Rett networked database: An integrated clinical and genetic network of rett syndrome databases†
Communicated by Nobuyoshi Shimizu
Abstract
Rett syndrome (RTT) is a neurodevelopmental disorder with one principal phenotype and several distinct, atypical variants (Zappella, early seizure onset and congenital variants). Mutations in MECP2 are found in most cases of classic RTT but at least two additional genes, CDKL5 and FOXG1, can underlie some (usually variant) cases. There is only limited correlation between genotype and phenotype. The Rett Networked Database (http://www.rettdatabasenetwork.org/) has been established to share clinical and genetic information. Through an “adaptor” process of data harmonization, a set of 293 clinical items and 16 genetic items was generated; 62 clinical and 7 genetic items constitute the core dataset; 23 clinical items contain longitudinal information. The database contains information on 1838 patients from 11 countries (December 2011), with or without mutations in known genes. These numbers can expand indefinitely. Data are entered by a clinician in each center who supervises accuracy. This network was constructed to make available pooled international data for the study of RTT natural history and genotype–phenotype correlation and to indicate the proportion of patients with specific clinical features and mutations. We expect that the network will serve for the recruitment of patients into clinical trials and for developing quality measures to drive up standards of medical management. Hum Mutat 33:1031–1036, 2012. © 2012 Wiley Periodicals, Inc.
Introduction
Rett syndrome (RTT; MIM# 312750) is a neurodevelopmental disorder, which becomes recognizable at 6–12 months [Neul et al., 2010]. In the majority of cases (99.5%), the disease is sporadic. At least four phenotypic variants have been described: (1) the more common classic form described by Rett in the 1960s [Rett, 1966]; (2) the less common preserved speech variant described by Zappella in the 1990s [Zappella, 1992]; (3) the rare early onset seizure variant [Hanefeld, 1985]; and (4) the very rare congenital variant [Rolando, 1985]. The penetrance is nearly complete. A few unaffected gene carriers have been described, who usually have skewed X-chromosome inactivation and are recognized as the healthy mothers of two affected siblings.
RTT is a rare disease, with incidence estimated at 1:10,000 [Leonard et al., 1997]. However, due to the high clinical variability, the disease frequency could be underestimated. The diagnosis of the mildest form, the Zappella variant, could be particularly challenging for nonexpert clinicians and these girls could be often misdiagnosed as autistic patients. In addition, sudden death in young still undiagnosed patients may also contribute to under reporting.
Classic RTT is characterized by a period of regression with the loss of purposeful hand use and spoken language and the development of gait abnormalities and hand stereotypies. After the regression, a stage of stabilization and potentially even improvement ensues, with some individuals partially regaining skills.
Atypical RTT syndrome is also characterized by a period of regression followed by recovery or stabilization, but the course of this form is dramatically different. The Zappella (preserved speech) variant is characterized by better preserved hand use, the recovery of some language after regression with the ability to use single words or phrases, and less severe intellectual disability (IQ up to 50). The early onset seizure variant is characterized by onset of seizures before 5 months of age, before regression usually commences, and often with frequent infantile spasms and refractory myoclonic epilepsy. The congenital variant is characterized by severe psychomotor delay from birth, severe postnatal microcephaly apparent before 4 months, and regression in the first 5 months.
This overlapping but wide clinical spectrum of diseases may result from mutations in one of three genes. In 1999, Amir et al. first found RTT caused by MECP2 gene (MIM# 300005) mutations [Amir et al., 1999]. In 2000, De Bona et al. found that the same gene was mutated in the Zappella variant [De Bona et al., 2000]. MeCP2 is a transcriptional regulator of specific genes and microRNAs. Phosphorylation of its C-terminal part leads to transcriptional induction of BDNF (MIM# 113505), dendrite branching, and neuronal maturation. Scala et al. reported CDKL5 (MIM# 300203) mutations associated with the early onset seizure variant [Scala et al., 2005]. CDKL5 is a kinase that associates in vivo with MeCP2 and mediates its phosphorylation in vitro. In 2008, the FoxG1 gene (MIM# 164874) was identified as a cause of the congenital variant form [Ariani et al., 2008]. FOXG1, like MECP2, is a transcriptional regulator that is expressed specifically in brain.
Several RTT databases have been established. The first one, RettBASE (http://mecp2.chw.edu.au/), collected predominantly molecular genetic data and includes mutations reported in English-language peer-reviewed journals and data generated by the Australian cohort of RTT patients [Christodoulou et al., 2003]. The other databases include both clinical and genetic data. One of them, InterRETT [Fyfe et al., 2003] was based on data collection by distributing a questionnaire to the families [Fyfe et al., 2001]. Most of the other databases operate within one country and are supervised by clinicians. The Italian Rett Database and Biobank included 357 patients and had 20 structured and 7 descriptive clinical items, and 17 structured genetic items (http://www.biobank.unisi.it) [Sampieri et al., 2007]. The SYRENE (SYndrome REtt NEtwork) included 232 French Rett patients and had 81 structured and 1 descriptive clinical item, and 12 structured genetic items (http://afsr.in2p3.fr/RETT/). The British Isles Rett Syndrome Survey (BIRSS) included 275 British Rett patients and had 271 structured and 94 descriptive clinical items, and 6 structured genetic items; it included also longitudinal data. The Barcelona Rett database included 388 Spanish Rett patients and had 44 structured and 4 descriptive clinical items, and 4 structured genetic items. Finally, a recent American survey collected data on the natural history of the disease [Percy et al., 2007]. Using an “adaptor approach” that allowed the preservation and integration of the original data, we have established the Rett Networked Database (http://www.rettdatabasenetwork.org/), designed to be a unified data repository allowing researchers and physicians to access comprehensive patient information.
Database Network Structure
Rett Networked Database Data Harmonization
The Rett Networked Database is a comprehensive, permissive, and flexible unified structure available online at http://www.rettdatabasenetwork.org/. To build it, a data harmonization process was required. Hundreds of items have been analyzed and a common database schema has been defined and approved by the Scientific Review Board (Appendix). To give an idea of the harmonization process, Table 1 shows the example of the item “Head score” in the domain “Head” (Table 1). This item is one of the 293 clinical items and 16 genetic items that were generated through a data harmonization process (Supp. Table S1, column B). All these items were grouped in 31 domains (Supp. Table S1, column A).
| Barcelona Rett Database | British Isles Rett Syndrome Survey (BIRSS) | Italian Rett Database and Biobank | SYndrome REtt NEtwork (SYRENE) | Rett Database Network | |||
|---|---|---|---|---|---|---|---|
| Stagnation head | Acquired microcephaly | OFC fall | OFC at present | Head | PC deceleration of head growth | PC at evaluation | Head score |
| Yes | Yes | 1 = yes, any evidence of fall from original centile | percentile ≤ 3 | 2 = postnatal microcephaly | yes | percentile ≤ 3 | 2 = postnatal microcephaly |
| Yes | No | percentile > 3 | 1 = deceleration of head growth | yes | percentile > 10 | 1 = deceleration of head growth | |
| yes | percentile 3–10 | ||||||
| No | No | 2 = no evidence of fall from original centile | 0 = no deceleration | no | 0 = no deceleration | ||
- OFC: Occipitofrontal circumference.
Rett Networked Database Minimum Data Set
In March 2010, the Scientific Review Board of Rett Networked Database met in Paris and established a minimum data set, which has been reviewed and integrated on August 18, 2011. It consists of 63 clinical items and 7 genetic items (Supp. Table S1, column B, gray values). The specific values of each of these items are illustrated in Supp. Table S1, column C.
Regarding the genetic items, mutation names comply with the accepted guidelines proposed by the Human Genome Variation Society (www.hgvs.org/mutnomen) [den Dunnen and Antonarakis, 2000].
Rett Networked Database Web Server Interface Implementation
The Rett Networked Database Web server is intended to facilitate the access to the data stored in the database and to provide tools for its analysis. The Web server interface consists of HTML pages dynamically generated using VBScript and Active Server Pages (ASP) technologies. The scripting programs run on an Internet Information Server and use the MySQL (http://www.mysql.com) relational database system.
The Web site was shaped in such a manner that on the first page only the minimum data set of items is displayed. Within each group of items (each domain), more detailed entries accommodate the total of 293 clinical and 16 genetic items. All the items are grouped in 31 domains (30 clinical and 1 genetic). The values of most items (192/309, 62%) are assigned by a drop-down menu. Most of the drop-down menus have two (yes/no) or three values as, for example, for “Weight score” (1) below 3rd percentile; (2) 3rd to 25th percentile; and (3) above 25th percentile (Fig. 1A). In some cases (51/309, 17%), the value is a number as in the case of “occipitofrontal circumference (OFC) at birth in centimeters” or “Regression age in months” (Fig. 1B). In 64 out of 309 (21%), the value is represented by a text field such as “Regression speech Note” or “Behavioral Disturbances” (Fig. 1B). A group of 23 clinical items contains longitudinal information. The values of 3 out of 16 genetic items, namely, “Mutated gene”, “Nucleotide change” and “AA change” are assigned by a dynamic drop-down menu (Fig. 1C). Values are inserted as a text field that generates a drop-down menu.
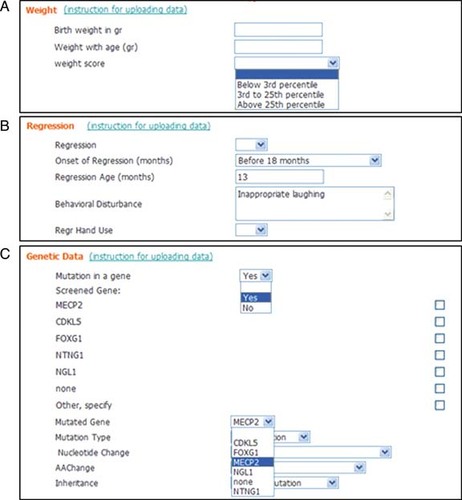
Types of values of the items. A: Example of a static drop-down menu for item “weight score” and of a longitudinal item “weight with age (gr)” in the “weight” domain. B: Example of values present in the “regression” domain: numerical field value “regression age (months)” and text field value “behavioral disturbance” C: Example of yes/no drop-down menu for the item “mutation in a gene” and a dynamic drop-down menu for the item “mutated gene,” present in the “genetic data” domain.
The data are accessible to the participants through the use of a protected password. In addition, data are accessible to the scientific community according to rules that guarantee transparency and equity. There are three different levels of access: (1) Public access. General information and the description of the database content is available to the public; (2) Aggregated data access. Registered users have access to aggregated data only. Registered users have to disclose their identity and affiliation and have to agree to the data protection policy (Human Biobanks and Genetics Research Databases guidelines); (3) Full access. Access to individual data and statistical analysis is given upon the submission of a research proposal to the Scientific Review Board composed of the major contributors (Appendix). The Scientific Review Board may decide the conditions of access, for example, scientific collaboration or acknowledgment or citation of the paper that describes the database (key contributors will be coauthors). The approval of the local research ethics committee of the University of the Coordinator of the database will be required too.
Rett Networked Database Data Storage
The database network was created following an “adaptor approach.” This approach allowed us to preserve the original data as stored in each contributing database and to integrate them in a comprehensive, permissive, and flexible unified structure. The system uses a set of predefined import procedures to extract, decode, and convert data from preexisting databases and to insert them into the new, central database so that the new archive represents a unified repository (Fig. 2).
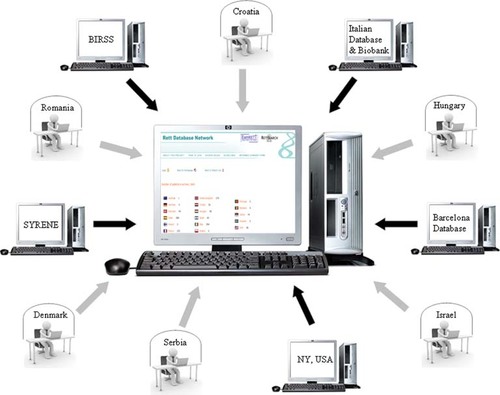
Scheme representing Rett Networked Database. In the central part of the figure, the dynamically generated Web server interface is represented. Dark gray arrows: data flow from preexisting databases to the new central one. Light gray arrows: direct data insertion from local centers.
The data are stored in a MySQL server. The interface has been developed using a common scripting language (VB/ASP). This application allows data storage for users who do not have a local computerized data management system. In such a case, there are two options: (1) to insert data directly into the main archive (geographical and institutional identifications will be displayed); or (2) to construct a local or national archive to be connected with the main one. The archive is permissive so that patients with only partially filled items can be inserted. For users who have an active national database, periodic data update is performed by an information interchange system.
Maintenance- and hardware/software-qualified assistance are guaranteed by an informatic structure 24 hr a day and 7 days a week. The structure ensures availability and redundancy of connectivity, immediate power supply in case of energy interruption, constant temperatures and humidity within the data rooms, and security to avoid unauthorized access. Furthermore, data security is ensured by scheduled backups.
Rett Networked Database Analysis Tools
Basic data mining operations have been applied that cut across applications and develop scalable algorithms for their execution. The data mining interface is developed to ensure maximum flexibility so that users can perform any search they want. This approach allows them to better identify novel genotype–phenotype correlations, to better select subgroups of patients for clinical trials, and to improve the efficiency of the modifier genes study.
Data mining is possible using the “search” button placed in the homepage after the log in (Fig. 3). It is possible to search for patients within the whole archive or to select a specific country or center. Patient data can be searched for features in 46 items belonging to 25 different domains (e.g. head, height, etc.) of the minimum data set. It is possible to perform multiple searches at once. The maximum number of items that can be selected simultaneously is 10.
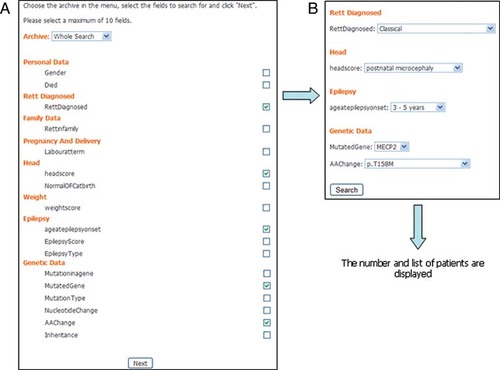
The “search tool.” A: Example of research of patients from the whole archive having specific values for five items (RettDiagnosed, headscore, Ageatepilepsy, MutatedGene, AAChange) belonging to eight domains (personal data, Rett diagnosed, family data, pregnancy and delivery, head, weight, epilepsy, genetic data) (modified from the longer list available at www.rettdatabasenetwork.org/Search.asp). By clicking on the “next” button, a second page, where specific values for the selected items can be chosen, is visualized (B). By clicking on the “search” button, the system will return the total number and the list of all patients fulfilling the selected criteria.
Rett Networked Database Present Shape
Beside the data imported from the four preexisting databases, several other countries started to insert patients directly in the network: Israel (60 patients), Croatia (26 patients), Serbia (44 patients), Denmark (64 patients), Hungary (58 patients), and Romania (9 patients). Furthermore, an additional local database consisting of 96 patients has been connected (Tri-State Rett Syndrome Center, New York). Additional requests to join the network have been made by Germany, Sweden, and Finland.
As of December 2011, this database contains detailed clinical and molecular information on 1838 patients, representing the largest cohort of RTT patients collected up to now.
Among them, 1252 have mutations in MECP2, 64 in CDKL5, 6 in FOXG1. Among the MECP2 mutations, 40% introduce an amino acid change in either methyl binding domain or transcriptional repressor domain (TRD), 39% an early premature termination codon before the TRD domain, 14% a late premature termination codon after TRD domain, and 7% are gene deletions. Regarding CDKL5 mutations, 37% introduce an amino acid change in the catalytic domain, 26% an early premature termination codon within the catalytic domain, 25% a late premature termination codon after the catalytic domain, and 12% are gene deletions. Regarding FOXG1 mutations, most of them introduce an early premature termination codon before the fork-head domain.
Discussion
The Rett Networked Database was created in order to connect those databases already in existence and to create a unified data repository following an “adaptor approach.” The Rett Networked Database now allows the collection of standardized and easily comparable clinical and genetic data of a large number of Rett patients, representing the largest collection worldwide, and it avoids the dissemination of information on many different databases. This network represents a unique attempt to create a connection of locus-specific databases that for many rare diseases remain dispersed, and distinct and we would expect that other locus-specific databases will follow the same approach in the future.
Presently, 14 centers from 11 countries of Europe, the Middle East, and the United States are connected in the network. Data are entered by each center by a clinician who supervises the accuracy of stored information. This characteristic makes the network unique in the sense that each patient can be contacted anytime for clinical update and research or clinical studies.
Although patients are anonymized, the system is able to recognize if clinicians attempt to insert the same patient twice. In such a case, the system stops the “new” reinsertion and permits the updating of a patient's data even if the patient has originally been entered by another center, preserving original data. This mechanism will be particularly useful when more centers from the same country participate in the network, while this is less likely at present.
The system is comprehensive, including 309 items (293 clinical and 16 genetic) grouped in 31 domains (30 clinical and 1 genetic). The values of most items (62%) are assigned by a drop-down menu with two (yes/no) or three values. In some cases (17%), the value is a number indicating a measure or an age. In as few items as possible (21%), the value is represented by a text field since these are less useful for statistical analysis. A consistent number of clinical items (23) contain longitudinal information that would be particularly useful if the network was being used for clinical trials.
The system is also easily scalable allowing additional items/domains to be added in the future. At the beginning of the harmonization process, the values present in the drop-down menu for the item “age at epilepsy onset” did not include the range 0–1 year. Shortly after the inclusion of the CDKL5 patients, the need to add the additional value 0–1 year was felt and the system easily allowed this change.
Finally, the system is permissive since patients with incomplete data can be inserted, although at least the minimum dataset (70 items: 63 clinical and 7 genetic) is recommended. In case of a first evaluation, a clinician can immediately insert clinical data without genetic data that will be updated later on.
Access to the network is open to other, already existing databases worldwide that choose to join. The new archive represents a unified repository, in which additional national or local cohorts of patients may be inserted. The network started in 2009 with five centers (5 countries), grew to 11 (8 countries) in 2010, and to 14 (11 countries) in 2011; and given this trend, we anticipate further expansion.
Data are accessible to the participating database contributors (Appendix) and to the scientific community according to rules that assure transparency and equity. Data access will allow registered users to quickly and easily share novel information. This database will be of great help in diagnosing the disease and genetic counseling and will lead to novel insights, especially in the rare clinical variants concerning which there is often a lack of specific knowledge and experience. This database might also be of assistance when interpreting a novel genetic variant.
Data from the Rett Networked Database are available for the international community for (1) further natural history studies and (2) further genotype–phenotype studies. In addition, these data may be used: (3) to give an indication as to the proportion of patients of different ages who have specific symptoms (and who might therefore be interested in treatments targeting those symptoms) and minimum numbers of affected individuals who have specific mutations (and therefore for whom particular treatments might be applicable); (4) to identify which centers might therefore be useful for recruiting specific groups of patients (e.g., for clinical trials). Finally, another use could be: (5) to develop quality of care measures that would be useful to drive up standards of medical management.
In conclusion, this international effort will be of great value to both the scientific and the clinical RTT communities. It will secure RTT patients, worldwide, the best continuous research in the syndrome, which eventually can give them a better quality of life and it will inspire countries to gather information about their RTT population, which would further lead to foundation of new RTT centers.
Acknowledgements
We are grateful to the patients and their families who participated in this study. We thank Rossano Di Bartolomeo and Marco Maria D'Andrea, 3W Net Service, for their work in the construction of the database network. In addition, we are grateful to Kurt Zatloukal for suggesting the “adaptor approach” and to Gerard Nguyen, President of Rett Syndrome Europe for connecting with parents' associations. We thank the biobank “Cell lines DNA bank of Rett syndrome, X linked Mental Retardation and other genetic diseases” supported by Telethon grant GTB07001C to A.R. This project was supported by RettSearch (microgrants to AR), the E-RARE EuroRett network, Association Française du Syndrome de Rett, the Catalan Rett Association, and Associazione Italiana Rett. The BIRSS is grateful for support from Rett UK.
Appendix
Scientific Review Board of the Rett Networked Database as of December 2011: Alessandra Renieri (Coordinator), Francesca Mari (University of Siena, Siena, Italy), Laurent Villard (Université de la Méditerranée and Inserm, Marseille, France), Nadia Bahi-Buisson (University Paris V Descartes, Paris, France), Angus Clarke, Anna Hryniewiecka-Jaworska (Cardiff University, Wales, U.K.), Mercedes Pineda, Ana Roche Martinez, Judith Armstrong (Hospital Sant Joan de Déu, Barcelona, Spain), Bruria Ben-Zeev (Sheba Medical Center, Ramat-Gan, Israel). Members as of December 2011: Edvige Veneselli, Maria Pintaudi (University of Genova, Genova, Italy), Silvia Russo, Francesca Cogliati (Istituto Auxologico Italiano, Milan, Italy), Aglaia Vignoli (Ospedale San Paolo, Milano, Italy), Giorgio Pini (Versilia Hospital, Viareggio, Italy), Milena Djuric (University of Belgrade, Belgrade, Serbia), Anne-Marie Bisgaard (Glostrup, Denmark), Kirstine Ravn (Glostrup, Denmark), Vlatka Mejaški-Bošnjak (University of Zagreb, Zagreb, Croatia), Béla Melegh, Polgár Noémi (University of Pécs, Pécs, Hungary), Dana Craiu (Carol Davila University of Medicine, Bucharest, Romania), Aleksandra Djukic (Montefiore Medical Center, Albert Einstein College of Medicine, New York). At present, this last center contributed only with anonymous genetic data. Clinical data will be inserted as soon as the Institutional review board approval will be obtained.




