Diagnostic performance of one-step nucleic acid amplification for intraoperative sentinel node metastasis detection in breast cancer patients
Abstract
The purpose of this prospective multicenter study was to assess one-step nucleic acid amplification (OSNA) for intraoperative sentinel lymph node (SLN) metastasis detection in breast cancer patients, using final histology as the reference standard. OSNA results were also compared to intraoperative histology SLN evaluation and to standard clinicopathological risk markers. For this study, fresh SLNs were cut in four blocks, and alternate blocks were used for OSNA and histology. CK19 mRNA copy number was categorized as strongly positive, positive or negative. Positive histology was defined as presence of macrometastasis or micrometastasis. When discrepancies occurred, the entire SLNs were subjected to histological studies and the node lysates to additional molecular studies. Five hundred three SLN samples from 233 patients were studied. Mean time to evaluate two SLNs was 40 min. Sensitivity per patient was 91.4% (95% CI, 76.9–98.2%), specificity 93.3% (95% CI, 88.6–96.6%), positive likelihood ratio 13.7 and negative likelihood ratio 0.1. Sensitivity was 63.6% for frozen sections and 47.1% for touch imprint cytology. Both methods were 100% specific. Positive histology and positive OSNA were significantly associated with highest clinical stage, N1 status and vascular invasion; and OSNA results correlated with HER2/neu status and benefited patients with negative histology. These findings show that OSNA assay can allow detection of SLN metastasis in breast cancer patients intraoperatively with a good sensitivity, thus minimizing the need for second surgeries for axillary lymph node detection.
Sentinel lymph node (SLN) biopsy is now widely used as part of the staging procedure in patients with early-stage breast cancer. When the SLNs are free of metastasis, axillary lymph node dissection (ALND) and the associated morbidity can be avoided.1 SLN biopsy allows the pathologist to examine a small number of nodes (usually 1–3) in greater detail compared to the standard method for examining ALND specimens. Serial sectioning with hematoxylin-eosin staining and immunohistochemistry (IHC) has been reported to identify occult node metastasis in 7 and 20% of patients, respectively.2 Therefore, the SLNs are usually further examined postoperatively using step sectioning and IHC with cytokeratin (CK) antibodies. However, the methods and protocols vary widely.3
Intraoperative histological evaluation of SLNs allows ALND during the tumor excision procedure when the results are positive, thus obviating the need for a second surgical procedure. Nevertheless, intraoperative histological examination, performed by 60% of centers, lacks sensitivity (which varied between 60 and 90%) and no standardized protocol is available.4
To overcome the shortcomings of histopathological methods, molecular assays have been developed. Most of them are based on quantitative reverse transcription-PCR (QRT-PCR). QRT-PCR is a very sensitive method that can detect one cancer cell among 107 normal cells, which is 10–100 times more sensitive than IHC.5 Studies have evaluated several mRNA markers, including the epithelial cell marker cytokeratin 19 (CK19) and cancer-related markers such as mammoglobin.6-9 Two molecular-based intraoperative diagnosis of lymph node metastasis have been developed. The first one using the combination of CK19 and mammoglobin as markers are no longer available.10, 11 The second one is a one-step nucleic acid amplification test that amplifies CK19 mRNA (OSNA, Sysmex, Kobe, Japan).12 It was developed to accurately detect metastasis measuring 0.2 mm or more.13-17
Our aim was to assess the intraoperative diagnostic performance of OSNA versus extensive histological evaluation. We also assessed OSNA results according to intraoperative histology and to standard clinicopathological risk markers.
Material and Methods
Patients
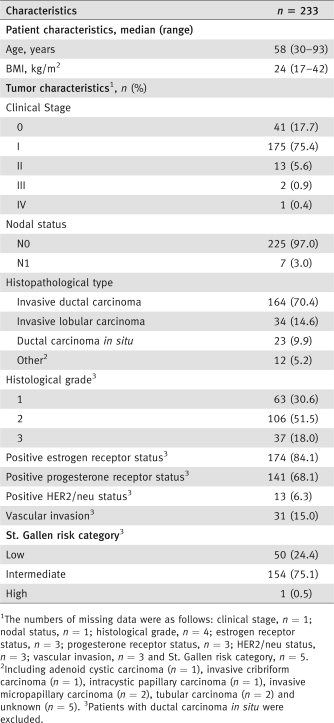
We conducted a multicenter prospective study in eight French clinical centers. The study was approved by an ethics committee (CPP Ile de France II; N° ID RCB, 2007-A00706-47). Between September 2007 and December 2007, all breast cancer patients scheduled for surgery with SLN biopsy were considered for enrolment. Tumors were staged according to the American Joint Committee on Cancer/International Union against Cancer (AJCC/UICC) classification.18, 19 We did not include patients who had other types of cancer with metastatic spread, patients given neoadjuvant therapy or patients younger than 18 years of age. All patients gave written informed consent. Patient characteristics, which are detailed in Table 1, included age, BMI (body mass index), clinical stage, nodal status, histopathological type, Scarff Bloom Richardson grade, estrogen and progesterone receptor status, HER2/neu status, peritumoral vascular invasion and the St. Gallen risk category.20
 |
Preparation of the sentinel lymph nodes
The excised SLNs were cut into four equal slices, as described previously.12, 15, 16 Two alternate slices (a and c) were prepared for OSNA and the other two slices (b and d) were fixed in 4% buffered formaldehyde and embedded in a paraffin block.
Histological examination
In five centers, the two slices (b and d) for the histological analysis were first used for intraoperative frozen section (one hematoxylin-eosin stained level) or touch imprint diagnosis, according to standard practice in those centers.
For the sections, five ribbons were cut with a 200-μm skip space. From each ribbon, three sections were prepared, one for hematoxylin and eosin staining and two for IHC with anti-human CK19 antibody (Clone RCK108, Dako, Denmark) and pan-CK antibody AE1/AE3 (Dako), respectively, as previously described.13, 16 Macrometastasis was defined as a tumor deposit larger than 2 mm and micrometastasis as a tumor deposit larger than 0.2 mm, but no greater than 2 mm.19 Tumor deposits no greater than 0.2 mm were categorized as isolated tumor cells (ITCs) and recorded as histologically negative pN0 (i+) in this study. All histological investigations were performed by pathologist experts in breast cancer tumors. OSNA results were blinded to pathologists performing histology.
One-step nucleic acid amplification assay for CK19 mRNA (OSNA)
Automated reverse transcription loop-mediated isothermal amplification (RT-LAMP)21 of CK19 mRNA in the RD-100i detection system (Sysmex) was performed, without prior mRNA isolation and purification, as recommended elsewhere.12 The assay was performed in duplicate on a pure sample and on a diluted sample (1/10). Homogenates were then stored at −80°C.
Results were automatically characterized by the CK19 mRNA copy number/μL of the original tissue homogenate. A strongly positive result (++; CK19 mRNA copy number greater than 5,000/μL) is associated with macrometastasis, a positive result (+; copy numbers between 250 and 5,000/μL) with micrometastasis, and a negative result (−, copy numbers no greater than 250/μL) with either ITCs or no tumor.12 Inhibition of amplification is a rare event detected as a positive result (+, micrometastasis) in the diluted sample, but not the pure sample.
Intensive investigation of discrepancies
Discordant cases between OSNA and histology could have occurred, because metastatic foci could be located only in one SLN sample piece dedicated for one or the other method. This is called tissue allocation bias (TAB). When OSNA was positive and histology negative, consecutive ribbons with 200-μm skip space were cut until exhaustion of the remainder of the paraffin-embedded SLN slices. The sections were stained with hematoxylin-eosin and immunostained with CK19 and AE1/AE3. Additionally, in all cases of discrepancies, the SLN homogenates were shipped to Sysmex (Norderstedt, Germany) and subjected to blind molecular analysis. Tests were performed as previously described.12, 13, 15-17 In brief, QRT-PCR was performed for CK19 and the breast-tissue specific markers SPDEF (SAM pointed domain containing ETS transcription factor) and FOXA1 (forkhead box A1). CK19 protein expression was assessed using Western blot.
OSNA and intensive molecular investigation showing the same results (both negative or both positive) were taken to indicate TAB, that is, presence of tumor deposit in either the b and d slices used for histology or the a and c slices used for OSNA. When TAB occurs, the two methods cannot be directly compared.
Statistical analysis
We performed an intention-to-diagnose analysis. To evaluate the diagnostic performance of OSNA, we computed sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), positive likelihood ratio (LR+) and negative likelihood ratio (LR−), using histology as the reference method.
We used both SLN samples (node or node fragment) and patients as unit of analysis. A patient was classified as having a positive OSNA result if OSNA on at least one SLN sample was (+) or (++) and as having a negative OSNA result otherwise. Histology was considered positive in patients with at least one macrometastasis or micrometastasis in a SLN sample and negative otherwise.
The diagnostic performance indexes were computed with their 95% confidence intervals (CI) before and after exclusion of the samples with TAB, and before and after exclusion of the patients with at least one sample with TAB.
We used logistic regression to evaluate associations linking OSNA results to standard outcome markers and to the St. Gallen risk category.20 We investigated its potential benefit in the subgroup of histology-negative patients.22 We constructed the models using all patients, and we included the outcome markers, the histological result and their interaction terms as a covariate. All p values and odds ratios (ORs) with their 95% CIs were derived from logistic regression models, except for tables with zero cells, for which we used Fisher's exact test and did not compute the 95% CI. All analyses were univariate and explanatory.
Finally, we determined the best CK19 mRNA copy-number cutoff for separating macrometastasis, micrometastasis and no metastasis in our data set (using Youden's index), comparatively with cutoffs established in previous studies.12 Samples with TAB or inhibition of amplification were excluded from this analysis.
SAS statistical software (release 9.1; SAS Institute, Cary, NC) was used for all analyses.
Abbreviations
ALND: axillary lymph node dissection; CI: confidence interval; CK: cytokeratin; IHC: immunohistochemistry; ITCs: isolated tumor cells; LR−: negative likelihood ratio; LR+: positive likelihood ratio; NPV: negative predictive value; OR: odds ratio; OSNA: one-step nucleic acid amplification; PPV: positive predictive value; QRT-PCR: quantitative reverse transcription-PCR; SLN: sentinel lymph node; TAB: tissue allocation bias
Results
Patients, description of sentinel lymph node samples and OSNA assay
Among 234 included patients, two had no SLNs identified and one had bilateral surgical interventions and was counted as two patients resulted in 233 patients analyzed (Table 1). Both OSNA and histology were performed on 503 samples derived from 456 SLNs. The median numbers of samples and SLNs per patient were both two (range, 1–4).
The median time needed for the OSNA assay was 33, 40, 48 and 54 min for 1, 2, 3 and 4 SLN samples, respectively.
Diagnostic performance of OSNA and histology per sample (Table 2 and Fig. 1)
Of the 63 samples containing histological metastases (45 macrometastases and 18 micrometastases), 51 were OSNA-positive [43 (++) and 8 (+)], yielding a sensitivity of 80.9% (95% CI, 69.0–89.8%). Of the 440 histology-negative samples (28 ITC and 412 negative), 413 (27 + 386) had negative OSNA results, yielding a specificity of 93.9% (95% CI, 91.2–96.0%). PPV was 65.4% (95% CI, 53.7–75.8%) and NPV 97.2% (95% CI, 95.1–98.6%). Samples positive by OSNA were 13 times (LR+, 13.2) more likely to contain metastases (true positives) than to contain no metastasis (false positives). Samples negative by OSNA were five times (LR−, 0.20) more likely to be free of metastasis than to contain metastases.
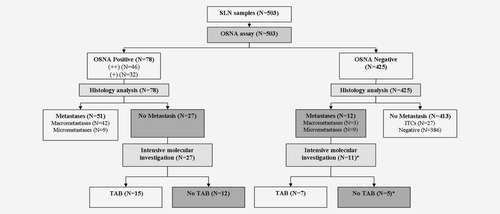
Flow chart of the 503 SLN samples analyzed by both OSNA and histology. SLN: sentinel lymph node; TAB: tissue allocation bias; ITCs: isolated tumor cells. *One sample with no discordant case investigation (not considered as a TAB case). Cells with gray background indicate discrepancies between OSNA and histological results (OSNA-positive/histology-negative or OSNA-negative/histology-positive) and the remaining discordant cases (no TAB) after intensive investigation of discrepancies.
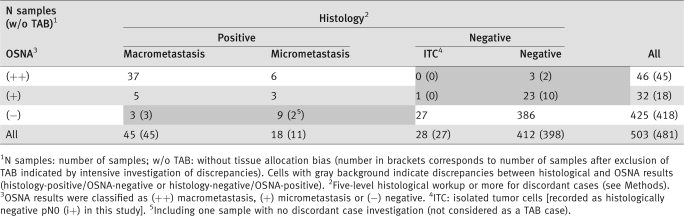 |
Thirty-nine discordant cases (histology-negative/OSNA-positive or histology-positive/OSNA-negative) were encountered (Table 2 gray cells). Twenty-seven were histology-negative/OSNA-positive [three OSNA (++) samples and 24 OSNA (+) samples including nine with inhibition]. All 27 samples remained histologically negative after extension to all levels in slices b and d, whereas 15 samples (including two with inhibition) had OSNA-positive lysates of the a and c slices, suggesting TAB (Fig. 1). Twelve histology-positive/OSNA-negative samples were found. One was not investigated by further molecular analysis and seven were negative by further molecular analysis, suggesting TAB (Fig. 1). After exclusion of the 22 samples with possible TAB, sensitivity was 91.1% (95% CI, 80.3–97.1%), specificity 97.2% (95.1–98.6%), PPV 80.9% (95% CI, 69.0–89.8%), NPV 98.8% (97.2–99.7%), LR+ 32.3 and LR− 0.1.
Diagnostic performance of OSNA and histology per patient (Table 3)
Of the 42 patients with metastases (26 with macrometastases and 16 with micrometastases), 33 were OSNA-positive [28 (++) and 5 (+)], yielding a sensitivity of 78.6% (95% CI, 63.1–89.7%). Of the 191 histology-negative patients (20 with ITC and 171 with all samples negative), 168 had negative OSNA results, yielding a specificity of 88.0% (95% CI, 82.4–92.3%). PPV was 58.9% (95% CI, 44.9–71.9%), NPV 94.9% (95% CI, 90.5–97.7%), LR+ 6.5 and LR− 0.2.
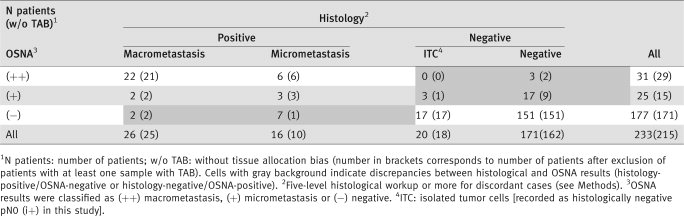 |
The 22 samples with possible TAB were found in 18 patients (four had TAB in all SLN samples). After exclusion of these 18 patients, sensitivity was 91.4% (95% CI, 76.9–98.2%), specificity 93.3% (95% CI, 88.6–96.6%), PPV 72.7% (95% CI, 57.2–85.1%), NPV 98.2% (95% CI, 94.9–99.7%), LR+ 13.7 and LR− 0.1.
Results of intraoperative histology
Of the 503 SLN samples, 350 (70%) were investigated intraoperatively by frozen section (n = 187) or touch imprint cytology (n = 163). Compared to the final five-level histological study, frozen section detected 14 of 17 macrometastases and none of the five micrometastases. Sensitivity was 63.6% (95% CI, 40.6–82.8%), specificity 100% (95% CI, 97.7–100.0%), PPV 100% (95% CI, 76.8–100.0%) and NPV 95.4% (95% CI, 91.0–98.0%). Touch imprint cytology detected eight positive cases corresponding at definitive histology to seven of the nine macrometastases and one of the eight micrometastases. Sensitivity was 47.1% (95% CI, 22.9–72.2%), specificity 100% (95% CI, 97.5–100.0%), PPV 100% (95% CI, 63.0–100.0%) and NPV 94.2% (95% CI, 89.2–98.4%).
Benefit of the OSNA method in specified subgroups (Table 4)
Both positive histology and positive OSNA were significantly associated with highest clinical stage, N1 nodal status and vascular invasion. HER2/neu-positive patients were more likely to have positive OSNA results (OR = 5.1; 95% CI, 1.6–16.6; p = 0.006), and intermediate or high St. Gallen risk patients were more likely to have positive histological results (OR = 5.3; 95% CI, 1.5–17.9; p = 0.008).
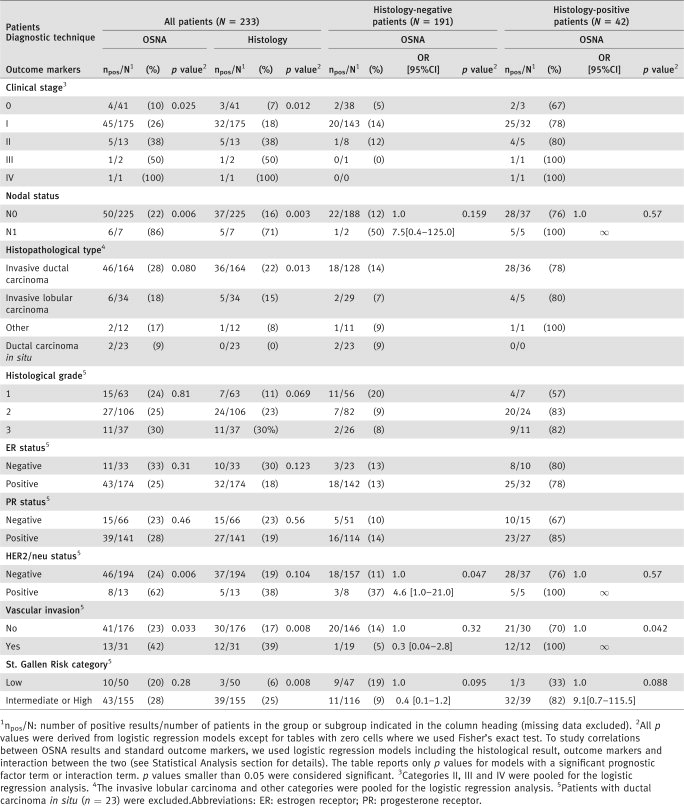 |
In the subgroup of histology-positive patients, a positive OSNA result significantly increased the probability of vascular invasion (p = 0.042) and increased the risk of N1 nodal status and vascular invasion (nonsignificant differences, but all results were OSNA-positive). In the subgroup of histology-negative patients, HER2/neu-positive patients were more likely to be OSNA-positive (OR = 4.6; 95% CI, 1.0–21.0; p = 0.047). The direction of association between the OSNA result and the St. Gallen risk category differed according to histological results.
CK19 mRNA cutoffs (Fig. 2)
Of the 503 samples, 13 were OSNA-positive (+) with inhibition, including four with metastases (three macrometastases and one micrometastasis) and nine with negative histology (including one with ITCs). Figure 2 reports the level of CK19 mRNA expression in the 490 OSNA assays without inhibition (as copy number could not be accurately measured in samples with inhibition), according to the histological result and to whether TAB occurred.
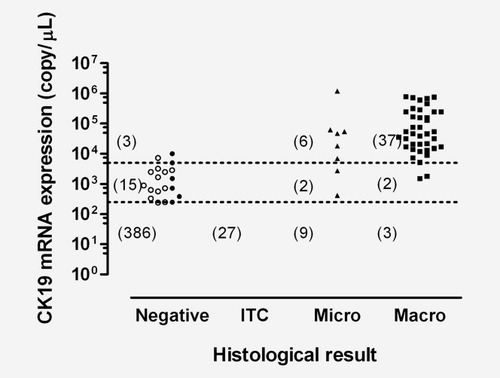
CK19 mRNA expression in the 490 OSNA assays. The 490 OSNA assays were those without inhibition. The 424 SLN samples with copy numbers less than 250/μL are not shown (385 histology-negative, 27 ITCs, nine micrometastases and three macrometastases). The open symbols represent the samples with tissue allocation bias: 12 were histology-negative/OSNA(+), one was histology-negative/OSNA(++), none contained ITCs, seven were micrometastases/OSNA(−) (not shown, with 0 copy/μL) and none contained macrometastasis. The broken line at the top corresponds to 5 ×·103 copies/μL (cutoff between macrometastasis and micrometastasis according to Tsujimoto et al.12) and the broken line at the bottom to 2.5 × 102 copies/μL (cutoff between micrometastasis and nonmetastasis according to Tsujimoto et al.). Numbers in brackets are the numbers of SLN samples with copy numbers greater than or equal to 5 × 103 copies/μL, between 2.5 × 102 and 5 × 103 copies/μL and lower than or equal to 2.5 × 102 copies/μL.
The statistical analysis showed that the best CK19 mRNA cutoff for separating positive and negative histological samples was 380 copies/μL (Youden's index, 89.4%). Nevertheless, the reference cutoff of 250 copies/μL12 produced a similar Youden's index of 89.2%, because no samples had CK19 mRNA values between 250 and 380 copies/μL. The best cutoff for separating macrometastasis from micrometastasis was 720 copies/μL (Youden's index, 90.5%). With the reference cutoff value of 5,000 copies/μL, Youden's index was 86.5%. However, using the 720 copies/μL cutoff changed the OSNA results for only four samples.
Discussion
We evaluated the test OSNA intraoperatively in 456 SLNs from 233 patients against the final histological evaluation. The median time needed for OSNA testing of two SLNs was 40 min, confirming the suitability of OSNA for intraoperative use. The test OSNA is characterized by the detection of only CK19 mRNA expression. CK19 is highly sensitive and is, consequently, the most widely used marker for tumor cell detection in patients with breast cancer. Testing for CK19 expression is often combined with testing for mammoglobin, which is more specific for breast cancer.5, 7, 9, 23 However, mammoglobin expression varies widely in breast cancer (61–93% of the tumors) and is preferably lost in high-grade tumors (G3).24 Results of a study of 43 potential markers suggested that a two-marker assay might be optimal, its sensitivity being 94%.6 After exclusion of the sample for which a manipulation error occurred, sensitivity per SLN sample in our study was 94.4%, similar to the sensitivity obtained by sectioning the entire SLNs at 250-μm intervals.11 Furthermore, Hughes et al.6 encountered the same sensitivity when using CK19 and mammoglobin compared with CK19 alone. Nevertheless, OSNA evaluates only CK19, which is a luminal CK marker. Indeed, absence of CK19 protein expression by IHC has been reported in young women with triple-negative phenotype breast cancer.25 Therefore, a reasonable strategy may consist in testing the diagnostic breast biopsy sample for CK19 using IHC before deciding to use OSNA.
As no markers specific for breast cancer cells are available and because these markers can be expressed by normal tissue, a cutoff above which expression is deemed abnormal must be defined for each marker. With molecular biology testing, the proportion of positive nodes is increased by 14–40% compared to nodes positive by histology.5, 8, 23, 26-29 Nevertheless, in a study of 144 nodes from pN0 patients, specificity of the OSNA technique was 100%.12 Specificity was 97.1% in a study of 104 histology-negative nodes.14 The OSNA CK19 copy-number cutoff corresponds to the presence of 5,000 tumor cells. Therefore, it is unlikely that positive results might occur because of epithelial displacement or illegitimate transcription, which involve only 500–1,000 non-tumor cells.30 Finally, the presence of ectopic breast tissue is rare, with only seven cases found in a study of more than 3,500 axillary lymph node biopsies.31 Furthermore, in our study, the cutoff for separating positive (++ or +) OSNA results from negative OSNA results according to histological results was computed and found to be very close to the cutoff defined by OSNA. Using this new cutoff did not change the classification of patients as having positive or negative SLNs.
In this study, SLNs were cut into four equal slices and alternate slices were prepared for OSNA and histology, which was considered as the gold standard. As different parts of the node were used for each method because each technique required different tissue preparation, discrepancies between OSNA and histological results were expected.32 It was the reason why further intensive molecular analyses have been performed when discordant results occurred. Nevertheless, these additional molecular tests as a tool to investigate the OSNA result also had their limit. Because of storage and RNA isolation procedure, RNA content in the sample could be diminished as compared to the original OSNA run. When OSNA result was confirmed by further molecular testing, it was considered as a TAB.
In this study, after excluding samples with possible TAB, sensitivity of OSNA per patient was 91.4%. OSNA was positive in 24 of the 26 patients with macrometastasis and in nine of the ten patients with micrometastasis. In one of these patients, who had two SLNs sampled including one with a macrometastasis and the other with negative histology, there was a manipulation error. The other patient with a macrometastasis had negative OSNA results, but CK19 highly expressed in the molecular tests and this discrepancy remained unexplained. Finally, in the patient with a micrometastasis, QRT-PCR values for CK19, SPDEF and FOXA1 were close to the cutoffs. Specificity of OSNA per patient was 93.3% (after exclusion of TAB) with 12 of the 215 patients histology-negative/OSNA-positive. After exclusion of TAB, sensitivity of OSNA per SLN sample was 91.1% and specificity 97.2%. Several studies have been performed using OSNA technique12-17 on axillary lymph nodes,13-15 SLNs16, 17 or both12 in breast cancer patients. In all but two studies,12, 14 an intensive investigation of discrepancies with further molecular analysis was performed as in our study in case of discordance between OSNA and histological results. Sensitivity of OSNA technique per node sample after discordance case investigation, if performed, varied between 82.717 and 100%,13 and specificity between 94.114 and 97.7%.17 In Feldman et al.'s study,17 sensitivity was quite lower than in Snook et al.'s study16 (91.7%) and than in ours (91.1%), whereas they have all been performed on SLNs. Nevertheless, in the American study the protocol of node cutting was slightly different and the number of micrometastases was higher that might explain this lower sensitivity.
Finally, in our study, sensitivity of intraoperative methods was 63.6% for frozen section histology and 47.1% for touch imprint cytology that was lower than OSNA technique sensitivity. Both methods had very low micrometastasis detection rates, but were 100% specific.
Several studies evaluated the potential clinical significance of histology-negative/RT-PCR-positive nodes. With conventional RT-PCR, all the studies26-28, 33 but two8, 29 found that having histology-negative/RT-PCR-positive nodes correlated with some histological outcome markers. Only two studies sought correlations with clinical outcomes and encountered conflicting results.8, 29 Real-time RT-PCR was assessed in only two studies,5, 23 both of which found a significant association between having histology-negative/RT-PCR-positive nodes and the St. Gallen risk category. In our study, in the subgroup of histology-negative patients, HER2/neu-positive patients were more likely to be OSNA-positive.
OSNA does not detect isolated ITCs, but their clinical relevance remains debated perhaps in part because with current guidelines the distinction between ITCs and micrometastasis is unreliable in a substantial number of cases.34 In a recent study of 954 patients staged according to the AJCC, N0(i−) and N0(i+) patients had no statistically significant difference in survival or recurrence-free survival.35 A prospective study of 1,259 breast cancer patients suggested that ALND might be unnecessary in patients with ITCs.36 On the other hand, among women with favorable early-stage breast cancer who did not receive adjuvant therapy, ITCs or micrometastases in regional lymph nodes were associated with a lower 5-year disease-free survival rate.37
In conclusion, the use of the OSNA assay can be expected to decrease the number of women who require a second surgical procedure for ALND, because its sensitivity is higher than intraoperative histological evaluation. It may allow standardization of the SLN evaluation with a more extensive study of each node compared to the currently used step sections. Nevertheless, as CK19 is the only marker used in the OSNA assay, we recommend testing CK19 expression, at least by IHC, on the initial diagnostic breast biopsy sample. Finally, the clinical relevance of histology-negative/OSNA-positive SLNs needs to be assessed by long-term studies.
Acknowledgements
The authors are grateful to Noël Lucas (medical coordinator), Dominique Mariolle (financial account manager) and Jean-François Leforestier (data manager) from the Unité de Recherche Clinique of the Hôpital Européen GeorgesPompidou, a publicly funded teaching hospital in Paris, France. Sysmex Corporation contributed to providing the RD-100i system, funding of laboratory consumables for the OSNA assay, but had no role in data interpretation and writing of the report.




