ECHS1 Mutations Cause Combined Respiratory Chain Deficiency Resulting in Leigh Syndrome
Contract grant sponsor(s): Grants-in-Aid for Research on Intractable Diseases (Mitochondrial Disease) from the Ministry of Health, Labor and welfare of Japan; Research Grant for Nervous and Mental Disorders from the National Center of Neurology and Psychiatry (21A-6, 24-8) and JSPS KAKENHI (25670275).
Communicated by David Rosenblatt
ABSTRACT
The human ECHS1 gene encodes the short-chain enoyl coenzyme A hydratase, the enzyme that catalyzes the second step of β-oxidation of fatty acids in the mitochondrial matrix. We report on a boy with ECHS1 deficiency who was diagnosed with Leigh syndrome at 21 months of age. The patient presented with hypotonia, metabolic acidosis, and developmental delay. A combined respiratory chain deficiency was also observed. Targeted exome sequencing of 776 mitochondria-associated genes encoded by nuclear DNA identified compound heterozygous mutations in ECHS1. ECHS1 protein expression was severely depleted in the patient's skeletal muscle and patient-derived myoblasts; a marked decrease in enzyme activity was also evident in patient-derived myoblasts. Immortalized patient-derived myoblasts that expressed exogenous wild-type ECHS1 exhibited the recovery of the ECHS1 activity, indicating that the gene defect was pathogenic. Mitochondrial respiratory complex activity was also mostly restored in these cells, suggesting that there was an unidentified link between deficiency of ECHS1 and respiratory chain. Here, we describe the patient with ECHS1 deficiency; these findings will advance our understanding not only the pathology of mitochondrial fatty acid β-oxidation disorders, but also the regulation of mitochondrial metabolism.
Introduction
Mitochondrial fatty acid β-oxidation provides carbon substrates for gluconeogenesis during the fasting state and contributes electrons to the respiratory chain for energy production. Once a fatty acid is activated to the acyl-coenzyme A (CoA) form and enters the mitochondrial fatty acid β-oxidation pathway, it undergoes the four following enzymatically catalyzed reaction steps during each β-oxidation cycle (Supp. Table S1): (1) dehydrogenation, (2) hydration, (3) a second dehydrogenation step, and finally (4) a thiolytic cleavage that generates one acetyl-CoA or, in certain cases, one propionyl-CoA and an acyl-CoA that is two carbons shorter than the acyl-CoA precursor. Each individual step involves specific enzymes encoded by different genes with different substrate preferences (Supp. Table S1). The first dehydrogenation reaction is catalyzed mainly by four enzymes—short-, medium-, long-, and very long chain acyl-CoA dehydrogenases (SCAD, MCAD, LCAD, and VLCAD)—with substrate optima of C4, C8, C12, and C16 acyl-CoA esters, respectively, still each dehydrogenase can utilize other suboptimal substrates [Ikeda et al., 1983, 1985a, 1985b; Ensenauer et al., 2005]. The short-chain enoyl-CoA hydratase (ECHS1) catalyzes the next step and has substrate optima of C4 2-trans-enoyl-CoA, also called crotonyl-CoA. Although ECHS1 also catalyzes hydration of medium chain substrates, longer acyl chains (e.g., C16-intermediates) are hydrated by mitochondrial trifunctional protein (MTP) [Uchida et al., 1992; Kamijo et al., 1993]. MTP consists of an alpha-subunit with long-chain enoyl-CoA hydratase and long-chain 3-hydroxyacyl-CoA dehydrogenase (LCHAD) activities and a beta-subunit with long-chain 3-ketothiolase activity.
Mitochondrial fatty acid β-oxidation disorders generally cause impaired energy production and accumulation of partially oxidized fatty acid metabolites. They are clinically characterized by hypoglycemic seizures, hypotonia, cardiomyopathy, metabolic acidosis, and liver dysfunction [Kompare and Rizzo, 2008]. The most common genetic defect in MTP is LCHAD deficiency [MIM #609016]; deficiency involving reduced activity of all three MTP enzymes [MIM #609015] is reported much less frequently and is often associated with infantile mortality secondary to severe cardiomyopathy [Spiekerkoetter et al., 2004]. Deficiency of SCAD [MIM #201470], which catalyzes the first dehydrogenation reaction and has similar substrate optima with regard to carbon chain as ECHS1, have been studied for years, and the range of associated phenotypes includes failure to thrive, metabolic acidosis, ketotic hypoglycemia, developmental delay, seizures, and neuromuscular symptoms such as myopathy and hypotonia [Jethva et al., 2008].
Here, we describe a patient with ECHS1 deficiency who presented with Leigh syndrome [MIM #256000] accompanied by hypotonia, metabolic acidosis, and developmental delay. Additionally, the patient presented with combined respiratory chain deficiency, which is not commonly described in most clinical reports of mitochondrial fatty acid β-oxidation disorders. Finally, we discuss the pathology of ECHS1 deficiency and possible interactions between mitochondrial fatty acid β-oxidation and the respiratory chain, which are two important pathways in mitochondrial energy metabolism.
Materials and Methods
This study was approved by the ethical committee of National Center of Neurology and Psychiatry. All the samples in this study were taken and used with informed consent from the family.
Whole-mtDNA Genome Sequence Analysis
Long and accurate PCR amplification of mtDNA followed by direct sequencing was performed according to the previous publication with a slight modification [Matsunaga et al., 2005].
Targeted Exome Sequencing
Almost all exonic regions of 776 nuclear genes (Supp. Table S2), in total 7,368 regions, were sequenced using the Target Enrichment System for next-generation sequencing (HaloPlex; Agilent Technologies, Santa Clara, California, USA) and MiSeq platform (Illumina, San Diego, California, USA). Sequence read alignment was performed with a Burrows–Wheeler Aligner (version 0.6.1) to the human reference genome (version hg19). Realignment and recalibration of base quality scores was performed with the Genome Analysis Toolkit (version 1.6.13). Variants were detected and annotated against dbSNP 135 and 1000 Genomes data (February 2012 release) by Quickannotator.
Sanger Sequencing
Sanger sequencing of candidate genes was performed with the BigDye Terminators v1.1 Cycle Sequencing kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) as per manufacturer's protocol. Details of primers and conditions are available upon request. DNA sequences from the patients were compared against the RefSeq sequence and the sequences of a healthy control or parents those were sequenced in parallel.
Cell Culture
The patient-derived primary myoblasts were established from the biopsy of patient's skeletal muscle and cultured in DMEM/F-12 (Thermo Fisher Scientific) supplemented with 20% (v/v) heat-inactivated fetal bovine serum (FBS, Thermo Fisher Scientific). DLD-1 (human colon carcinoma) cells were provided by Taiho pharmaceutical company (Tokyo, Japan) and cells were cultured in RPMI-1640 (Thermo Fisher Scientific) supplemented with 10% (v/v) heat-inactivated FBS (Thermo Fisher Scientific). All cells were cultured in 5% CO2 at 37°C.
Preparation of Mitochondrial Fraction
Mitochondrial fractions from patient's skeletal muscle and patient-derived myoblasts were prepared according to the literature with a slight modification [Frezza et al., 2007].
Immunoblotting
Mitochondrial fraction and protein lysates were prepared from patient's skeletal muscle and patient-derived Myoblasts. Thirty micrograms of protein of mitochondrial fraction or 50 micrograms of protein lysate was separated on 4%–12% Bis-Tris gradient gels (Thermo Fisher Scientific) and transferred to polyvinylidene fluoride membranes. Primary antibodies used were against ECHS1 (Sigma-Aldrich, St. Louis, Missouri, USA), complex II 70 kDa subunit (Abcam, Cambridge, England), β-actin (Santa Cruz, Biotechnology, Dallas, Texas, USA), HA (Wako, Tokyo, Japan), and AcGFP (Thermo Fisher Scientific).
Enzyme Assays
Enzyme activities of mitochondrial respiratory complexes I–V and citrate synthase (CS) were measured in mitochondrial fraction prepared from patient's specimens. The assays for complexes I–IV and CS were performed as described previously [Shimazaki et al., 2012]. The assay for complex V was carried out following the method by Morava and his colleagues with modifications [Morava et al., 2006]. The enoyl-CoA hydratase activity was assayed by the hydration of crotonyl-CoA by a slight modification of the procedure described earlier [Steinman and Hill, 1975]. Five micrograms of protein of the mitochondrial fraction prepared from patient-derived myoblasts was added to 0.3 M Tris–HC1, pH 7.4, containing 5 mM EDTA (Ethylenediaminetetraacetic acid). The reaction was started by the addition of 200 μM crotonyl-CoA and the decrease in absorbance at 280 nm was monitored at 30°C.
Construction of the Immortalized Patient-Derived Myoblasts
The patient-derived myoblasts and control myoblasts were transfected with pEF321-T vector (A kind gift from Dr. Sumio Sugano, University of Tokyo) and the cells were cultured serially for more than ten population doublings until the morphological alteration was observed [Kim et al., 1990].
Expression Vector Preparation and Transfection
For construction of a mammalian expression vector, full-length ECHS1 (GenBank accession number NM_004092.3) was amplified from a cDNA prepared from control subject using PrimeSTAR GXL DNA polymerase (TaKaRa, Tokyo, Japan). The PCR product was cloned into pEBMulti-Pur (Wako) and the clone was verified by Sanger sequencing. The empty expression vector or an ECHS1 expression vector was transfected into immortalized patient-derived myoblasts using Lipofectamine LTX Reagent (Thermo Fisher Scientific). Each of the two missense variants, c.2T>G; p.M1R and c.5C>T; p.A2V, was independently introduced into the clone by PCR-based site-directed mutagenesis. Each insert with C-terminal HA tag was cloned into pIRES2-AcGFP1 (Clontech Laboratories, Mountain View, California, USA) and the clones were verified by Sanger sequencing. WT and mutant ECHS1 expression vector were transfected into DLD-1 cells using Lipofectamine LTX Reagent (Thermo Fisher Scientific). Twenty-four hours later, the cell lysate was subjected to immunoblotting.
Results
The patient reported here was a boy born to unrelated, healthy parents after a 40-week pregnancy (weight 3,300 g, length 52 cm, occipitofrontal circumference (OFC) 34.5 cm). Auditory screening test at 2 months of age revealed hearing impairment, and he began to use a hearing aid at 6 months of age. Psychomotor developmental delay was noted at 5 months of age; he could not sit alone, or speak a meaningful word as of 4 years of age. Nystagmus was noted at 10 months of age. Muscle hypotonia, spasticity, and athetotic trunk movement became prominent after 1 year of age. His plasma (20.2 mg/dl) and a cerebrospinal fluid lactate were elevated (25.3 mg/dl, control below 15 mg/dl). Urinary organic acid profiling reveals significantly elevated excretion of glyoxylate (Table 1). Analysis of blood acylcarnitines showed no abnormalities. Brain magnetic resonance scan image showed bilateral T2 hyperintensity of the putamen, typical for Leigh syndrome (Fig. 1A). Because Leigh syndrome is generally caused by defects in the mitochondrial respiratory chain or the pyruvate dehydrogenase complex, we performed a muscle biopsy to measure enzyme activities of mitochondrial respiratory complexes in the patient. Mitochondrial fractions prepared from patient or control specimens were used for all activity measurements. Activity of each respiratory complex was normalized relative to CS activity; normalized values for complexes I, III, and IV activity were decreased to 39%, 34%, and 64% of control values, respectively (Fig. 1B). Moreover, we performed blue native PAGE (BN-PAGE) to examine if the assembly of respiratory complexes were altered in the patient. As a result, there were no clear difference between the patient and the control (Supp. Fig. S1).
| Patient RPA (%) | Controls RPA (%) | |
|---|---|---|
| TCA cycle intermediates | ||
| α-Ketoglutarate | 4.52 | 3.00–102.90 |
| Aconitate | 20.37 | 15.10–86.10 |
| Isocitrate | 8.98 | 8.30–29.00 |
| Other metabolites | ||
| Lactate | 11.83a | <4.70 |
| Pyruvate | 3.18 | <24.10 |
| 3-Hydroxyisobutyric acid | 1.95 | <9.00 |
| Methylcitric acid | 0.14a | Less than trace amount |
| p-Hydroxy-phenyllactic acid | 40.05a | <7.00 |
| Glyoxylate | 37.71a | <6.10 |
- a Values outside the normal range.
- RPA(%), relative peak area to the area of internal standard (heptadecanoic acid, HDA).
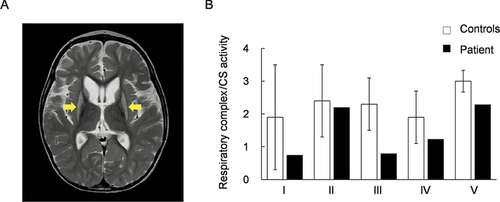
T2-weighted magnetic resonance scan image and enzyme activities of mitochondrial respiratory complexes. A: T2-weighted magnetic resonance scan image (MRI) shows bilaterally symmetrical hyperintensities in the putamen (arrows in the image); these are characteristic of Leigh syndrome. B: Enzymatic activities of five mitochondrial respiratory complexes (I, II, III, IV, and V) were measured in mitochondrial fractions prepared from the patient's skeletal muscle. Respiratory complexes activities were normalized to citrate synthase activity. Black bars show patient values and white bars show control values. Control values were mean values obtained from five healthy individuals. Patient activity values for complexes I, III, and IV were 39%, 34%, and 64% of the control values, respectively. Error bars represent standard deviations.
Mitochondrial respiratory chain defects can be due to pathogenic mutations in mitochondrial DNA (mtDNA) or nuclear DNA (nDNA) coding for mitochondrial components. Initially, long and accurate PCR amplification of mtDNA followed by direct sequencing was performed and no mutations known to be associated with Leigh syndrome were identified, but previously reported polymorphisms were found (Supp. Table S3). Therefore, to identify the responsible mutations in nDNA, targeted exome sequencing was performed. Coverage was at least 10× for 86.2% of the target regions, and 30× or more for 73.4%. In all, 5,640 potential variants were identified; these included 811 splice-site or nonsynonymous variants. Among those 811 variants, 562 were on the mismapping reads that contained multiple apparent mismatches to the reference DNA sequence. Of the remaining 249 variants, nine that were on target regions with less than 10× coverage were eliminated because data reliability was low. Filtering against dbSNP 135 and 1000 Genomes data, this number was reduced to 13 including compound heterozygous variants in the ECHS1 [MIM #602292] and 11 heterozygous variants in 11 separate genes (Supp. Table S4). Those variants have been submitted to dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). Because most mitochondrial diseases caused by known nDNA mutations are inherited in an autosomal recessive manner, we focused on the compound heterozygous variants in ECHS1—c.2T>G; p.M1R and c.5C>T; p.A2V—as primary candidates.
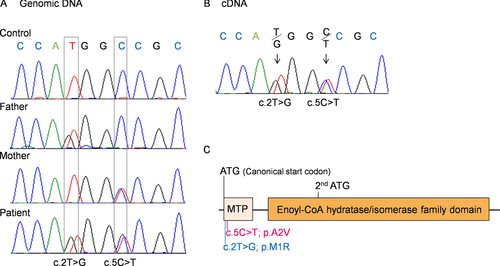
To confirm the targeted exome sequencing results, we performed Sanger sequencing of genomic ECHS1 DNA and ECHS1 cDNA from the patient and his parents. We identified both variants, c.2T>G and c.5C>T, and the respective normal alleles in genomic DNA and cDNA from the patient (Fig. 2A and B) and no other ECHS1 variants were detected except for common SNPs in the open reading frame. Analysis of genomic DNA from the patient's parents showed that patient's father was heterozygous for only one variant, c.2T>G, and the patient's mother for only the other variant, c.5C>T (Fig. 2A). These results indicated that the patient inherited each variant separately and that both mutant alleles were expressed in the patient (Fig. 2B). Each variant was nonsynonymous and in the region encoding the mitochondrial transit peptide (1–27 amino acids) of ECHS1 [Hochstrasser et al., 1992]; moreover, c.2T>G; p.M1R was a start codon variant (Fig. 2C).
Next, immunoblotting with primary antibodies against ECHS1 was performed to assess protein expression. Mitochondrial fractions prepared from patient and control skeletal muscle were used; whole-cell lysates or mitochondrial fractions prepared from patient-derived or control myoblasts were also used. All experiments using these specimens showed that the expression level of ECHS1 protein of the patient was too low to detect by immunoblotting even though the expression level of SDHA was almost the same as controls (Fig. 3A–C). These findings indicated that c.2T>G; p.M1R and c.5C>T; p.A2V mutations caused a remarkable reduction in ECHS1 protein expression. Notably, patient-derived and control myoblasts were similar with regard to ECHS1 mRNA expression (Fig. 3D), indicating that the mutations apparently affected ECHS1 protein expression directly. Next, we measured ECHS1 enzyme activity in mitochondrial fractions prepared from patient-derived and control myoblasts. ECHS1 activity was normalized to CS activity, and activity in patient-derived myoblasts was 13% of that in control myoblasts (Fig. 3E). Therefore, the mutations caused a severe depletion of ECHS1 protein expression thereby decreasing ECHS1 enzyme activity.
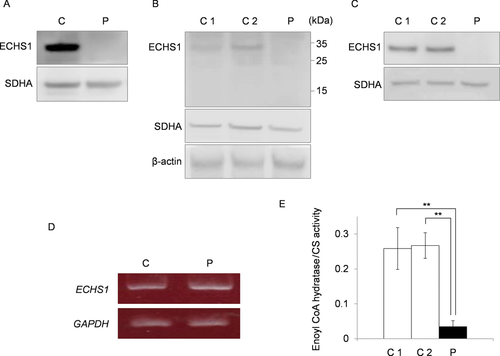
ECHS1 expression and enzyme activity. ECHS1 expression was analyzed by immunoblotting. C1/2, control; P, patient. Mitochondrial fraction prepared from patient's skeletal muscle (A) or whole-cell lysate (B) and mitochondrial fraction (C) prepared from the patient-derived myoblasts were analyzed via immunoblotting. All findings indicated that ECHS1 levels in patient samples were too low to detect by immunoblotting. D: RT-PCR was used to assess ECHS1 mRNA levels in the patient. Notably, patient-derived myoblasts and control myoblasts did not differ with regard to ECHS1 mRNA level. E: Mitochondrial fractions prepared from patient-derived myoblasts were used to estimate ECHS1 enzyme activity in the patient. All ECHS1 activity measurements were normalized to CS activity; ECHS1 activity in patient-derived samples was 13% of that in control samples. The experiments were performed in triplicate. Error bars represent standard deviations. (**P < 0.005 Student's t-test).
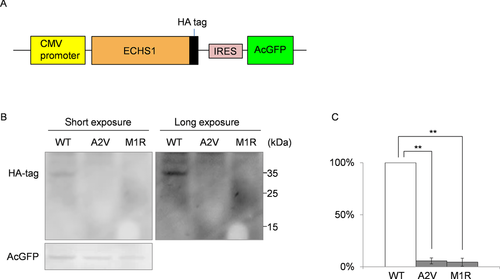
To examine the stability of each mutated protein, we constructed three pIRES2-AcGFP1 expression plasmids, each expressed a different HA-tagged protein: wild-type, M1R-mutant, or A2V-mutant ECHS1. The expression of AcGFP was used as a transfection control. After the transfection into DLD-1 cells, immunoblotting of whole-cell lysate with anti-HA and GFP antibodies showed markedly higher expression of wild-type ECHS1 than of either mutant protein; all ECHS1 expression was normalized to AcGFP expression (Fig. 4, Supp. Fig. S2). This result indicated that ECHS1 protein expression was significantly reduced in the patient because of each mutation.
To confirm that the patient had ECHS1 deficiency, we performed a cellular complementation experiment. Patient-derived myoblasts had to be immortalized for these experiments because nonimmortalized cells exhibited poor growth and finite proliferation. The patient-derived myoblasts and control myoblasts were transfected with pEF321-T vector (a kind gift from Dr. Sumio Sugano, University of Tokyo). We then ascertained that ECHS1 protein expression and activity were lower in immortalized patient-derived myoblasts than in controls (Fig. 5A and B). We then transduced an empty expression vector, pEBMulti-Pur (Wako), or a pEBMulti-Pur construct containing a full-length, wild-type ECHS1 cDNA into the immortalized patient-derived myoblasts; cells with the vector only or the ECHS1-expression construct are hereafter called vector-only and rescued myoblasts, respectively. ECHS1 protein expression level and enzyme activity were analyzed in mitochondrial fractions prepared from rescued myoblasts. Relative expression level of ECHS1 in rescued myoblasts was 11 times higher than that in vector-only myoblasts (Fig. 5A), and ECHS1 activity normalized to CS activity in rescued myoblasts was 49 times higher than that in vector-only myoblasts (Fig. 5B). From these cellular complementation experiments, we concluded the patient had ECHS1 deficiency.

Since the patient showed the combined mitochondrial respiratory chain deficiency in the skeletal muscle as mentioned above, we used a cellular complementation experiment to determine whether wild-type ECHS1 rescued the respiratory chain defect in patient-derived myoblasts. First, we measured enzyme activities of each mitochondrial respiratory complex in mitochondrial fractions prepared from immortalized patient-derived myoblasts. CS activity normalized values for complexes I, IV, and V activity in immortalized patient-derived myoblasts were decreased to 17%, 39%, and 43% of the mean values of immortalized control myoblasts (Fig. 5C). Then, we measured enzyme activity in mitochondrial fractions prepared from rescued myoblasts and found that each activity of complexes I, IV, and V was mostly restored relative to that in vector-only myoblasts. In rescued myoblasts, CS activity normalized values of complexes I, IV, and V were 3.5, 1.3, and 2.2 times higher than those in vector-only myoblasts (Fig. 5C). Mitochondrial respiratory complex activity was mostly restored in rescued myoblasts, suggesting that there was an unidentified link between deficiency of ECHS1 and respiratory chain.
Discussion
Here, we described a patient harboring compound heterozygous mutations in ECHS1. Immunoblotting analysis revealed that ECHS1 protein was undetectable in patient-derived myoblasts; moreover, these cells showed significantly lower ECHS1 enzyme activity than controls. Exogenous expression of two recombinant mutant proteins in DLD-1 cells showed c.2T>G; p.M1R and c.5C>T; p.A2V mutations affected ECHS1 protein expression. Cellular complementation experiment verified the patient had ECHS1 deficiency.
The c.2T>G; p.M1R mutation affected the start codon and therefore was predicted to impair the protein synthesis from canonical initiation site. In the reference ECHS1 sequence, the next in-frame start codon is located in amino acids 97 (Fig. 2C). Even if translation could occur from this second start codon, the resulting product would lack the whole transit peptide and part of the enoyl-CoA hydratase/isomerase family domain (Fig. 2C). The c.5C>T; p.A2V mutation was located in the mitochondrial transit peptide and the mutation may affect the mitochondrial translocation of ECHS1. Surprisingly, the MitoProt-predicted mitochondrial targeting scores for the wild-type and A2V-mutant proteins were 0.988 and 0.991, respectively [MitoProt II; http://ihg.gsf.de/ihg/mitoprot.html; Claros and Vincens, 1996] and not markedly different from each other. Nevertheless, mislocalized mutant protein may have been degraded outside of the mitochondria. Consistent with this speculation was the finding that immunoblotting of lysate from patient-derived myoblasts (Fig. 3B) or from transfected cells that overexpressed the recombinant p.A2V-mutant ECHS1 (Fig. 4B, Supp. Fig. S2) did not show upper shifted ECHS1 bands that indicated ECHS1 with the transit peptide. Another possible explanation is that the mutation affected the translation efficiency because it was very close to the canonical start codon. It can change secondary structure of ECHS1 mRNA or alter the recognition by the translation initiation factors. As stated above, even if there was a translation product from the second in-frame start codon, that product would probably not function.
This patient presented with symptoms that are indicative of fatty acid oxidation disorders (e.g., hypotonia and metabolic acidosis), but he also presented with neurologic manifestations, including developmental delay and Leigh syndrome, that are not normally associated with fatty acid β-oxidation disorders. Interestingly, developmental delay is also found in cases of SCAD deficiency [Jethva et al., 2008]. In the absence of SCAD, the byproducts of butyryl-CoA—including butyrylcarnitine, butyrylglycine, ethylmalonic acid (EMA), and methylsuccinic acid—accumulate in blood, urine, and cells. These byproducts may cause the neurological pathology associated with SCAD deficiency [Jethva et al., 2008]. EMA significantly inhibits creatine kinase activity in the cerebral cortex of Wistar rats but does not affect levels in skeletal or heart muscle [Corydon et al., 1996]. Elevated levels of butyric acid modulated gene expression because excess butyric acid can enhance histone deacetylase activity [Chen et al., 2003]. Moreover, the highly volatile nature of butyric acid as a free acid may also add to its neurotoxic effects [Jethva et al., 2008].
On the other hand, it is very rare for fatty acid β-oxidation disorders causing Leigh syndrome. Therefore, the most noteworthy manifestation in this patient was Leigh syndrome. Leigh syndrome is a neuropathological entity characterized by symmetrical necrotic lesions along the brainstem, diencephalon, and basal ganglion [Leigh, 1951]. It is caused by abnormalities of mitochondrial energy generation and exhibits considerable clinical and genetic heterogeneity [Chol et al., 2003]. Commonly, defects in the mitochondrial respiratory chain or the pyruvate dehydrogenase complex are responsible for this disease. This patient's skeletal muscle samples exhibited a combined respiratory chain deficiency, and this deficiency may be the reason that he presented with Leigh syndrome. Although it remained unclear what caused the respiratory chain defect, cellular complementation experiments showed almost complete restoration, indicating there was an unidentified link between ECHS1 and respiratory chain. One of the possible causes of respiratory chain defect is the secondary effect of accumulation of toxic metabolites. For example, an elevated urine glyoxylate was observed in this patient. Although the mechanism of this abnormal accumulation is not clear at the moment, it was shown that glyoxylate inhibited oxidative phosphorylation or pyruvate dehydrogenase complex by in vitro systems [Whitehouse et al., 1974; Lucas and Pons, 1975]. Therefore, we speculate that in our patient, ECHS1 deficiency induced metabolism abnormality including glyoxylate accumulation, and glyoxylate played a role in decreased enzyme activities of respiratory chain complexes. Interestingly, a recent paper describing patients with Leigh syndrome and ECHS1 deficiency showed decreased activity of pyruvate dehydrogenase complex in fibroblasts [Peters et al., 2014], (Supp. Table S5). BN-PAGE showed the assembly of respiratory complex components in the patient was not clearly different from the control (Supp. Fig. S1). This result suggests that the respiratory chain defect in the patient is more likely because of the secondary effect of accumulation of toxic metabolites. On the other hand, many findings indicate interplays between mitochondrial fatty acid β-oxidation and the respiratory chain. For example, Enns et al. [2000] mentioned the possibility of the physical association between these two energy-generating pathways from overlapping clinical phenotypes in genetic deficiency states. More recently, Wang and his colleagues actually showed physical association between mitochondrial fatty acid β-oxidation enzymes and respiratory chain complexes (Wang et al., 2010). Similarly, Narayan et al. demonstrated interactions between short-chain 3-hydroxyacyl-CoA dehydrogenase (SCHAD) and several components of the respiratory chain complexes including the catalytic subunits of complexes I, II, III, and IV via pull-down assays involving several mouse tissues. Considering the role of SCHAD as a NADH-generating enzyme, this interaction was suggested to demonstrate the logical physical association with the regeneration of NAD through the respiratory chain [Narayan et al., 2012]. Still more recently, mitochondrial protein acetylation was found to be driven by acetyl-CoA produced from mitochondrial fatty acid β-oxidation [Pougovkina et al., 2014]. Because the activities of respiratory chain enzymes are regulated by protein acetylation [Zhang et al., 2012], this finding indicated that β-oxidation regulates the mitochondrial respiratory chain. Remarkably, acyl-CoA dehydrogenase 9 (ACAD9), which participates in the oxidation of unsaturated fatty acid, was recently identified as a factor involved in complex I biogenesis [Haack et al., 2010; Heide et al., 2012]. Cellular complementation experiments that involve overexpression of wild-type ACAD9 in patient-derived fibroblast cell lines showed restoration of complex I assembly and activity [Haack et al., 2010]. Accumulating evidence indicates that there are complex regulatory interactions between mitochondrial fatty acid β-oxidation and the respiratory chain.
ECHS1 has been shown to interact with several molecules outside the mitochondrial fatty acid β-oxidation pathway [Chang et al., 2013; Xiao et al., 2013] and the loss of this interaction can affect respiratory chain function in a patient. Further functional analysis of ECHS1 will advance our understanding of the complex regulation of mitochondrial metabolism.
Acknowledgments
We acknowledge the technical support of Dr. Ichizo Nishino, Dr. Ikuya Nonaka, Dr. Chikako Waga, Takao Uchiumi, Yoshie Sawano, and Michiyo Nakamura. We also thank Dr. Sumio Sugano (the University of Tokyo) for providing the pEF321-T plasmid.
Disclosure statement: The authors have no conflict of interest to declare.




