.27 The protective effect observed in subgroups of the ones who never smoke or ever drink seems contradictory and interesting. It is supposed that the toxic substances from tobacco and alcohol are transported or metabolized by many other enzymes such as CYP1A1, except the transporter SLC22A3. On the other hand, it's maybe only a result of the limitation of the size of our samples. The reasons above may explain the statistical significance found in the subgroup of no family history in a certain degree. In a word, it needs further investigation with a larger sample size.
In our study, we carried out functional annotation of the 923 SNPs in the websites of HaploReg v4.1 according to linkage disequilibrium (LD) of r2 > 0.6. In total, 14 tag SNPs were selected for the after study. Subsequently, rs420038, rs9456537, rs2048328, rs3124784 and rs7765803 were associated with the risk of colorectal cancer. Next, eQTL analysis revealed that the intron rs420038 and exon rs7765803 had effects on the mRNA levels of SLC22A3 and LPA, respectively, and the other three loci did not affect their gene expression. The results may be explained by the limitations of the sample size and lack of two-phase validation. In addition, we cannot rule out that these sites may induce the development of tumors by changing the amino acid types to affect the structure and function of proteins. Moreover, we did not investigate rs7765803 because of the inconsistent trend of effects that CC, CG and GG genotypes of rs7765803 exerted on the expression of LPA, and LPA was mainly researched in the development of coronary heart disease with very low expression level in intestinal tissue.
Interestingly, when reviewing the 923 variants, we found all the 6 missenses were located at gene of LPA. The SIFT or PolyPhen2 predictions (http://sift.jcvi.org/, http://genetics.bwh.harvard.edu/pph2/) of them are shown in the Supporting Information Table S8. The results showed that rs1801693, rs3124784 and rs3798220 would be probably to affect the protein SLC22A3. On the other hand, we found that SLC22A3 was widely expressed in various organs (Supporting Information Fig. S2a), while LPA was mainly expressed in the liver with very low expression in other organs. (Supporting Information Fig. S2b). Additionally, the eQTL analysis determined that rs420038 and rs7765803 were related with gene expression. Considering hard detection of Lp (a) encoded by LPA in colorectal tissues and cells, we thus chose rs420038 of SLC22A3 as the study object in a further study.
SLC22A3 is a gene in the chromosome 6q26-q27 region. It encodes a protein of the solute carrier family 22 family that might play a crucial role in the occurrence and development of colorectal cancer. Functional annotations have suggested that rs420038 may affect the expression of SLC22A3 by influencing its promoter activity, a finding that was verified in a reporter gene assay. Consequently, the contribution of rs420038 to colorectal cancer development may result from its alteration of the transcriptional activity of SLC22A3. We queried available ENCODE data for potential regulatory elements. It was predicted that rs420038 played a role in regulatory chromatin states in colon and rectal smooth muscle cell lines. Further evidence from proteins bound by ChIP (ENCODE) suggested that rs420038 was associated with the binding ability with transcription factor TCTF in HMEC, HeLa-S3 and NHEK cell lines(http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr6%3A160807898%2D160808398&hgsid=699125323_iXYILyRGcEoALa7an00S4kv25z1f). Further investigation about the internal mechanism would be expected by other researchers in the future. SLC22A3, a multispecific organic cation transporter, participates in complex metabolic processes in vivo, including transport, inactivation and excretion of various endogenous and exogenous carcinogens.28-30 It has been reported that the protein can not only transport antitumor drugs such as oxaliplatin and irinotecan,26, 31-33 but also is associated with some endogenous substances, e.g., dopamine, epinephrine, norepinephrine, histamine, and 5-hydroxytryptamine.19, 29, 34 Norepinephrine and adrenergic receptor signaling pathways were reported to be associated with the development and progression of colorectal cancer. Yokoo et al. showed that the transporter played a significant role in oxaliplatin-induced killing of colorectal cancer cells.31 Mohelnikova-Duchonova et al. found that the expression of SLC22A3 in 32 pancreatic cancer tissues was significantly higher than that in the corresponding normal tissues. And combined with prognostic data analysis, patients with a high expression of SLC22A3 showed longer overall survival when receiving the treatment of nucleotide analogs.35 However, there is no study exploring the relationship between SLC22A3 and the occurrence, development or drug sensitivity of colorectal cancer. In our study, we found that mRNA expression level of SLC22A3 was higher in the colorectal cancer tissues than that in the corresponding adjacent normal tissues. However, there is a limitation that we could not find a correlation of SLC22A3 protein with the mRNA level by linear correlation analysis since the matched samples for mRNA and protein were not acquired.
Moreover, our in vitro experiments indicated that the overexpression of SLC22A3 notably promoted cell proliferation and increased the ability of migration and invasion, whereas the knockdown of SLC22A3 showed opposite results. However, no significant difference was found regarding the overexpression of SLC22A3 resulting in a decrease in DLD1 cell apoptosis compared to the control group, but a consistent trend was demonstrated that the knockdown of SLC22A3 resulted in an increase in SW620 cell apoptosis compared to that in the control group. Additional experiments in multiple colorectal cancer cells are required to verify the results.
In our research, the patients received oxaliplatin- or irinotecan-based chemotherapy after operating. It is regrettable that we have not acquired all the survival information of the individuals included since the follow-up time of some cases are very short, even less than 6 months. It's a limitation of our study. However, we will continue and strengthen postoperative observation and regularly follow up to obtain the complete information about the survival in the future study. If there was an association of our studied SNPs with disease-free survival of the patients through survival analysis, then additional experiments involved anticancer drugs such as oxaliplatin or irinotecan would be conceived and designed.
In conclusion, we identified a novel susceptibility locus, rs420038, for colorectal cancer located in the SLC22A3 gene in the Chinese population. SNP rs420038 A allele may involve in downregulating the expression of SLC22A3, which plays a role in decreasing colorectal cancer risk. Further investigation is needed to elucidate the underlying molecular mechanism of SLC22A3 in colorectal cancer.
Acknowledgements
We would like to thank Meilin Wang (Nanjing Medical University) for laboratory assistance.
Authors’ contributions
L.Z. and M.D. conceived and designed the study; A.R. and S.L. performed the experiments and analyzed the data; T.C. and Y.S. contributed analysis tools; A.R., S.S., S.L. and M.D. critically revised the study; S.S. and A.R. wrote the paper. All authors reviewed the study.




