Mutation Update and Review of Severe Methylenetetrahydrofolate Reductase Deficiency
Contract grant sponsors: Rare Disease Initiative Zurich (radiz), a clinical research priority program for rare diseases of the University of Zurich, Switzerland and the Swiss National Science Foundation (SNSF 31003A_138521 and 31003A_156907); RVO VFN 64165 and PRVOUK-P24/LF1/3.
Communicated by Jan P. Kraus
ABSTRACT
Severe 5,10-methylenetetrahydrofolate reductase (MTHFR) deficiency is caused by mutations in the MTHFR gene and results in hyperhomocysteinemia and varying severity of disease, ranging from neonatal lethal to adult onset. Including those described here, 109 MTHFR mutations have been reported in 171 families, consisting of 70 missense mutations, 17 that primarily affect splicing, 11 nonsense mutations, seven small deletions, two no-stop mutations, one small duplication, and one large duplication. Only 36% of mutations recur in unrelated families, indicating that most are “private.” The most common mutation is c.1530A>G (numbered from NM_005957.4, p.Lys510 = ) causing a splicing defect, found in 13 families; the most common missense mutation is c.1129C>T (p.Arg377Cys) identified in 10 families. To increase disease understanding, we report enzymatic activity, detected mutations, and clinical onset information (early, <1 year; or late, >1 year) for all published patients available, demonstrating that patients with early onset have less residual enzyme activity than those presenting later. We also review animal models, diagnostic approaches, clinical presentations, and treatment options. This is the first large review of mutations in MTHFR, highlighting the wide spectrum of disease-causing mutations.
Introduction
Methylenetetrahydrofolate reductase (MTHFR; MIM #607093; E.C. 1.5.1.20) is a cytoplasmic enzyme that catalyzes the physiologically irreversible reduction of 5,10-methylenetetrahydrofolate (methyleneTHF) to 5-methyltetrahydrofolate (methylTHF), a reaction that requires NADPH as electron donor and FAD as cofactor. Severe deficiency of MTHFR, first described in 1972 [Mudd et al., 1972; Shih et al., 1972], is inherited in an autosomal-recessive manner. It remains the most common metabolic disorder of folate metabolism [Watkins and Rosenblatt, 2014], although the incidence is likely very rare, for example, two individuals were detected in >400,000 retrospective newborn screening (NBS) samples [Tortorelli, et al., 2010]. It is characterized by hyperhomocysteinemia, with elevated plasma S-adenosylhomocysteine, homocystinuria, increased plasma cystathionine, as well as low/normal plasma methionine and S-adenosylmethionine (AdoMet). These metabolic imbalances result from decreased flux through methionine synthase (EC 2.1.1.13; MIM #156570), which depends on the methylTHF generated by MTHFR to serve as methyl donor for the methylation of homocysteine to methionine. Methionine in turn is converted to AdoMet, which is the vital cellular methyl donor [Watkins and Rosenblatt, 2014] as well as an allosteric regulator of homocysteine metabolism.
Human MTHFR protein is a homodimer with each subunit consisting of an N-terminal catalytic domain (amino acids, aa ∼1–356)—which binds methyleneTHF, NADPH, and FAD—and a C-terminal regulatory domain (aa ∼363–656), connected by a short linker region (aa 357–362) (Fig. 1). The catalytic domain appears to be sufficient to carry out the entire enzymatic reaction. In the absence of a eukaryotic MTHFR protein structure, structural studies have mainly focused on the E. coli homologue MetF, which consists only of the catalytic domain. Nevertheless, these studies have revealed residues critical for binding of FAD [Guenther, et al., 1999] and methylTHF [Pejchal, et al., 2005; Lee et al., 2009]. The regulatory domain binds AdoMet, resulting in allosteric inhibition of the enzyme [Sumner, et al., 1986], an effect that can be reversed by adenosylhomocysteine binding [Daubner and Matthews, 1982; Yamada, et al., 2001]. Mutations in the regulatory domain may affect AdoMet-mediated inhibition, either increasing or decreasing the inhibitory constant, depending on the nature and location of the mutation [Burda, et al., 2015b]. MTHFR can also be regulated by phosphorylation, resulting in decreased enzymatic activity. Recombinant expression of human MTHFR in insect cells [Yamada, et al., 2005] and yeast [Marini, et al., 2008] produces both phosphorylated and unphosphorylated protein, a phenomenon also found for endogenous MTHFR of human cancer cell lines [Zhu, et al., 2014]. Substitution of threonine to alanine on residue 34 (p.Thr34Ala) appears to block this phosphorylation [Yamada, et al., 2005; Marini, et al., 2008].
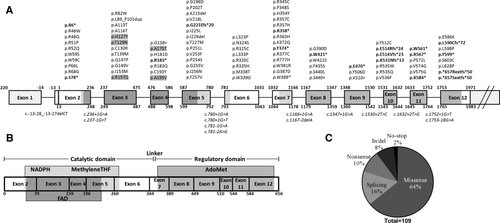
In humans, the MTHFR gene resides on chromosome 1p36.3 and was originally described as containing 11 exons [Goyette, et al., 1998]. Later, variable 5′ and 3′ untranslated regions and splice variants were identified, resulting in transcript isoforms of 7.5 - 9.5 kb and protein of 70 - 77 kDa [Tran, et al., 2002]. Current assemblies (e.g. NCBI Annotation Release 107) describe the major MTHFR isoform to consist of 12 exons (NM_005957.4, ENST00000376590), the first of which is non-coding (Fig. 1), resulting in a protein of 656 aa and 74.6 kDa (NP_005948.3, ENSP00000365775). The most important difference between these new assemblies and the original description is that in Goyette et al. [1998] the starting “A” of the ATG initiation codon was designated +13, while the current nomenclature correctly designates it as +1. This had led to confusion in the literature as most published papers cite the mRNA sequence according to the original labelling of Goyette and coworkers. However, since variation databases such as ClinVar and dbSNP use the updated labelling, we have preferred to use it here. For clarity, we have described each variation using labelling in both styles in Table 1, and in the text we primarily refer to changes to the protein.
| Nucleotide change [Goyette et al., 1998]a | Nucleotide change (HGVS; NM_005957.4)a | Exon/ Intron | Predicted amino acid change | Number of alleles | Number of families | Variant reference | Literature reference |
|---|---|---|---|---|---|---|---|
| N-terminal catalytic domain | |||||||
| c.41-40delCT | c.-13-28_-13-27delCT | Intron 1 | 1 | 1 | rs786204005, CD155816 | Burda et al. [2015b] | |
| c.28A>T | c.16A>T | Exon 2 | p.Arg6* | 2 | 1 | CM013904, CM155646 | Tonetti et al. [2001] |
| c.148C>T | c.136C>T | Exon 2 | p.Arg46Trp | 3 | 2 | rs138189536, CM155648 | Burda et al. [2015b] |
| c.149G>A | c.137G>A | Exon 2 | p.Arg46Gln | 1 | 1 | rs776483190 | Burda et al. [2015b] |
| c.164G>C | c.152G>C | Exon 2 | p.Arg51Pro | 2 | 2 | rs201618781, CM960994 | Goyette et al. [1996]; Wong et al. [2015] |
| c.167G>A | c.155G>A | Exon 2 | p.Arg52Gln | 10 | 8 | CM950818 | Goyette et al. [1995]; Tonetti et al. [2002, 2003]; Prasad et al. [2011]; D'Aco et al. [2014]; Burda et al. [2015b] |
| c.188G>C | c.176G>C | Exon 2 | p.Trp59Ser | 2 | 1 | rs786204007, CM155512 | Burda et al. [2015b] |
| c.189G>T | c.177G>T | Exon 2 | p.Trp59Cys | 4 | 2 | rs767789270 | This study |
| c.209C>T | c.197C>T | Exon 2 | p.Pro66Leu | 2 | 1 | rs796064512 | This study |
| c.214C>G | c.202C>G | Exon 2 | p.Arg68Gly | 1 | 1 | rs763539350, CM155593 | Burda et al. [2015b] |
| c.245C>G | c.233C>G | Exon 2 | p.Ser78* | 2 | 1 | CM035839 | Tonetti et al. [2003] |
| c.248+1G>A | c.236+1G>A | Intron 2 | Splice site | 2 | 1 | SCV000258401 | Wong et al. [2015] |
| c.249-1G>T | c.237-1G>T | Intron 2 | Splice site | 3 | 3 | CS961627 | Goyette et al. [1996]; Burda et al. [2015b] |
| c.256C>T | c.244C>T | Exon 3 | p.Arg82Trp | 1 | 1 | rs786204009, CM155649 | Burda et al. [2015b] |
| c.276_314dup | c.264_302dup | Exon 3 | p.Leu89_Pro101dup | 1 | 1 | rs786204010 | Burda et al. [2015b] |
| c.349G>A | c.337G>A | Exon 3 | p.Ala113Thr | 4 | 3 | rs147257424, CM155514 | Burda et al. [2015b] |
| c.358G>A | c.346G>A | Exon 3 | p.Ala116Thr | 2 | 2 | CM000525 | Sibani et al. [2000]; Arai and Osaka [2011] |
| c.391C>T | c.379C>T | Exon 3 | p.His127Tyr | 1 | 1 | rs769381688, CM155515 | Burda et al. [2015b] |
| c.398C>A | c.386C>A | Exon 3 | p.Thr129Asn | 1 | 1 | CM127685 | Crushell et al. [2012] |
| c.400T>C | c.388T>C | Exon 3 | p.Cys130Arg | 1 | 1 | rs786204012, CM155650 | Burda et al. [2015b] |
| c.428C>T | c.416C>T | Exon 3 | p.Thr139Met | 1 | 1 | SCV000258402 | Yano et al. [2004] |
| c.452A>C | c.440A>C | Exon 3 | p.Gln147Pro | 6 | 3 | rs786204013, CM155516 | Burda et al. [2015b] |
| c.458_459delinsTT | c.446_447delinsTT | Exon 3 | p.Gly149Val | 6 | 4 | CX962709 | Arai and Osaka [2011]; Goyette et al. [1996]; Yano et al. [2004]; Tsuji et al. [2011] |
| c.471C>G | c.459C>G | Exon 3 | p.Ile153Met | 2 | 2 | CM030908 | Sibani et al. [2003]; Burda et al. [2015b] |
| c.482G>A | c.470G>A | Exon 3 | p.Arg157Gln | 9 | 6 | rs121434295, CM941069 | Goyette et al. [1994]; Michot et al. [2008]; Burda et al. [2015b]; This study |
| c.486A>T | c.474A>T | Exon 4 | p.Gly158=(splicing) | 4 | 2 | CS129635 | Ben-Shachar et al. [2012] |
| c.535G>A | c.523G>A | Exon4 | p.Ala175Thr | 2 | 1 | CM102999 | Forges et al. [2010]; Burda et al., 2015a |
| c.553C>G | c.541C>G | Exon 4 | p.His181Asp | 1 | 1 | CM035838 | Tonetti et al., 2003 |
| c.559C>T | c.547C>T | Exon 4 | p.Arg183* | 10 | 6 | rs121434294, CM941070 | Goyette et al., 1994 ; Cappuccio et al., 2014; Burda et al., 2015b |
| c.560G>A | c.548G>A | Exon 4 | p.Arg183Gln | 4 | 1 | rs574132670, CM155651 | Burda et al., 2015b |
| c.590G>A | c.578G>A | Exon 4 | p.Cys193Tyr | 1 | 1 | CM035842 | Tonetti et al., 2003 |
| c.596C>T | c.584C>T | Exon 4 | p.Ala195Val | 2 | 1 | CM108907 | Bathgate et al., 2012 |
| c.599G>A | c.587G>A | Exon 5 | p.Gly196Asp | 1 | 1 | rs786204014, CM155616 | Burda et al., 2015b |
| c.616C>A | c.604C>A | Exon 5 | p.Pro202Thr | 2 | 1 | SCV000258403 | Schiff et al., 2011 |
| c.655_657delAAG | c.643_645delAAG | Exon 5 | p.Lys215del | 1 | 1 | rs746353274, CD155530 | Burda et al., 2015b |
| c.664G>T | c.652G>T | Exon 5 | p.Val218Leu | 2 | 1 | CM106684 | Urreizti et al., 2010 |
| c.674delG | c.662delG | Exon 5 | p.Gly221Glufs*20 | 2 | 1 | SCV000258404 | Wong et al. [2015] |
| c.685A>C | c.673A>C | Exon 5 | p.Ile225Leu | 2 | 1 | rs200100285 | Tamura et al. [2014]; Burda et al. [2015b] |
| c.689_691delTCA | c.677_679delTCA | Exon 5 | p.Ile226del | 1 | 1 | rs786204016, CD155620 | Burda et al. [2015b] |
| c.692C>T | c.680C>T | Exon 5 | p.Thr227Met | 2 | 1 | CM950820 | Goyette et al. [1995] |
| c.764C>T | c.752C>T | Exon 5 | p.Pro251Leu | 2 | 1 | CM950821 | Goyette et al. [1995] |
| c.769G>T | c.757G>T | Exon 5 | p.Val253Phe | 1 | 1 | CM127686 | Crushell et al. [2012]; Burda et al. [2015a, 2015b] |
| c.772C>T | c.760C>T | Exon 5 | p.Pro254Ser | 2 | 1 | rs786204017, CM155652 | Burda et al. [2015b] |
| c.776G>T | c.764G>T | Exon 5 | p.Gly255Val | 1 | 1 | rs786204018, CM155517 | Burda et al. [2015b] |
| c.779T>A | c.767T>A | Exon 5 | p.Ile256Asn | 1 | 1 | rs373398993, CM155517 | Burda et al. [2015b] |
| c.781T>G | c.769T>G | Exon 5 | p.Phe257Val | 1 | 1 | rs786204019, CM155513 | Burda et al. [2015b] |
| c.792+1G>A | c.780+1G>A | Intron 5 | Splice site | 4 | 2 | CS951475 | Goyette et al. [1995]; Schiff et al. [2011] |
| c.792+1G>T | c.780+1G>T | Intron 5 | Splice site | 1 | 1 | rs786204020, CS155526 | Burda et al. [2015b] |
| c.793-1G>A | c.781-1G>A | Intron 5 | Splice site | 1 | 1 | CS147900 | D'Aco et al. [2014] |
| c.793-2A>G | c.781-2A>G | Intron 5 | Skips exon 6 | 2 | 1 | CS035858 | Tonetti et al. [2003] |
| c.980T>C | c.968T>C | Exon 6 | p.Leu323Pro | 2 | 2 | rs121434297, CM960995 | Goyette et al. [1996]; Sahai et al. [2014] |
| c.983A>G | c.971A>G | Exon 6 | p.Asn324Ser | 2 | 1 | rs267606887, CM983381 | Kluijtmans et al. [1998] |
| c.985C>T | c.973C>T | Exon 6 | p.Arg325Cys | 2 | 1 | rs371085894, CM950822 | Goyette et al. [1995] |
| c.1010T>C | c.998T>C | Exon 6 | p.Leu333Pro | 2 | 1 | CM016157 | Homberger et al. [2001] |
| c.1015C>T | c.1003C>T | Exon 6 | p.Arg335Cys | 1 | 1 | CM950823 | Goyette et al. [1995] |
| c.1016G>A | c.1004G>A | Exon 6 | p.Arg335His | 1 | 1 | rs543016186, CM155653 | Burda et al. [2015b] |
| c.1025T>C | c.1013T>C | Exon 6 | p.Met338Thr | 6 | 5 | rs368321176, CM030909 | Sibani et al. [2003]; Cappuccio et al. [2014]; Burda et al. [2015b] |
| c.1027T>G | c.1015T>G | Exon 6 | p.Trp339Gly (splicing) | 12 | 5 | rs267606886, CM983382 | Kluijtmans et al. [1998]; Sibani et al. [2003]; Burda et al. [2015b] |
| c.1045C>T | c.1033C>T | Exon 7 | p.Arg345Cys | 1 | 1 | SCV000258405 | This study |
| c.1054C>T | c.1042C>T | Exon 7 | p.Pro348Ser | 1 | 1 | rs786204021, CM155623 | Burda et al. [2015b] |
| c.1072C>T | c.1060C>T | Exon 7 | p.His354Tyr | 2 | 1 | rs786204022, CM155626 | Burda et al. [2015b] |
| Putative linker region | |||||||
| c.1081C>T | c.1069C>T | Exon 7 | p.Arg357Cys | 9 | 2 | CM950824 | Goyette et al. [1995]; Tonetti et al. [2000] |
| c.1082G>A | c.1070G>A | Exon 7 | p.Arg357His | 2 | 2 | CM088302 | Birnbaum et al. [2008]; Kim et al. [2013] |
| c.1084C>T | c.1072C>T | Exon 7 | p.Arg358* | 2 | 1 | rs377443637, CM983383 | Kluijtmans et al. [1998] |
| C-terminal regulatory domain | |||||||
| c.1100G>A | c.1088G>A | Exon 7 | p.Arg363His | 2 | 1 | rs786204023, CM155629 | Burda et al. [2015b] |
| c.1126A>G | c.1114A>G | Exon 7 | p.Lys372Glu | 1 | 1 | rs786204024, CM155630 | Burda et al. [2015b] |
| c.1134C>G | c.1122C>G | Exon 7 | p.Tyr374* | 1 | 1 | CM000526 | Sibani et al. [2000] |
| c.1141C>T | c.1129C>T | Exon 7 | p.Arg377Cys | 15 | 10 | rs121434296, CM960996 | Goyette et al. [1996]; Sibani et al. [2003]; Tonetti et al. [2003]; Strauss et al. [2007]; Burda et al. [2015b] |
| c.1142G>A | c.1130G>A | Exon 7 | p.Arg377His | 6 | 2 | CM147021 | Lossos et al. [2014]; Burda et al. [2015b] |
| c.1153C>T | c.1141C>T | Exon 7 | p.Trp381Arg | 2 | 1 | CM147019 | Lossos et al. [2014] |
| c.1172G>A | c.1160G>A | Exon 7 | p.Gly387Asp | 2 | 2 | CM000527 | Sibani et al. [2000]; Burda et al. [2015b] |
| c.1178G>A | c.1166G>A | Exon 7 | (p.Trp389*) Skips exon 7 | 2 | 1 | CS103000 | Forges et al. [2010]; Burda et al. [2015a, 2015b] |
| c.1178+1G>A | c.1166+1G>A | Intron 7 | Skips exon 6 | 3 | 2 | CS035859 | Tonetti et al. [2002, 2003] |
| c.1179-2delA | c.1167-2delA | Intron 7 | (Skips exon 8) p.Trp389Trpfs*1 | 5 | 4 | rs780014899, CD155532 | Burda et al. [2015b] |
| c.1181G>A | c.1169G>A | Exon 8 | p.Gly390Asp | 1 | 1 | CM1314017 | Kim et al. [2013] |
| c.1274G>A | c.1262G>A | Exon 8 | p.Trp421* | 1 | 1 | CM030910 | Sibani et al. [2003] |
| c.1274G>C | c.1262G>C | Exon 8 | p.Trp421Ser | 2 | 2 | rs200137991, CM016159 | Homberger et al. [2001]; Burda et al. [2015b] |
| c.1316T>C | c.1304T>C | Exon 8 | p.Phe435Ser | 3 | 2 | CM106685 | Urreizti et al. [2010]; Munoz et al. [2015] |
| c.1331C>T | c.1319C>T | Exon 8 | p.Ser440Leu | 2 | 1 | CM035840 | Tonetti et al. [2002] |
| c.1332G>A | c.1320G>A | Exon 8 | p.Ser440=(splicing) | 2 | 2 | rs367585605, CS155527 | Burda et al. [2015b] |
| c.1359+1G>A | c.1347+1G>A | Intron 8 | splice site | 2 | 2 | CS155636 | Prasad et al. [2011];b Burda et al. [2015b] |
| c.1420G>T | c.1408G>T | Exon 9 | p.Glu470* | 10 | 6 | CM016158 | Homberger et al. [2001]; Sibani et al. (2003]; Urreizti et al. [2010]; Burda et al. [2015b] |
| c.1528T>G | c.1516T>G | Exon 9 | p.Tyr506Asp | 1 | 1 | rs786204026, CM155615 | Burda et al. [2015b] |
| c.1542G>A | c.1530G>A | Exon 9 | p.Lys510=(splicing) | 25 | 13 | CM135284, CS1412548 | Richard et al. [2013]; Broomfield et al. [2014]; Burda et al. [2015b] |
| c.1542+2T>C | c.1530+2T>C | Intron 9 | p.Tyr512Trpfs*3 | 1 | 1 | rs786204027, CS155635 | Burda et al. [2015b] |
| c.1547A>G | c.1535A>G | Exon 10 | p.Tyr512Cys | 2 | 1 | CM147020 | [Lossos et al. [2014] |
| c.1551dupA | c.1539dupA | Exon 10 | p.Glu514Argfs*24 | 3 | 2 | CI44310 | Michot et al. [2008]c; Munoz et al. [2015] |
| c.1553_1554delAG | c.1541_1542delAG | Exon 10 | p.Glu514Valfs*23 | 4 | 4 | CD031063 | Sibani et al. [2003]; Sahai et al. [2014]d; Wong et al. [2015]; Rummel et al. [2007]e |
| c.1605delG | c.1593delG | Exon 10 | p.Lys531Asnfs*13 | 2 | 1 | SCV000258406 | Schiff et al. [2011] |
| c.1615C>T | c.1603C>T | Exon 10 | p.Arg535Trp | 4 | 2 | CM013905 | Tonetti et al. [2001]; Schiff et al. [2011] |
| c.1616G>A | c.1604G>A | Exon 10 | p.Arg535Gln | 1 | 1 | CM088303 | Birnbaum et al. [2008] |
| c.1618G>T | c.1606G>T | Exon10 | p.Val536Phe | 1 | 1 | rs786204028, CM155625 | Burda et al. [2015b] |
| c.1644+2T>G | c.1632+2T>G | Intron 10 | Skips exon 10 | 4 | 3 | rs749765738, CS155607 | Burda et al. [2015b] |
| c.1695G>A | c.1683G>A | Exon 11 | p.Trp561* | 1 | 1 | rs786204030, CM155520 | Burda et al. [2015b] |
| c.1711C>T | c.1699C>T | Exon 11 | p.Arg567* | 8 | 5 | rs140277700, CM983384 | Kluijtmans et al. [1998]; Sibani et al. [2000]; Tonetti et al. [2003] |
| c.1727C>T | c.1715C>T | Exon 11 | p.Pro572Leu | 3 | 3 | rs144508139, CM000528 | Sibani et al. [2000, 2003]; Burda et al. [2015b] |
| c.1733T>G | c.1721T>G | Exon 11 | p.Val574Gly | 2 | 1 | CM106686 | Urreizti et al. [2010]; Burda et al. [2015a, 2015b] |
| c.1736T>G | c.1724T>G | Exon 11 | p.Val575Gly | 2 | 1 | rs786204031, CM155632 | Burda et al. [2015b] |
| c.1762A>T | c.1750A>T | Exon 11 | p.Lys584* | 2 | 1 | CM000529 | Sibani et al. [2000] |
| c.1764+1G>T | c.1752+1G>T | Intron 11 | Skips exon 11 | 4 | 2 | rs747846362, CS155637 | Burda et al. [2015b) |
| c.1765-18G>A | c.1753-18G>A | Intron 11 | p.Asp585Glyfs*14 | 7 | 4 | rs777661576, CS155622 | Burda et al. [2015b] |
| c.1768G>A | c.1756G>A | Exon 12 | p.Glu586Lys | 1 | 1 | CM000530 | Sibani et al. [2000] |
| c.1780delC | c.1768delC | Exon 12 | p.Leu590Cysfs*72 | 2 | 1 | CD106687 | Urreizti et al. [2010]; Burda et al. [2015a, 2015b] |
| c.1805T>C | c.1793T>C | Exon 12 | p.Leu598Pro | 2 | 1 | rs786204034, CM155644 | Burda et al. [2015b] |
| c.1809_1810delinsGT | c.1797_1798delinsGT | Exon 12 | p.Tyr599* | 1 | 1 | rs786204035, CX155658 | Burda et al. [2015b] |
| c.1820C>G | c.1808C>G | Exon 12 | p.Ser603Cysf | 1 | 1 | rs758206023 | Burda et al. [2015b] |
| c.1895T>C | c.1883T>C | Exon 12 | p.Leu628Pro | 1 | 1 | rs786204037, CM155645 | Burda et al. [2015b] |
| c.1981T>C | c.1969T>C | Exon 12 | p.*657Argextfs*50 | 2 | 1 | rs768434408, CM155522 | Burda et al. [2015b] |
| c.1982G>C | c.1970G>C | Exon 12 | p.*657Serextfs*50 | 5 | 2 | CM035841 | Tonetti et al. [2003]; Burda et al. [2015b] |
- a The nomenclature per Goyette et al. [1998] represents legacy reporting in the literature. In the text, numbering of the nucleotide changes and exons/introns follows the nomenclature of NM_005957.4 with +1 as the number of the A of the ATG initiation codon (codon 1).
- b Reported the mutation as c.1348+1G>A.
- c Reported the mutation as A ins 1551(+1).
- d Reported the mutation as c.1539_1542delAG, p.E514VfsX31.
- e Reported the mutation as c.1552_c.1553delGA [p.E514fsX536].
- f This mutation was detected in cis with the c.1809_1810delinsGT change in one patient in Burda et al. [2015b].
A large number of SNPs in MTHFR are known, and a summary of the literature on the effect of these polymorphisms on protein function and disease is beyond the scope of this report. Of the few common variants, p.Ala222Val has been particularly well studied, and the effect of this variation in lowering protein stability [Frosst, et al., 1995] and residual enzymatic activity when found in combination with severe mutations [Sibani, et al., 2003] needs to be borne in mind.
Variants
To date, 109 mutations in MTHFR from 171 families (192 patients) have been described as causing severe MTHFR deficiency (Table 1, Supp. Table S1, Fig. 1A). Included in these numbers are the 3 novel mutations (Table 1) and 6 new patients (Supp. Table S1) described for the first time here. As found in most recessively inherited autosomal diseases, missense mutations represent the predominant mutation type (n = 70, Fig. 1C). In addition, 17 mutations which primarily cause aberrant splicing (3 of which are synonymous), 11 nonsense mutations, seven small deletions, two no-stop mutations, one small duplication, and one large duplication are known. No large deletions or deep intronic mutations have been reported.
The majority of mutations are private. Seventy mutations have been found only in single families, 33 are found in between two and five families, and six have been found in six or more families. The most frequently seen mutation is c.1530G>A (p.Lys510 = ), which causes a splicing defect, found in 13 families (Table 1). This synonymous change results in skipping of exon 9 causing a frameshift and premature termination of translation [Burda, et al., 2015b]. The second most frequently seen mutation, and the most common missense mutation, is p.Arg377Cys (c.1129C>T), found in 10 families (Table 1). The same residue is changed to histidine (c.1130G>A, p.Arg377His) in two families, suggesting an important role for this residue, which is located within the regulatory domain. Four other frequently seen missense mutations are p.Arg52Gln (c.155G>A), p.Arg157Gln (c.470G>A), p.Met338Thr (c.1013T>C), and p.Trp339Gly (c.1015T>G), which were found in eight, six, five, and five families, respectively (Table 1). In cells, p.Trp339Gly has been shown to produce an alternatively spliced variant leading to a frameshift, which was found in equal abundance to the normally sized splice variant [Kluijtmans, et al., 1998], thus indicating that the deficiency caused by this mutation may be more severe than that derived solely from the missense change. Indeed, the activity in fibroblasts of three patients homozygous for this mutation is virtually absent (0.2%–0.6% of control [Burda, et al., 2015b].
The next most common mutations are of the nonsense type: p.Arg183* (c.547C>T; 6 families), p.Glu470* (c.1408G>T; 6 families), and p.Arg567* (c.1699C>T; 5 families). Fibroblasts from patients homozygous for p.Arg183* [Rosenblatt, et al., 1992; ; Goyette, et al., 1994; Burda, et al., 2015b] and p.Glu470* [Burda, et al., 2015b] have virtually undetectable MTHFR activity. Anomalously, a patient homozygous for p.Arg567* was reported to have residual activity in fibroblasts of between 20% and 50% of healthy controls ([Kluijtmans, et al., 1998], Supp. Table S1). However, analysis of lymphocytes of the only homozygous patient revealed MTHFR activity of less than 10% of mean normal activity (Supp. Table S1) [Engelbrecht, et al., 1997], and patients compound heterozygous for this and a missense mutation showed activity in fibroblasts of <8% (Supp. Table S1). Thus, very low activity is more likely to be associated with this mutation, especially given the nature of the mutation and the early and severe presentation of this patient [Engelbrecht, et al., 1997; Kluijtmans, et al., 1998]. The rest of the mutations identified in severe MTHFR deficiency and summarized in Table 1 are found in a maximum of four families.
Mutations are found in every exon except for number 1, which does not have any coding sequence (Fig. 1A). However, the distribution of missense mutations across the MTHFR gene is not uniform. Although the catalytic and regulatory domains are comparable in size (∼356 aa and ∼293 aa, respectively) the catalytic domain hosts more than twice as many missense mutations as the regulatory domain (46 vs. 22, two in the linker). This is consistent with its requirement for enzymatic function since by comparison with bacterial homologues, all substrate and cofactor binding sites are expected to be hosted within the catalytic domain.
Inspection of the location of missense mutations reveals that the first arises at residue 46 (p.Arg46Trp: Table 1, Fig. 2). Amino acids N-terminal to this residue are poorly conserved (Fig. 2), with an unusually high proportion of serine residues (11 of the first 40 aa in human). Given the finding of phosphorylation within this region as a mean of MTHFR activity regulation in eukaryotic expression systems [Yamada, et al., 2005; Marini, et al., 2008] and human cancer cell lines [Zhu, et al., 2014], it is probable that this serine-rich region represents an evolutionarily new sequence for activity modulation, which is not present in E. coli, yeast, or C. elegans (Fig. 2).
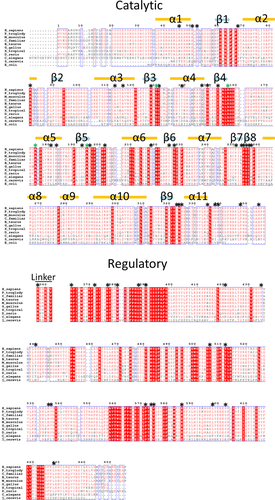
Within the catalytic domain, the only missense mutation changing a weakly conserved residue is p.Gln147Pro (Fig. 2). It is likely that mutation to a proline in this position is deleterious because it breaks the alpha-helix (α4) to which p.Gln147 belongs (Fig. 2). Within the catalytic domain, residues found to be involved in FAD binding in MetF [Guenther, et al., 1999] and also found to be mutated in FAD-responsive cell lines [Burda, et al., 2015b]) include: p.Thr129, p.Arg157, p.Ala175Thr, and p.Ala195, all of which are strictly conserved (Fig. 2). Interestingly, there is a 65 aa region close to the end of the catalytic domain within which no missense mutations have been found (c.770-967, Pro258-Thr322 inclusive; Figs. 2 and 3), whereas apparent mutational hotspots occur on either side of this region (N-terminal: p.Pro251Lue, p.Val253Phe, p.Pro254Ser, p.Gly255Val, P.Ile256Asn, p.Phe257Val; C-terminal: p.Leu323Pro, p.Asn324Ser, p.Arg325Cys; Table 1, Fig. 2). The lack of mutations and poor conservation within the Pro258-Thr322 region suggests that although these amino acids are localized in the catalytic domain, they are not critical for enzymatic activity. This is supported by the fact that 21 SNPs causing missense changes have been identified in this region (Ensembl, build 83), but none of these variations, to our knowledge, has been associated with disease. In contrast to this assertion, Leu268 (E. coli Phe223) was found to be in contact with the methylTHF moiety in E. coli crystal structures [Pejchal, et al., 2005] and its substitution to another amino acid modified the catalytic rate [Lee, et al., 2009].

The largest accumulation of mutations in the protein occurs directly following this region, at the junction of the catalytic and regulatory domains, which includes the linker region (c.1001–1100 in Fig. 3). The large number of mutations in this region suggests that it might be particularly important and/or susceptible to mutation. Indeed, there is support for both possibilities. It is fairly well conserved (Fig. 2) and in our investigation [Burda, et al., 2015b] we found that cell lines harboring mutations within this region showed increased sensitivity to AdoMet inhibition, supporting evolutionary selection. It also appears to be susceptible to mutagenesis, since half of the mutations appear in CpG dinucleotides (Fig. 3), which are highly mutable because they can spontaneously mutate to form TpG or CpA [Cooper and Youssoufian, 1988].
Finally, although arginine residues account for only 5.8% of amino acids of the MTHFR protein, 23 out of the 109 mutations (21%) occur on arginine residues (19/70 missense). Therefore, nearly half (16/38) of the arginine residues in MTHFR have been found to be mutated. Arginine is often the most mutated residue in proteins because 4/6 arginine codons lie on CpG dinucleotides. Indeed, 31 mutations in MTHFR were found to exist on CpG dinucleotides, of which 21 involved arginine residues.
Database
All of the variants in Table 1 were submitted to the freely accessible NCBI ClinVar Database http://www.ncbi.nlm.nih.gov/clinvar/ using the standard HGVS nomenclature.
Biological Significance
Thus far, investigation into severe MTHFR deficiency at the organism level has depended on mouse models of the disease. In 2001, Chen et al. [2001] published the first Mthfr knock-out mouse. Offspring of heterozygous matings showed the expected 1:2:1 Mendelian ratio, suggesting it is not neonatally lethal [Chen, et al., 2001]. However, 83% of offspring homozygous for the null mutation (nullizygotes) died at an average age of 9 days postpartum [Schwahn, et al., 2004]. Those nullizygotes that survived showed ∼10 times increased plasma homocysteine compared with their wild-type littermates as well as decreased weight gain over the first 2 weeks, although their weight later normalized [Chen, et al., 2001]. Changing the genetic background from BALB/c to C57Bl/6 enhanced the 5-week survival rates (26.5% vs. 81%) despite an increase in plasma homocysteine levels [Lawrance, et al., 2011]. The survivability of BALB/c nullizygotes also increased when dams were supplemented with betaine (74% 5-week survival) [Schwahn, et al., 2004] or mefolinate (synthetic methylTHF; 64% 3-week survival) [Li, et al., 2008] during gestation and lactation. No effect on the surviving nullizygotes was found if betaine supplementation was withdrawn after weaning, suggesting the critical need for homocysteine remethylation occurred during the first few weeks after birth [Schwahn, et al., 2004].
In addition to perturbation of plasma homocysteine levels, Mthfr knock-out mice had decreased methionine (∼50%), methionine/homocysteine ratio, and dimethylglycine (63%) [Schwahn, et al., 2003], as well as significantly decreased AdoMet and/or increased adenosylhomocysteine, with global DNA hypomethylation [Chen, et al., 2001]. This is reflective of the patient situation, who show elevated (∼10-fold) homocysteine, low to low–normal methionine and sometimes decreased AdoMet levels [Bönig, et al., 2003; Huemer, et al., 2016]. Plasma total folate levels were approximately 25% of wild type, of which only 40% was methylTHF, as opposed to at least 80% in controls [Ghandour, et al., 2004].
Tissue analysis showed fatty infiltration of the liver in 3-week-old MTHFR deficient mice as well as lipid deposition in the proximal aorta, cerebellar pathology, and developmental retardation [Chen, et al., 2001]. Indeed, Mthfr knock-out mice have dramatically reduced cerebellum and cerebral cortex size, but cerebellar abnormalities are partially ameliorated when dams are supplemented with betaine [Chen, et al., 2005]. A decreased expression of Ser/Thr protein phosphatase 2A and leucine carboxyl methyltransferase (LCMT1) due to hypomethylation was observed in the hippocampus, cerebellum, and, to a lesser extent, the cortex of young Mthfr knock-out mice [Sontag, et al., 2014]. Cerebellar morphology was improved with mefolinate treatment [Li, et al., 2008]. Mthfr knock-out mice are also more susceptible to the adverse effects of mild neonatal stress, indicated by less exploration and activity in females and increased stress-related parameters compared with wild-type controls, whereas males have modified behavior in a social setting [Kezurer, et al., 2013]. In male mice (BALB/c), abnormal spermatogenesis and infertility was found, which improved when betaine supplementation was begun during pregnancy and maintained in the affected male [Kelly, et al., 2005]. By contrast, C57Bl/6 mice were fertile, but showed decreased sperm numbers and altered testicular histology [Chan, et al., 2010].
Clinical Significance
Clinical Spectrum
Patients with MTHFR deficiency are usually born at term following an uncomplicated pregnancy. Patients may, after uneventful adaptation, develop first signs of the disease as early as the first day of life [Huemer, et al., 2016]. A classical presentation suggestive of MTHFR deficiency is a neonate with generalized muscular hypotonia, feeding problems and/or failure to thrive, lethargy, apnoea, and eventually microcephaly [Thomas and Rosenblatt, 2005; Diekman, et al., 2014]. Less common symptoms are enlarged ventricles or hydrocephalus and eye disease (nystagmus, optic atrophy) [Huemer, et al., 2016]. Due to the variable combination of mainly nonspecific clinical manifestations, delay in diagnosis can be significant [Thomas and Rosenblatt, 2005; Huemer, et al., 2016]. Untreated patients show progressive developmental delay and mental retardation, whereas some patients may also present with epilepsy and neurological disease (abnormal gait, spasticity) [Huemer, et al., 2016]. Unlike classical homocystinuria, caused by cystathionine β-synthase deficiency, vascular pathology is a rare complication. Patients may also present with late-onset disease, mainly with behavioral problems and an unspecific spectrum of psychiatric symptoms often accompanied by cognitive impairment and neurological abnormalities, compatible with myelopathy or ataxia and spasticity [Lossos, et al., 2014; Huemer, et al., 2016]. Brain atrophy and white matter disease are present in almost all cases [Thomas and Rosenblatt, 2005]. Indeed, MTHFR deficiency primarily and severely affects the central nervous system (CNS), probably by causing reduced cerebral methylation as suggested by decreased AdoMet levels in cerebrospinal fluid [Surtees, et al., 1991; Smith, et al., 2012; Strauss, et al., 2007]. The impact of the generally decreased concentrations of methylTHF in the CNS [Crushell, et al., 2012; Schiff and Blom, 2012] on clinical course and outcome are incompletely understood at present [Thomas and Rosenblatt, 2005]. Plasma total homocysteine levels do not generally correlate with the clinical pattern or onset.
Treatment Strategies
Since causal treatment for MTHFR deficiency is not available, betaine is the mainstay of symptomatic treatment. Early treatment with betaine has recently been shown to prevent neurocognitive decline in patients with MTHFR deficiency [Diekman, et al., 2014; Strauss, et al., 2007]. Brain edema has been reported as a side effect of betaine treatment in two patients with classical homocystinuria and excessively high methionine levels [Yaghmai, et al., 2002; Devlin, et al., 2004; Lawson-Yuen and Levy, 2006], suggesting patients should be carefully monitored.
Supplemental methionine may add clinical benefit [Abeling, et al., 1999]. At least in theory, riboflavin could be an additional therapeutic option in patients with in vitro FAD responsiveness. Folinic acid [Holme, et al., 1989; Diekman, et al., 2014] or methylTHF [Clayton, et al., 1986; Schiff and Blom, 2012] have not been convincingly shown to improve the clinical course in MTHFR deficiency. Folic acid should be avoided in MTHFR deficiency since it may aggravate the deficit of methylTHF in the CNS [Hyland, et al., 2010]. Nitrous oxide, which inhibits methionine synthase, may cause acute clinical deterioration and is contraindicated in patients with MTHFR deficiency [Selzer, et al., 2003].
Genotype–Phenotype Correlation
Due to the large number of private mutations, genotype–phenotype correlations or correlations between type or location of mutation and clinical course have so far not been established in MTHFR deficiency. Here, we present a comprehensive list of patients identified from the literature and this study, along with their residual activity and age of onset (early: <1 year; late >1 year) (Supp. Table S1). This list includes the six new patients described in this study, as well as the 35 others to which we have contributed clinical (age of onset) or biochemical data. This not only allows a more precise estimate of the number of published patients with severe MTHFR disease, previous estimates varying wildly (100 to >200), but also the ability to stratify patients for genotype–phenotype correlations. Even with a thorough search of the literature, this would not have been possible without the help/advice of the physicians who cared for these patients (see Acknowledgements; for patient inclusion criteria, see footnote in Supp. Table S1).
We have identified 171 families (192 patients) with confirmed MTHFR deficiency with or without mutations, of which 143 families (162 patients, including one asymptomatic sibling) have confirmed mutational analysis (Supp. Table S1). Data on residual activity and time of clinical presentation were available for 121 patients, of which 65 presented with early onset and 56 with late onset. Mean residual activity was significantly lower in early-onset patients (3.4% of control) compared with late-onset patients (14.5% of control), and the calculated effect size of this difference was large (Fig. 4A). However, when comparing these differences, one must keep in mind the different types of assays used (physiological or reverse direction) and the types of cells in which the activity was measured (e.g., fibroblasts, lymphocytes, liver). These differences may have an effect on the level of activity found, and sometimes there are discrepancies in activity values even for the same patient (Supp. Table S1). Nevertheless, these data clearly support and consolidate the previously proposed theory that residual MTHFR activity is likely to be an important factor in disease severity [Watkins and Rosenblatt, 2014]. Full data (mutations, activity, onset) was available for 115 patients, which we split into seven categories according to the type and localization of the mutation in each allele (Fig. 4B). The majority of these patients (n = 33) carried two missense mutations in the catalytic domain. Early- and late-onset were equally distributed in all categories except when patients harbored two missense mutations in the regulatory domain (n = 9), which was significantly associated with late-onset disease, or when they had two splicing mutations in the regulatory domain (n = 14), which was associated with early-onset disease (Fig. 4B). These differences are also likely to be due to differences in the level of residual activity since patients with two missense mutations within the regulatory domain had a mean of 23% control activity, whereas patients with two splicing mutations within the regulatory domain, half of whom were homozygous for the common p.Lys510 = (c.1530G>A) mutation, had a mean of 1.8% control activity. Finally, analysis of 11 families with more than one patient showed no significant difference of residual enzyme activities between siblings (Mann–Whitney test).

Diagnostic Strategies
Elevated plasma total homocysteine is the first pointer toward the group of homocystinurias to which MTHFR belongs. Within this group, certain plasma metabolite and biochemical differences can be helpful in the diagnosis of MTHFR deficiency. For example, plasma methionine is high in classical homocystinuria but low or low–normal in patients with MTHFR deficiency and other remethylation defects. In contrast, plasma cystathionine determined by sufficiently sensitive methods is decreased in CBS deficiency, whereas it is elevated in remethylation defects including MTHFR deficiency. Elevated methylmalonic acid in plasma or urine is characteristic of combined inborn errors of intracellular cobalamin metabolism (i.e., affecting both adenosylcobalamin and methylcobalamin synthesis, e.g., cblC defect) or nutritional vitamin B12 deficiency, but not of MTHFR deficiency and other remethylation defects. Megaloblastic anemia is often present in deficiencies of methionine synthase such as cblE and cblG, as well as MTHFD1 deficiency [Burda, et al., 2015a], but not in MTHFR deficiency. Although these diagnostic steps should quickly lead to initiation of treatment, the definitive diagnosis requires enzymatic studies in cultivated fibroblasts and/or molecular genetic testing. It is important to note, however, that the large number of SNPs within MTHFR makes mutational analysis problematic, especially given that most mutations are private and synonymous mutations may also cause disease. Further, some patients retain high enzymatic activity. Therefore, combined mutation and kinetic analysis remains indispensable for reliable diagnosis of MTHFR deficiency.
Newborn Screening
From a clinical point of view, NBS for MTHFR resulting in early betaine treatment would likely be beneficial for affected individuals [Diekman, et al., 2014]. However, data on efficacy and feasibility of NBS for MTHFR deficiency is currently lacking. NBS for MTHFR deficiency and other remethylation defects may be possible by detection of low methionine and methionine/phenylalanine ratio in dried blood spots with total plasma homocysteine serving as a second tier marker [Huemer, et al., 2015].
Future Prospects
To study enzyme kinetics and the dysfunction caused by missense variants, MTHFR has been expressed in E. coli [Frosst, et al., 1995; Goyette and Rozen, 2000; Sibani, et al., 2003; Yano, et al., 2004], yeast [Shan, et al., 1999; Marini, et al., 2008], insect cells [Yamada, et al., 2001], and COS-1 cells [Martin, et al., 2006]. Although some expressed mutants give activities similar to those found endogenously, in many cases discrepancies have been found in the percent of control activity between endogenous and overexpressed mutant proteins [Sibani, et al., 2003; Yano, et al., 2004] and even supranormal activity found in overexpression of some mutants [Shan, et al., 1999; Goyette and Rozen, 2000], an effect also seen in related diseases (e.g., CBS deficiency [Melenovska, et al., 2015]). Thus, it remains undetermined whether overexpression of human MTHFR protein in nonhuman cells can faithfully reproduce the activity dysfunction caused by these mutations in vivo. Therefore, the best measure of functional deficiency determination is MTHFR activity determined in patient fibroblasts. MTHFR activity was previously measured in the reverse direction (i.e., methylTHF to methyleneTHF) only, but more recently has been determined in the forward physiological direction [Suormala, et al., 2002; Smith, et al., 2013; Burda, et al., 2015b]. This has allowed the elucidation of more complex kinetics, including determination of the Km for methyleneTHF, NADPH, and FAD, as well as the Ki for AdoMet in fibroblasts of patients [Rummel, et al., 2007; Burda, et al., 2015b] This more complete enzymatic characterization has helped to explain the potential dysfunction in those patient cell lines that retain relatively high (>25% of wild type) enzymatic activity, levels that would otherwise not be expected to be pathogenic. It has also led to some unexpected results, such as the finding of an altered Km for NADPH in all patient cell lines that carried a missense mutation in the regulatory domain [Burda, et al., 2015b]. This latter finding highlights our lack of understanding of the precise role of the regulatory domain and its interaction with the catalytic domain, which cannot be ascertained from bacterial models. Further enzymatic and structural investigation of mammalian MTHFR protein is clearly warranted.
Thus far, a very well-characterized knock-out mouse has been used to study severe MTHFR deficiency. This mouse displays many aspects of the human phenotype: they are healthy at birth, but then show early clinical abnormalities including neurological deficits and elevated homocyst(e)ine, and they are responsive to betaine. These similarities to the human situation suggest that the observed positive effects of maternal betaine supplementation during pregnancy with increased offspring survival and decreased male infertility in this mouse model may also translate to humans, potentially supporting such an experimental trial. Nevertheless, the generation of further mice disease models might still be of value. For example, a hypomorphic mouse with an FAD-responsive missense mutation may be useful to study milder disease and/or the potential impact of flavins in therapy, whereas a tissue-specific knock-out may assist in monitoring the importance of metabolites and activity in, for example, brain dysfunction. Likewise, other organisms (e.g., zebrafish) might help to establish the role of MTHFR in embryonic development, keeping in mind that in humans MTHFR deficiency does not manifest until after birth.
Since MTHFR deficiency is a potentially treatable disease, early diagnosis is crucial and betaine treatment should be started as early as possible. It is therefore imperative for clinicians to be aware of the clinical patterns of early- and late-onset forms of the disease. Although NBS has not yet been widely implemented for MTHFR deficiency, it is feasible and should be pursued. Our findings here suggest that disease severity is correlated to residual enzymatic activity, so that determination of enzymatic activity in cultured cells remains a cornerstone of diagnosis. Furthermore, the results of this and other studies emphasize the need for detailed enzymatic (i.e., determination of Km for cofactor and NADPH) and mutational investigation in order to distinguish subjects with disease but high residual activity (up to 50% of control activity), from obligate heterozygotes with decreased residual activity (as low as ∼20% of control activity) [Kang, et al., 1991a; Tonetti, et al., 2001; Tonetti, et al., 2003] and apparently no disease. Finally, this extensive report of more than 100 mutations in over 170 patients with severe MTHFR deficiency provides an important basis for future investigations aimed at improved diagnosis and treatment of this debilitating disease.
Acknowledgments
We thank Dr. Jaime Barea (University of California, San Diego, CA) for sending patient material and data, as well as Dr. Alexander Broomfield (Saint Mary's Hospital, Manchester, UK), Dr. Alexander Lossos (Hebrew-University-Hadassah Medical Center, Jerusalem, Israel), Dr. Manuel Schiff (Robert Debré University Hospital, Paris, France), and Dr. Bernd Schwahn (Saint Mary's Hospital, Manchester, UK) for useful discussion and help with identification of individual patients in the literature. We also thank Ms. Hana Vlášková and Jitka Sokolová for reviewing the nomenclature of genetic variants and database entries.
Disclosure statement. The authors declare no conflict of interest.