Expanding the genotypes and phenotypes for 19 rare diseases by exome sequencing performed in pediatric intensive care unit
Juan Liu and Yu Zheng contributed equally to this study as co-first authors.
Abstract
Phenotypes of some rare genetic diseases are atypical and it is a challenge for pediatric intensive care units (PICUs) to diagnose and manage such patients in an emergency. In this study, we investigated 58 PICU patients (39 deceased and 19 surviving) in critical ill status or died shortly without a clear etiology. Whole exome sequencing was performed of 103 DNA samples from their families. Disease-causing single-nucleotide variants (SNVs) and copy number variants (CNVs) were identified to do genotype-phenotypes analysis. In total, 27 (46.6%) patients received a genetic diagnosis. We identified 34 pathogenic or likely pathogenic SNVs from 26 genes, which are related to at least 19 rare diseases. Each rare disease involved an isolated patient except two patients caused by the same gene ACAT1. The genotypic spectrum was expanded by 23 novel SNVs from gene MARS1, PRRT2, TBCK, TOR1A, ECE1, ARX, ZEB2, ACAT1, CPS1, VWF, NBAS, COG4, and INVS. We also identified two novel pathogenic CNVs. Phenotypes associated with respiratory, multiple congenital anomalies, neuromuscular, or metabolic disorders were the most common. Twenty patients (74.1%) accompanied severe infection, 19 patients (70.1%) died. In summary, our findings expanded the genotypes and phenotypes of 19 rare diseases from PICU with complex characteristics.
1 INTRODUCTION
Critical diseases caused by undiagnosed genetic conditions are challenging for pediatric intensive care units (PICUs). For most PICU patients, the symptoms are usually complicated by emergency admission, and some of them are in critical condition with unexplained liver failure, renal trauma, or consciousness disturbances. It is now known that genetic disorders (congenital defects) are the major reasons for hospitalization and mortality in infants in neonatal intensive care units (NICUs) or PICUs (Weiner et al., 2011). In the past 6 years, by using exome sequencing (ES) or whole-genome sequencing (WGS) in 1185 children in PICUs/NICUs, the diagnostic rate of genetic disorders reached about 19%–69.7% globally (Australian Genomics Health Alliance Acute Care Flagship, 2020; Clark et al., 2019; Daoud et al., 2016; Elliott et al., 2019; Farnaes et al., 2018; French et al., 2019; Gubbels et al., 2020; James et al., 2020; Kingsmore et al., 2019; Kingsmore et al., 2020; Meng et al., 2017; Petrikin et al., 2018; Ritter et al., 2020; Sanford et al., 2019; Schofield et al., 2019; L. D. Smith et al., 2015; Soden et al., 2014; Stark et al., 2017, 2019, 2016; N. B. Tan et al., 2019; T. Y. Tan et al., 2017; van Diemen et al., 2017; H. Wang, Lu, et al., 2020; H. Wang, Qian, et al., 2020; Willig et al., 2015; Wu et al., 2019). This dramatically improved the diagnosis, treatment, and prognosis in clinical work (Arts et al., 2019; Australian Genomics Health Alliance Acute Care Flagship, 2020; Farnaes et al., 2018; French et al., 2019; Meng et al., 2017; Mestek-Boukhibar et al., 2018; Ritter et al., 2020; Sanford et al., 2019; Schofield et al., 2019; Soden et al., 2014; Stark et al., 2017; Stark et al., 2019; Z. A.-O. Stark et al.; N. B. Tan et al., 2019; T. Y. Tan et al., 2017; H. Wang, Lu, et al., 2020; H. Wang, Qian, et al., 2020; Willig et al., 2015; Wu et al., 2019).
In western developed countries, it was estimated that more than 40 million children have rare genetic disorders, and the mortality rate reached about 30% in patients under 5 years old (Kuehne et al., 2019). In developing countries, with the gradual popularization of next-generation sequencing technology, rapid detection of genetic disorders in PICU patients has become possible. In the developed area of China, three studies from the Children's Hospital of Fudan University in Shanghai revealed that 85 of 163 pediatric patients had genetic findings (H. Wang, Lu, et al., 2020; H. Wang, Qian, et al., 2020), and 44 in 223 deceased newborns received genetic diagnosis (Yang et al., 2020). However, many rare diseases have atypical phenotypes. French et al. (2019) stated that 90% of the molecular diagnosis-positive patients had phenotypic descriptions that were atypical, and about 10% of the patients had a genetic diagnosis whose phenotype precisely predicted underlying genes. In the central south of China, before the clinical ES, WGS, and CNV analysis were available, patients usually had a long turn-around time in different hospitals with more complex clinic features. This increases the difficulty for PICU clinicians to diagnose precisely and manage shortly. Therefore, this study was aimed to identify the primary disease-causing factors underlying the complex and severe symptoms from PICU, and thus to obtain clues for precise diagnosis and improvement of clinical management of some rare genetic diseases.
In this study, we retrospectively investigated the deceased or critically ill children without clear etiology in one central PICU by WES. By screening single-nucleotide variants (SNVs), short insertions or deletions (InDels), and copy number variants (CNVs) simultaneously, we then identified disease-causing variants and further investigated the genotypes and phenotypes of the patients with genetic findings.
2 MATERIALS AND METHODS
2.1 Clinical patient selection
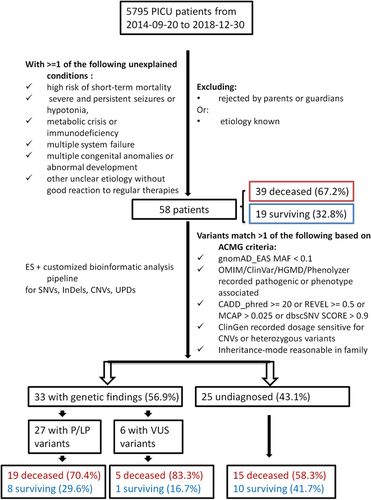
Hunan Children's Hospital is a central hospital in pediatrics in Hunan province. From September 20, 2014 to December 30, 2018, 5795 patients were transferred to the PICU department from other hospitals in the local prefectural area. Initially, we recruited the PICU patients who were in dying status or without good reactions to regular therapies and who suspected a genetic disorder but had not undergone any genetic testing. Finally, after excluding patients with known etiology or rejected by parents or guardians, 58 patients participated having at least one of the following conditions: (1) high risk of short-term mortality but lacking medical explanation; (2) severe unexplained seizures or hypotonia; (3) metabolic crisis of unknown cause; (4) immunodeficiency of unknown origin; (4) multiple system failures of unknown pathogenesis; (5) multiple congenital anomalies or abnormal development; and (6) other uncertain etiologies (Borghesi et al., 2017). Figure 1 shows the flow diagram of the selection of the studied patients.
The investigated patients included 33 males and 25 females with the age between one month and 15 years old. The average age was 2.2 years old. Appropriate informed consent was obtained from the parents/guardians of all participants before sample collection. We collected detailed information on disease history, family history, and clinical features of each patient. Phenotypes were standardized using human phenotype ontology (HPO) (Köhler et al., 2017) and the Chinese Human Phenotype Ontology Consortium.
2.2 Sample collection
Peripheral blood (2–3 ml) was collected from each participant using edetate disodium, including 23 trios, 28 probands only, three families (proband + parents + siblings), and four paired WES (proband + father/mother).
In total, we collected 117 samples from 58 patients and 30 family controls, and the genomic DNA of each sample was extracted using the standard phenol-chloroform protocol.
2.3 Exome sequencing
One μg genomic DNA samples were fragmented to an average size of 180–280 bp and subjected to exome capture using the Agilent SureSelect Human All Exon V6 Kit (Agilent Technologies) according to the manufacturer's instructions. The prepared libraries were then sequenced with 2 × 150 bp reads using the Illumina NovaSeq. 6000 system (Illumina) following the manufacturer's instructions. Each sample yielded more than 10 GB of raw data with more than 90% bases having a Phred quality score > 30. The mean coverage was ×100 of the genome and the minimum coverage of ×10 was about 99%.
2.4 Bioinformatic analysis and interpretation
The raw reads were preprocessed using trim_galore (version 0.6.4, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove adapter-contaminated ends and low-quality bases with Phred scores <20. The polished reads with a length ≥ of 80 bp were subsequently mapped to the human reference genome (version: GRCh38) using Burrows–Wheeler Aligner (BWA, version 0.7.17-r1188) software (H. Li & Durbin, 2010). The duplicate reads were removed using Picard (version 2.21.1, https://broadinstitute.github.io/picard/). The pipeline of the best practice of Genome Analysis Toolkit (GATK, version 3.8) (DePristo et al., 2011) was employed to refine the alignment and call variants. The obtained SNVs and InDels were then annotated using ANNOVAR (K. Wang et al., 2010) and InterVar (version: v20180118) (Q. Li & Wang, 2017) to classify pathogenicity based on population frequency, functional impact, and so forth. For each case, we calculated the phenotype-relevant score of each identified gene with variants using Phenolyzer (Yang et al., 2015) according to the corresponding HPO words. Public databases such as OMIM (https://www.omim.org/), HGMD (Stenson et al., 2020), ClinVar (Landrum et al., 2016), and ClinGen (Rehm et al., 2015) were also screened for the associated disease and pathogenicity of each gene variant.
All variants were interpreted and classified according to the American College of Medical Genetics and Genomics (ACMG) guidelines (Richards et al., 2015). The filtering criteria of the detected variants are presented in the workflow in Figure 1. The annotated variants with deleterious effects or with population frequency less than 0.01 in East Asians in gnomAD exome or genome databases were kept and then prioritized according to the phenotype correlation score. Stopgain, InDels, missense variants with CADD (Rentzsch et al., 2019) Phred score ≥ 20 or REVEL (Ioannidis et al., 2016) score > 0.5 or M-CAP (Jagadeesh et al., 2016) score > 0.025, and variants predicted splicing alternation with dbscSNV ADA score and RF score > 0.9 were kept in high priority. wInterVar (https://wintervar.wglab.org/) and varsome (version: 9.5.3, https://varsome.com/) were further queried to confirm the ACMG variants classification criteria of these variants.
2.5 Sanger sequencing
Pathogenic or likely pathogenic SNVs and InDels were further validated using Sanger sequencing, if (1) depth < 20, or (2) minor allele fraction ≤ 0.3 or ≥ 0.7. PCR primers were designed using the Primer-BLAST program (Ye et al., 2012). All variants were validated by independent PCR amplification and bidirectional DNA sequencing performed on an ABI 3130 DNA analyzer (Applied Biosystems). The segregation pattern was analyzed in the pedigree to confirm the mode of inheritance.
2.6 Compound heterozygous variants validation
For the patients with parents sequenced, paternal or maternal inheritances of the two heterozygous variants and whether they were in trans were confirmed. For the patients without parents' DNA samples, if the chromosomal distance between the two heterozygous variants was below 150 Bp (patient P6), the Integrative Genomics Viewer (IGV) (Thorvaldsdottir et al., 2013) was employed to view the variants-located chromosomal region of the patient's bam file to confirm in translocation; if the distance between the two heterozygous variants was blow 3 Kb (patient P2 and P14), PCR primers were designed using the Primer-BLAST program (Ye et al., 2012) containing both two variants and then the PCR products were performed TA cloning (Trower & Elgar, 1994) to confirm whether the two variants were in trans.
2.7 CNV analysis
For the patients without pathogenic or likely pathogenic SNVs or InDels identified, CNV analysis was further performed. The refined bam files generated in previous “Bioinformatic analysis and interpretation” step were used. CNVkit (version 0.9.6) (Talevich et al., 2016) was employed to detect germline CNVs in each patient. If the patient had family unaffected controls sequenced, the family controls were used for the reference. If only probands were sequenced without family controls, the 45 family control samples in this study were divided into male group and female group respectively to be used for the reference. The corresponding bed file associated with Agilent SureSelect Human All Exon V6 kit was used as target regions. CNVs were called using threshold parameter “-t=−1.1,−0.4,0.3,0.7” for germline variations. The called CNVs were then annotated with OMIM collected genes and ClinGen dosage-associated genes with haploinsufficiency and triplosensitivity descriptions. The CNVs were then interpreted based on the ACMG and ClinGen technical standards (Erin Rooney Riggs et al., 2019). DECIPHER database (Firth et al., 2009) was used to query any overlapped CNVs and associated phenotypes. Pathogenic or likely pathogenic CNVs were further validated via chromosomal microarray analysis (CMA) using the Infinium Asian Screening Array-24+ v1.0 (Illumina).
2.8 Multidisciplinary team confirmation
Geneticists and clinicians (including PICU experts, neurologists, hematologists, etc.) at Hunan Children's Hospital cooperated to confirm the genetic diagnosis of each identified patient. The validated variants were classified into de novo, compound heterozygous, or homozygous according to the genotype of family members. We selected the inheritance-pattern-matched and phenotype-explainable genomic variants and evaluated the genotype-phenotype relationship for diagnosis.
2.9 Statistical analysis
We divided the 58 patients into two groups: 33 patients with genetic findings and 25 patients without genetic findings. According to the survival status, there were 39 deceased patients versus 19 surviving patients. Clinical features were classified according to age, affected systems, and symptoms. Statistical analyses were performed using IBM SPSS software (version 18.0; IBM Corp.). Chi-square test and Fisher's exact test were applied to compare the genetically diagnosed and undiagnosed groups and the deceased and surviving groups. The calculation of all p values was two-sided.
3 RESULTS
3.1 Clinical profile of the selected patients
Between September 20, 2014 and December 30, 2018, 5795 patients were admitted to the PICU of Hunan Children's hospital. A total of 283 have died and the rate of death is about 4.9%. Among the studied 58 patients, 39 (67.2%) died and 19 survived (Figure 1). These patients had high mortality compared to the overall patients. The median hospital turn-around time was 18.9 days (range 5–25.5), and the median turn-around time in the PICU was 13.5 days (3.8–19). Twenty-six (44.8%, 26/58) patients had severe pneumonia. Twenty four patients had respiratory abnormalities and sepsis respectively, followed by 22 patients with metabolic disorders, 21 patients with multiple congenital anomalies (MCA), 20 patients with neuromuscular disorders, 11 patients were suspected of having immunodeficiency, nine patients with hepatic disorders, six patients with hemopathy, five patients with cardiac disorders, and four patients with renal disorders (Table S1). Severe infections, such as sepsis and pneumonia, were the most common complications associated with the found primary disorders (Table S1). Fifteen (25.9%) patients had a severe encephalic infection. Furthermore, 12 (20.7%) and 11 (19.0%) patients were suspected of having metabolic disorders or immunodeficiency combined with severe infection, respectively. The statistics of the clinical phenotypes, diagnoses, and management are shown in Table S1.
Two patients were identified pathogenic CNVs via WES and confirmed by CMA. Patient P9 had a copy loss del(7)(q36.3) with a size of 4.7 Mb and a copy gain dup(18)(q12.1q23) with a size of 48.8 Mb (Table 1). The two regions contain 40 genes and 355 genes respectively, including ClinGen haploinsufficiency gene SHH, MNX1, SETBP1, SMAD4, TCF4, and ASXL3. The detailed information of the identified CNVs was listed in Table S2. This patient presented micrognathia, glossoptosis, hypoplastic heart, urethral stenosis, inguinal hernia, malnutrition, and global developmental delay. Patient P10 had a copy loss del(20)(q13.13) with a size of 2.9 Mb (Table 1). This region contains 57 genes including ClinGen haploinsufficiency gene ADNP (Table S2). He manifested hypotelorism, abnormality of eye movement, cryptorchidism, inguinal hernia, and global developmental delay. He also has lymph node tuberculosis. These identified CNVs were classified as pathogenic according to the ACMG and ClinGen criteria, and they had not been reported before.
| Category | Patient | Sex | Age | Hospital turn-around time (days) | PICU Turn-around time (days) | Gene | Gene matched phenotypes | Additional features | Phenotype explained | Severe infection? | Prognosis | Possible clinical interventions |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RES | P1 | F | 3m | 44 | 44 | CFTR | Fibrocystic lung disease: chronic bronchopulmonary infection, pseudomonas colonization | Primary | Pneumonia | Deceased | Protective quarantine to avoid severe infection | |
| P2 | M | 2m | 9 | 9 | MARS1, DNAH11 (VUS) | Interstitial lung and liver disease: hepatomegaly, liver dysfunction, anemia, respiratory insufficiency, dyspnea, cough | Inguinal hernia, hydrocele testis | Primary | Pneumonia | Deceased | ||
| P3 | F | 6m | 18 | 6 | RTEL1 | Pulmonary fibrosis, IRDS, | Leukocytosis | Primary | Sepsis, pneumonia | Deceased | ||
| P4 | M | 15y | 20 | 20 | AQP5 | Palmoplantar keratoderma, Bothnian type | Seizures, hemophagocytosis | Partial | Sepsis, pneumonia, encephalitis | Deceased | ||
| NEU | P5 | F | 4m | 7 | 7 | PRRT2 | Seizures | Diarrhea | Primary | Surviving | Antiepileptic | |
| P6 | M | 3m | 40 | 40 | NFU1 | Multiple mitochondrial dysfunctions syndrome: pulmonary arterial hypertension, respiratory failure, lethargy, decreasing responsiveness, lactic acidosis, Infantile encephalopathy | Primary | Deceased | ||||
| P7 | M | 9m | 1 | 1 | TBCK | Seizures, periventricular white matter abnormalities | Partial | Sepsis | Deceased | |||
| MCA | P8a | F | 9y11m | 29 | 29 | del(X), TOR1A | Turner syndrome: short stature, webbed neck, hypotonia, recurrent otitis media, cystic hygroma | Brain neoplasm | Primary | Deceased | ||
| P9b | M | 9m | 17 | 4 | CNV: del(7)(q36.3), ins(18)(q12.1q23) | Congenital abnormalities, global developmental delay | Primary | Pneumonia | Surviving | |||
| P10 | M | 3m | 25 | 19 | CNV: del(20)(q13.13) | Congenital abnormalities, global developmental delay | Lymph node tuberculoses | Primary | Sepsis, pneumonia | Deceased | ||
| P11 | M | 6m | 18 | ECE1 | Hirschsprung disease, cardiac defects, and autonomic dysfunction: ventricular septal defect, atrial septal defect, failure to thrive in infancy, vomiting, sepsis | IRDS | Primary | Sepsis, pneumonia | Deceased | |||
| P12 | F | 1y2m | 23 | 5 | ARX | Agenesis of the corpus callosum | Gastroenteritis, acute kidney injury | Partial | Sepsis | Surviving | ||
| P13 | M | 1m | 7 | 7 | ZEB2 | Mowat-Wilson syndrome: aganglionic megacolon, abnormal facial shape, ventricular septal defect, patent ductus arteriosus, megacolon | Primary | Pneumonia, enterocolitis | Deceased | |||
| MET | P14 | F | 4m | 98 | 98 | ACAT1 | Beta-ketothiolase deficiency: ketoacidosis, infections may precipitate ketotic episodes | Primary | Sepsis | Surviving | Limitted protein intake and low leucine dietary | |
| P15 | M | 1y4m | 2 | 2 | ACAT1 | Beta-ketothiolase deficiency: metabolic acidosis, vomiting, intellectual disability, ketoacidosis | Primary | Pneumonia | Surviving | Limitted protein intake and low leucine dietary | ||
| P16 | M | 10m | 22 | 8 | CPS1 | Carbamoyl-Phosphate Synthase I Deficiency: coma, cerebral edema, respiratory alkalosis, hyperammonemia | Primary | Encephalitis | Deceased | Early intervention when hyperammonaemia | ||
| P17 | F | 1y3m | 35 | 35 | AGL | Glycogen Storage Disease: hepatomegaly, hepatic fibrosis, hypoglycemia, elevated transaminases, increased serum creatine kinase | Enteritis | Primary | Surviving | Dietary | ||
| HEM | P18 | F | 1y2m | 1 | 1 | VWF | Von Willebrand Disease Type 3: prolonged bleeding time, prolonged bleeding after surgery or trauma, intracranial hemorrhage | Primary | Deceased | VMF VIII supply at regular period | ||
| P19 | F | 1y11m | 22 | 10 | KRAS | Acute myelogenous leukemia | Primary | Sepsis, pneumonia | Deceased | |||
| HEP | P20 | F | 3y7m | 4 | 4 | NBAS | Infantile liver failure syndrome: vomiting, jaundice, hypoglycemia, lethargy, hyperammonemia, coagulopathy, and elevated liver enzymes during episodes, recurrent episodes of liver failure during intercurrent infections, complete recovery during intervals | Primary | Deceased | Liver transplanation | ||
| P21 | F | 9m | 1 | 1 | COG4 | Glycosylation IIj: hepatomegaly, liver failure, delayed psychomotor development, seizures, decreased coagulation factors, abnormal liver enzymes, microcephaly | Primary | Deceased | ||||
| REN | P22 | F | 1y2m | 12 | 12 | INVS | Kidney consumption, infantile nephronophthisis, senior-Loken syndrome: hypertension, respiratory failure, renal failure, heart failure, elevated serum creatinine, situs inversus and valvular or ventricular septal defects, global developmental delay | Pulmonary edema | Primary | Sepsis | Deceased | |
| CAR | P23 | F | 2m | 6 | 6 | TNNT2 | Cardiomyopathy: congestive heart failure,left ventricular dilation | Primary | Deceased | |||
| IMM | P24 | M | 6m | 53 | 46 | LIG4 | Immunodeficiency: recurrent severe infection, tuberculous lymphadenitis | Polydactyly | Primary | Sepsis, pneumonia | Deceased | Bone marrow transplanation |
| P25 | M | 5m | 5 | 5 | NLRP12 | Familial Cold Autoinflammatory Syndrome: splenomegaly, rash, fever | Hypoplastic heart, hydrocele testis, laryngomalacia, tracheomalacia, hyperpigmented nevi, Mongolian blue spot, global developmental delay | Partial | Pneumonia | Surviving | ||
| Other secondary findings | P26 | M | 5y8m | 11 | 11 | IDH1, SCN9A | Paroxysmal extreme pain disorder (Generalized epilepsy with febrile seizures plus, type 7); susceptibility to somatic glioma: seizures, febrile seizures, acute intracranial hypertension with abnormal compact image and bone destruction in CT | Partial | Encephalitis | Deceased | ||
| P27 | M | 8m | 12 | 12 | G6PD | Glucose-6-phosphate dehydrogenase (G6PD) deficiency | Protracted diarrhea, hypersensitivity drug reaction | Partial | Sepsis | Surviving | ||
| Other VUSs | P28 | F | 7y10m | 9 | 9 | PLCG2 | Autoinflammation, antibody deficiency, and immune dysregulation syndrome: interstitial pneumonitis, impaired B cell memory cells, recurrent infection | Primary | Sepsis | Deceased | ||
| P29 | M | 6m | 46 | 28 | PHOX2B, DSP | IRDS; congenital heart defects: alveolar hypoventilation, shallow breathing | Primary | Sepsis, pneumonia | Deceased | |||
| P30 | F | 11y8m | 1 | 1 | PARN | Interstitial pneumonitis: pulmonary fibrosis, immunodeficiency, anemia | Partial | Pneumonia | Deceased | |||
| P31 | M | 1m | 3 | 3 | MYH6 | Atrial septal defect, dilated cardiomyopathy, hypertrophic cardiomyopathy: ventricular septal defect, patent foramen ovale | Primary | Pneumonia | Surviving | Respiratory support | ||
| P32 | M | 1m | 24 | 24 | VCP, SCN5A | Amyotrophy, inclusion body myopathy with early-onset Paget disease and frontotemporal dementia 1: muscle weakness, dysphagia, dyspnea, motor neuron dysfunction | Laryngomalacia, cartilaginous trachea | Primary | Pneumonia | Deceased | ||
| P33 | M | 2y8m | 7 | 7 | ABCC2 | Dubin-Johnson syndrome: cholestasis, jaundice, conjugated hyperbilirubinemia | Primary | Pneumonia | Deceased |
- Abbreviations: CAR, cardiac disorder; HEM, hemopathy; HEP, hepatic disorder; IMM, immunodeficiency; IRDS, infantile respiratory distress syndrome; m, month for age; MET, metabolic disorder; NEU, neuromuscular disorder; REN, renal disorder; RES; respiratory disorder; VUS, variants uncertain significancy; y, year.
- a This patient died of acute tracranial hypertension and respiratory failure. She was further confirmed Turer syndrome after died via WES by CNV analysis and homozygous SNVs analysis. Her associated phenotypes including short stature, webbed neck, etc.
- b This patient was identified an abnormal karyotype 7q+ firstly when admitted to our hospital, and was further detected CNV of del(7)(q36.3) and ins(18)(q12.1q23).
3.2 Patients received a genetic diagnosis with SNVs
Of 58 patients, 25 patients (43.1%) had pathogenic or likely pathogenic (P/LP) SNVs with relevant phenotypes and were confirmed to have genetic disorders. The features of the diagnosed patients were detailed in Table 1. The variants were from 24 genes, which primarily explained the phenotypes of 18 patients (72.0%, 18/25) and partially explain that of seven patients (Table 1; Table S1). Fourteen patients had recessively inherited variants, including 11 compound heterozygous, two homozygous, and one X-linked recessive variation (Table 2). Eleven patients had autosomal dominant inherited variants, of which five were confirmed de novo. The ACMG criteria applied for the identified variants were detailed in Table S3.
| Patient ID | ES family | Inheritance/Zygosity | Gene name | HGVS transcript coding change | HGVS predicted protein change | Allele origion | Variant classificationa | Novel or reported |
|---|---|---|---|---|---|---|---|---|
| P1 | proband | het | CFTR | NM_000492.3:c.3196C>T and c.595C>T | p.(Arg1066Cys) and p.(His199Tyr) | Unknown | LP | HGMD and ClinVar recorded |
| P2 | proband | compHet | MARS1 | NM_004990.3:c.1547A>G and c.1793G>A | p.(Tyr516Cys) and p.(Arg598His) | Unknown | LP | Novel, p.(Arg598Cys) reported in PMID:32833345. |
| P2 | proband | compHet? | DNAH11 | NM_001277115.1:c.6934T>C and c.11352C>A | p.(Ser2312Pro) and p.(Phe3784Leu) | Unknown | VUS | Novel |
| P3 | proband | het | RTEL1 | NM_032957.4:c.709C>T | p.(Arg237Trp) | Unknown | LP | HGMD damaging, PMID:26022962 |
| P4 | proband | het | AQP5 | NM_001651.3:c.526G>A | p.(Gly176Arg) | Unknown | LP | Novel |
| P5 | proband | het | PRRT2 | NM_145239.2:c.971dup | p.(Val325SerfsTer16) | Unknown | P | Novel |
| P6 | proband | compHet | NFU1 | NM_015700.3:c.473+5G>A and c.473G>A | p.? and p.(Arg158Gln) | Unknown | P | HGMD and ClinVar damaging |
| P7 | trio + 2 siblings | hom | TBCK | NM_001163436.2:c.389T>A | p.(Ile130Asn) | Paternal + maternal | LP | Novel |
| P8 | proband | het | TOR1A | NM_000113.2:c.214C>T | p.(Gln72Ter) | Unknown | P | Novel |
| P8 | proband | het | APOE | NM_000041.3:c.784G>A and c.787G>A | p.(Glu262Lys) and p.(Glu263Lys) | Unknown | VUS | ClinVar and HGMD pathogenic |
| P11 | proband + mother | het | ECE1 | NM_001397.2:c.1966G>A | p.(Gly656Arg) | Unknown | LP | Novel |
| P12 | trio | de novo | ARX | NM_139058.2:c.947del | p.(Gly316AlafsTer9) | de novo | P | Novel |
| P13 | trio | de novo | ZEB2 | NM_014795.3:c.2717del | p.(Pro906LeufsTer24) | de novo | P | Novel |
| P14 | proband | compHet | ACAT1 | NM_000019.3:c.131T>C and c.263A>C | p.(Ile44Thr) and p.(Glu88Ala) | Unknown | LP | Novel |
| P15 | trio | compHet | ACAT1 | NM_000019.3:c.1117A>T and c.252del | p.(Lys373Ter) and p.(Glu85LysfsTer2) | Paternal + maternal | P | Novel |
| P16 | trio | compHet | CPS1 | NM_001875.4:c.1145C>T and c.1164+2T>C | p.(Pro382Leu) and p.? | Paternal + maternal | LP, P | Novel |
| P17 | trio | comphet | AGL | NM_000646.2:c.1687+1G>T and c.2633+5G>C | p.? | Maternal + paternal | P, LP | HGMD and ClinVar damaging, novel |
| P18 | trio | compHet | VWF | NM_000552.4:c.2635G>A and c.6963del | p.(Asp879Asn) and p.(Glu2322SerfsTer9) | Maternal + paternal | P, P | Reported, Novel |
| P19 | trio | de novo | KRAS | NM_004985.4:c.35G>A | p.(Gly12Asp) | de novo | P | ClinVar and HGMD |
| P20 | trio | compHet | NBAS | NM_015909.3:c.2630G>T and c.4288C>T | p.(Gly877Val) and p.(Gln1430Ter) | Maternal + paternal | LP, P | Novel |
| P21 | trio | compHet | COG4 | NM_015386.2:c.1255G>T and c.941G>A | p.(Glu419Ter) and p.(Cys314Tyr) | Paternal + maternal | P, LP | Novel |
| P22 | proband | hom | INVS | NM_014425.3:c.2887C>T | p.(Gln963Ter) | biparental? | P | Novel |
| P23 | trio | de novo | TNNT2 | NM_000364.3:c.650_652del | p.(Lys217del) | de novo | P | Reported |
| P24 | trio | compHet | LIG4 | NM_002312.3:c.833G>T and c.1144_1145del | p.(Arg278Leu) and p.(Leu382GlufsTer5) | Paternal + maternal | P, P | HGMD damaging, ClinVar |
| P25 | proband | het | NLRP12 | NM_144687.3:c.1742G>A | p.(Trp581Ter) | Unknown | P | Reported |
| P26 | proband | het | IDH1 | NM_005896.3:c.890G>T | p.(Cys297Phe) | Unknown | LP | Novel |
| P26 | proband | het | SCN9A | NM_002977.3:c.296G>A | p.(Arg99His) | Unknown | VUS | ClinVar VUS |
| P27 | trio | XLR | G6PD | NM_000402.4:c.1114C>T | p.(Leu372Phe) | Maternal | LP | HGMD damaging |
| P28 | proband | het | PLCG2 | NM_002661.4:c.64C>A | p.(Leu22Met) | Unknown | VUS | Novel |
| P29 | proband | het | PHOX2B | NM_003924.3:c.829C>A | p.(Pro277Thr) | Unknown | VUS | Novel |
| P29 | proband | het | DSP | NM_004415.3:c.38C>T | p.(Thr13Ile) | Unknown | VUS | HGMD damaging |
| P30 | proband | het | PARN | NM_002582.3:c.1385A>G | p.(Tyr462Cys) | Unknown | VUS | Novel |
| P31 | proband | het | MYH6 | NM_002471.3:c.4496C>A | p.(Thr1499Asn) | Unknown | VUS | Novel |
| P32 | proband | het | SCN5A | NM_000335.4:c.3067C>T | p.(Arg1023Cys) | Unknown | VUS | HGMD damaging |
| P32 | proband | het | VCP | NM_007126.4:c.697A>G | p.(Ile233Val) | Unknown | VUS | ClinVar VUS |
| P33 | proband | compHet? | ABCC2 | NM_000392.5:c.4239_4240dup and c.2755T>A | p.(His1414LeufsTer18) and p.(Ser919Thr) | Unknown | P, VUS | ClinVar pathogenic, ClinVar uncertain |
| P9 | proband | CNV | P | Novel | ||||
| P10 | trio | de novo | CNV | P | Novel |
- Note: compHet: compound heterozygotes; comphet?: supposed to be compound heterozyous variants but have not been confirmed in trans. P15–P18, P20, P21, and P24 were confirmed in trans by parental sequences (trio). P2 and P14 were confirmed in trans by TA cloning (distance < 3 Kb). P6 was confirmed in trans by IGV (distance < 150 bp).
- a Please see Table S3 for the matched details of ACMG variant classification criteria.
Notably, the identified genetic diseases were quite sporadic in the studied patients. Only two unrelated patients P14 and P15 carried compound heterozygous variations from the same gene ACAT1. P14 carried missense variants c.131T>C (p.Ile44Thr) and c.263A>C (p.Glu88Ala). P15 had a stopsense variant c.1117A>T (p.Lys373Ter) and a frameshift variant c.252del (p.Glu85LysfsTer2). Both patients presented with severe metabolic acidosis, hyponatremia, and hypokalemia (Table 1). Additional phenotypes included sepsis in P14, severe pneumonia, diarrhea, and vomiting in P15. They were surviving at the time of discharge and follow-up visit.
Patient P8 manifested mastoiditis of the middle ear, intracranial cystic lesion, hydrocephalus, subdural effusion, subarachnoid space hemorrhage, short stature, and webbed neck. She was identified two large copy losses on chromosome X expanding almost the p arm (chrX:10500-58517128) and q arm (chrX:62740577-156025374). We further analyzed her SNVs on chromosome X and found that the size of homozygous region was over 146.8 Mb, which indicated the loss of one chromosome X. Considering her phenotype of short stature and webbed neck, we thus speculated that she had Turner syndrome with a karyotype (45,X) (Table 1). As this patient was dead shortly, no blood sample was collected for karyotype analysis. In addition, she had pathogenic variant c.214C>T (p.Gln72Ter) of TOR1A (Table 2). This patient died of central respiratory failure with brain neoplasm.
Excepting the two novel CNVs, there was 23 novel SNVs identified among patients with a genetic diagnosis (Table 2). They were located in 15 genes. Some were located in genes with very limited genetic variants reported previously, such as COG4 and ECE1. For example, patient P21 was found compound heterozygous variants c.1255G>T (p.Glu419Ter) and c.941G>A (p.Cys314Tyr) in COG4. The first variant was inherited from her father, and the second was inherited from her mother. Biallelic mutations of COG4 were revealed to cause a congenital glycosylation disorder (Type IIj) with phenotypes of varying severity, such as compound development delay, recurrent infection, hypotonia, seizures, hepatosplenomegaly, liver cirrhosis, and coagulopathy (Ng et al., 2011; Reynders et al., 2009). Patient P21 presented similar phenotypes, such as persistent jaundice, decreased liver function, abnormal coagulation, and status epilepticus. She also had disseminated intravascular coagulation and a history of recurrent bronchopneumonia and seizures. The patient finally died of liver and respiratory failure.
Some identified P/LP SNVs could not explain the patients' primary phenotypes. These SNVs were considered as secondary findings, such as SNVs from P26 and P27. The two patients both had a severe infection. Patient P26 had c.296G>A (p.Arg99His) in gene SCN9A with uncertain significance. He also had a likely pathogenic variant c.890G>T (p.Cys297Phe) in gene IDH1 which has susceptibility to somatic glioma. The patient presented acute intracranial hypertension, fever, bilateral convulsive seizures, loss of consciousness. Computerized tomography (CT) revealed strips of dense shadows under the cortex of the frontal lobe on both sides and the globus pallidus on both sides. The large wing of the right sphenoid bone and the outer wall of the orbit could see a round-shaped bone destruction area with a clear boundary, about 1.2 × 1.2 × 1.4 cm (CT value is about 26–35HU). Patient P27 had a likely pathogenic variant c.1114C>T (p.Leu342Phe) in gene G6PD (Table 2). This variant was reported relating to glucose phosphate dehydrogenase deficiency (Vulliamy et al., 1993). This patient was a boy and was confirmed to have G6PD deficiency. He presented sepsis, diarrhea, fever, skin rash, hypersensitivity drug reaction, anemia, and malnutrition. No hemolytic anemia was found during his hospitalization period. This variant was inherited from his mother and was considered a second finding.
3.3 Six patients with VUS variants
In addition to P/LP variants, six patients (P28-P33) had VUS variants (Table 1, Table 2). P28 had one VUS in gene PLCG2 associated with autosomal dominant autoinflammation disease (MIM# 614878), and the patient presented interstitial pneumonitis, impaired B cell memory cells, and recurrent infection. P29 had one VUS in gene PHOX2B related to autosomal dominant central hypoventilation syndrome (MIM# 209880) and he presented idiopathic respiratory distress syndrome, alveolar hypoventilation, and shallow breathing. The patient also had one VUS in DSP related to autosomal dominant arrhythmogenic right ventricular dysplasia 8 (MIM# 607450), dilated cardiomyopathy (MIM# 615821), and keratosis palmoplantaris striata II (MIM# 621908), and he presented congenital heart defects. P30 had one VUS in gene PARN related to autosomal dominant pulmonary fibrosis and/or bone marrow failure (MIM# 616371). She manifested pulmonary fibrosis, immunodeficiency, and anemia. P31 had one VUS in gene MYH6 related to the atrial septal defect (MIM# 614089), autosomal dominant dilated cardiomyopathy (MIM# 613252), and hypertrophic cardiomyopathy (MIM# 613251). The patient presented ventricular septal defect and patent foramen ovale. P32 had one VUS in gene VCP related to autosomal dominant inclusion body myopathy with early-onset Paget disease and frontotemporal dementia (MIM# 167320). The patient presented muscle weakness, dysphagia, dyspnea, and motor neuron dysfunction. He also had one VUS in SCN5A. P33 had compound heterozygous variants in gene ABCC2, and one is likely pathogenic and the other is VUS. ABCC2 is related to autosomal recessive Dubin–Johnson syndrome. The patient manifested cholestasis, jaundice, and conjugated hyperbilirubinemia. These VUS-associated genes can primarily explain the phenotypes of five patients (P28, P29, P31–P33). As the lack of the DNA samples of their parents, whether these variants were de novo could not be determined. Their pathogenicity needs to be further investigated.
3.4 Phenotype characteristics of patients with genetic findings
Of the 33 patients with genetic findings, 24 (72.7%) patients died, and nine (27.3%) survived (Figure 1). Of the 27 patients who received a genetic diagnosis with P/LP variants, the genotypes explained the primary clinical phenotypes of 26 patients (78.8%, 26/33). In which, 13 (50%, 13/26) patients were complicated by severe infection and 10 were deceased. Only P14 and P15 with ACAT1 gene variants and P8 with CNV were surviving. Six patients (P2, P3, P10, P11, P22, P24) had phenotypes apart from the related phenotypes of the identified genetic diseases (Table 1). For example, P2 had an inguinal hernia and hydrocele testis, in addition to interstitial lung and liver disease caused by compound heterozygous mutation in the MARS1 gene.
In the six patients (P4, P7, P12, P25, P26, P27) who received a genetic diagnosis, their phenotypes could be partially explained by the identified genotypes. They all had severe infections. P4, P12, P25, and P27 had additional features apart from the identified genetic diseases. P4, P7, and P26 were deceased finally. In the six patients with VUS findings, only P31 was surviving with MYH6 variant accompanied pneumonia. Others died complicated by severe infection.
Overall, near half of the 27 patients with a genetic diagnosis had respiratory defects (44.4%) or severe pneumonia (48.1%) (Table S1). Nineteen patients (70.4%) had severe infections and 13 of them died (68.4%) (Table S4). Ten patients have phenotypes unexplained by the identified genotypes. Nineteen patients died finally.
4 DISCUSSION
Genetic diseases, particularly rare diseases, usually have early onset. It was estimated that 50%–75% of rare diseases affect children (Wright et al., 2018). Many of them impair multisystem and have a range of severe phenotypes. Rare diseases become an important reason for pediatrics hospital admission, and they account for 20%–35% of the early death in children less than 1 year old (Dodge et al., 2011; Kingsmore et al., 2020; Yang et al., 2020). In PICUs, a considerable proportion of patients may have a genetic etiology but have not undergone gene testing yet, particularly those who died without a clear etiology in some developing areas. This retrospective study employed WES integrating comprehensive SNV and CNV analyses, and revealed that 27 of the 58 regional PICU patients had genetic diseases. The identified disease-causing genes were quite sporadic. Our findings expanded the genotypes and phenotypes of at least 19 rare diseases, including 23 novel SNVs from 15 genes and two novel CNVs. 70.4% of the patients with genetic disorders deceased. Severe pneumonia or sepsis was a common accompanied symptom along with a genetic condition that resulted in high mortality.
This regional PICU represents a subgroup of patients who are born with a genetic condition and live in developing areas. In this study, the investigated patients were all transferred from local prefectural hospitals with specific disease progression. The median hospital turn-around time was long as 18.9 days. Some had complex clinical manifestations, and some phenotypes might be atypical. Of the investigated patients, a majority had multisystem abnormalities involving respiratory, neuromuscular, MCA, metabolic, hepatic, renal, cardiac, or hematological systems. WES and WGS provide a powerful early diagnosis method for patients with rare diseases with uncertain etiology, atypical clinical manifestations, or complex expression primarily (Johansen Taber et al., 2014; Kingsmore & Saunders, 2011; Pinxten & Howard, 2014; E. D. Smith et al., 2019). In our study, 56.9% of the participated patients had genetic findings and 46.6% can make a genetic diagnosis. The detected genetic diseases were various and only two patients were caused by the same gene.
In addition to atypical phenotypes and sporadically spread genetic disorders, severe infection (including sepsis, severe pneumonia, or encephalic infection) was found in the majority of our patients. These made the clinical manifestations more complex and critical. In a review by Hotchkiss et al. (2016), the mortality rates of sepsis and septic shock in the general population were 15%–25% and 30%–50%, respectively. Another review summarized that the hospital mortality of severe sepsis was 10.3% in US children (Watson & Carcillo, 2005). Among 123 patients with sepsis in the same PICU department in our hospital in 2019, the mortality rates of sepsis, severe sepsis, and septic shock were 5.5%, 25%, and 27.3%, respectively (unpublished data). The mortality rate of severe pneumonia was approximately 4.2% in 2015 in developing countries (McAllister et al., 2019). This study found high mortality (69%) in patients with severe infection, even excluding immunodeficient patients. In the patients who had severe infections with underlying genetic diseases primarily explained their phenotypes, the mortality increased to 76.9%. Previously, Watson and Carcillo (2005) summarized that the hospital mortality of US children with severe sepsis was 7.8% in previously healthy children, and increased to 12.8% in children with underlying disease. This probably indicates that severe infection concurrently contributes to the death of patients with genetic etiology.
An early precise diagnosis could improve the surviving rate and reduce the admission rate in the PICU. A recent study revealed that the mortality in infants with genetic diseases was 21.6% (205 of 948 infant deaths) (Kingsmore et al., 2020). Jacob et al. (2015) found that only 5% of 641 deceased NICU patients had genetic defects. In a retrospective study of 278 infants from the NICU, PICU, and cardiac ICU by WES, 48.1% of 81 deceased patients were diagnosed with genetic conditions (Meng et al., 2017). Our study revealed that 68.4% of deceased patients had genetic conditions, which is greater than the results of previous studies. This is probably due to the bias in selecting participants (67.2% deaths vs. 4.9% overall deaths during the same period). Respiratory system disorders, neuromuscular, and sepsis were more common in the deceased patients than in the surviving patients. Similarly, research on mortality in approximately 467,000 patients in a pediatric emergency department revealed that the main causes of death were respiratory, neuromuscular, or cardiac disorders and sepsis (Zhu et al., 2015). An earlier differential diagnosis and timely management could save lives or improve living quality.
Overall, the expanded genotypes and phenotypes in this study would facilitate precise diagnosis and subsequent treatments in clinical practice. For some diseases, early diagnosis can allow for interventions that reduce unnecessary testing and treatment. For some families, a clear diagnosis and poor prognosis are closely related to the parent's decision to pursue aggressive or palliative care. In our study, at least 11 children could have undergone appropriate clinical interventions according to the genetic diagnosis. Six lives could have been saved, and the quality of life of five children could have been improved. Furthermore, with known genetic variants and carrier status determined by genomic testing, it would be beneficial for the related families to undergo prenatal diagnosis and screening to avoid giving birth to babies with genetic defects. Thus, morbidity and mortality in newborns would be reduced, as well as the burden of disease.
Since this was a retrospective study, no instance data of clinical management could be obtained after receiving a molecular diagnosis. Other limitations include the following: first, complete data were not available from some patients to further evaluate the pathogenicity of the detected variants. A large number of detected variants were classified as having uncertain significance. For example, some cases lacked the samples of parents. Some lacked the related clinical data to confirm the genotype-phenotype correlations, including physical examinations, enzyme assays, imaging studies, or pathological detections. Some families of the patients were difficult to contact to obtain detailed follow-up information on disease progress and individual development. Second, WES-based technology cannot detect genomic variants in noncoding regions or structural variants, such as translocation, inversion, and mosaic variants. WGS could improve the diagnosis rate. For some undiagnosed neuromuscular cases, a special mitochondrial test would be a good choice. Third, the phenotypes of some patients might not be examined thoroughly due to limited time or facilities. Some phenotypes might be incomplete or late-onset due to individual differences. Forth, due to the limited sample number, there were a few patients with certain clinical criteria (such as hematopoiesis or renal diseases), and thus some statistical data might not be very strong. We believe further examinations and analyses could enhance genetic diagnosis more comprehensively and provide more sophisticated guidance for clinical practice.
5 CONCLUSION
WES integrated with CNV analysis helped to clarify the etiology of dozens of the regional PICU patients. Our findings expanded the genotypic and phenotypic spectrum over 19 rare diseases and will provide a basis for clinical practice in earlier preventing, diagnosis, and management of such rare diseases.
ACKNOWLEDGMENTS
This study was supported by the Key Laboratory of Emergency Medicine for Children (grant no.: 2018TP1028), and the Ministry of Science and Technology in China (grant no.:2012BAI04B02). The authors would like to thank the Pediatrics Research Institute of Hunan Province for its contribution of valuable assistance with the laboratory and field tests. They also would like to express our sincere gratitude to all those who helped during the writing of this thesis.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Conceptualization and investigation: Juan Liu, Yu Zheng, Yongjia Yang, Zhenghui Xiao, Yimin Zhu, and Xiulan Lu. Funding acquisition, investigation, supervision, resources acquisition, methodology, and validation: Zhenghui Xiao, Yimin Zhu, and Xiulan Lu. Original draft writing and editing, data analysis, and visualization: Yu Zheng and Juan Liu. Resources collection and data validation: Xiulan Lu, Yongjia Yang, Zhenqing Luo, and Yu Peng. Manuscript review and editing: Xiulan Lu, Zhenghui Xiao, Yimin Zhu, and Yongjia Yang. Jiaotian Huang, Desheng Zhu, and Ping Zang contributed to ideas refining, additional analyses, and finalization. All authors read and approved the final manuscript.
ETHICS STATEMENT
The samples were obtained appropriate informed consent from all participants.
Open Research
DATA AVAILABILITY STATEMENT
The novel variants found in this study have been submitted to ClinVar (Submission ID: SUB9827004, ClinVar accession: from SCV001737514 to SCV001737542). Other datasets used or analyzed during the current study are available from the corresponding author on reasonable request. Some or all data, models, or codes generated or used during the study are available from the corresponding author by request.




