The prevalence of 17 common respiratory viruses in patients with respiratory illness but negative for COVID-19: A cross-sectional study
Reyhaneh Sadeh Tehrani and Hanieh Mohammadjafari are co-first authors.
Abstract
Background and Aims
Second to COVID-19 pandemic, other viral respiratory infections are still important causes of human diseases or co-infections. Hence, the present study was carried out to investigate the common respiratory viruses in patients with respiratory illness diagnosed negative for severe acute respiratory syndrome coronavirus-2 in primary screening.
Methods
In a cross-sectional study, a real-time PCR was carried out using HiTeq. 17 Viro Respiratory pathogen One Step RT-PCR Kit (Genova, Bonda Faravar, Bioluence, Tehran, Iran).
Results
A total of 311 individuals (mean age ± SD: 48.2 ± 21.7 years, range: 1–97 years) underwent second PCR. Among these, 161 (51.7%) were female. In total, 55 (17.6%) cases (mean age ± SD: 45.7 ± 18.1 years) were found positive for respiratory viruses panel in the second PCR. The HCoV-OC43/HKU1 was in 5.4% (17/311), Flu A in 4.5% (14/311), HCoV-229E/NL63 in 2.8% (9/311), HMPV in 1.9% (6/311), HPiV 1, 2, 3 in 1.2% (4/311), HRSV in 0.9% (3/311), and HAdV in 0.6% (2/311) of the cases studies. Also, co-infection was detected in 4 samples (1.2%). In addition, sore throat (0.028), headache (p = 0.016), and body pain (p = 0.0001) were statistically the most significant symptoms in studied cases.
Conclusion
According to the findings of our study, respiratory virus infections and co-infections were 17.6% and 1.2% frequent, respectively. Interestingly, nearly half of our positive cases (47.2%) were identified by coronaviruses (ОС43, Е229, NL63, and HKUI), followed by influenza A virus (25.4%). However, for more comprehensive results, we recommend using greater sample size.
1 INTRODUCTION
Globally, acute respiratory infections (ARIs) are regarded as the fourth most common cause of mortality.1 Annually, as a frequent infection, upper respiratory tract infection (URI) occurs at least once for each person in the United States.2 In Iran, a variety of viruses can cause ARIs, such as influenza A and B viruses (Flu-A and Flu-B), rhinoviruses (RV), human metapneumovirus (hMPV), human adenoviruses (hAdV), (respiratory syncytial virus (RSV), and coronaviruses.3
Viruses are the main cause of URIs.4 During the COVID-19 pandemic in late 2019, other viral respiratory infections were still capable of infecting humans and even potentially co-infections happened.5, 6 Although respiratory viruses' co-infection is rare (0%–3%), monitoring and surveillance of common causes of viral respiratory infections are promising for studies investigating the etiology of pneumonia and respiratory illness.5, 6 More than 200 viruses could cause respiratory infection2 and some of them exhibit seasonal prevalence.2
The classical URI symptoms include sore throat, sneezing, rhinorrhea, nasal congestion, sinus pain, cough, headache, myalgia, loss of appetite, chills, and fever.2 Meanwhile, due to widespread COVID-19 infection, differentiating viral respiratory infection symptoms remains challenging.5, 6
Therefore, in the present study, we aimed to investigate the prevalence of 17 respiratory viruses in patients with respiratory illness who were diagnosed negative for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection in a primary screening via an in-house real-time PCR test.
2 MATERIALS AND METHODS
2.1 Population
Patients who were referred to Iran University of Medical Sciences, Tehran, Iran, from March 2021 to July 2022, and met the inclusion criteria were enrolled in the present study. Inclusion criteria were having apparent symptoms of respiratory illness and a negative PCR result for SARS-COV-2 and exclusion criteria were insufficient specimens for further use. Study protocols were approved by the Ethical Committee of Tehran Medical Sciences, Islamic Azad University, Tehran, Iran (code: IR.IAU.PS.REC.1401.343 and IR.IAU.PS.REC.1401.342). All experiments were performed in accordance with relevant guidelines and regulations. Informed consent was obtained from all participants and/or their legal guardians.
Sampling was done at initial diagnosis and COVID-19 PCR was carried out for each patient accordingly. A questionnaire including demographic information was filled out for each patient. A nasopharyngeal and throat swab was taken from each patient, put into an RNase-free tube (Progenlab, Germany) containing 1.5 cc streel Viral Transport Media (VTM; Parzhan Pazhoh Co., Tehran, Iran) and preserved at 4°C refrigerator for less than 24 h usage. RNA extraction was performed by RNA extraction kit (Favorgen Biotech Corporation) according to the protocols and then kept at −70°C freezer.
2.2 Respiratory viruses panel detection
HiTeq 17 Viro Respiratory pathogen One-Step RT-PCR Kit (Genova, Bonda Faravar, Bioluence, Tehran, Iran) was used for respiratory virus detection.7 Briefly, this multiplex real-time PCR method identifies 17 respiratory viruses using a mixture of primers and probes for their conserved genomic regions. These viruses include SARS-CoV-2, Influenza A (Flu A), Influenza A/H1N1 (Flu A/H1N1), Influenza B (Flu B), human coronavirus (HCoV) HKU1, HCoV-NL63, HCoV-229E, human metapneumovirus (HMPV), human respiratory syncytial virus (HRSV), human bocavirus -1, -2, -3 (HBoV), human parainfluenza -1, -2, -3 (HPiV), human adenovirus (HAdV), HCoV-OC43, and reaction mix of this kit was prepared in three separate tubes (Supporting Information: Table 1). The human RNase P gene was used as an internal control in the reaction mixture. Interpretation of the results is according to Supporting Information: Table 1. The 20 µL reaction mixture contains 10 µL of ready-to-use master mix plus 10 µL of sample RNA or control. A RotorGene Q MDx 5 plex real-time PCR system (RotorGene) was used via the following program: one step holding at 50°C 15 min, 95°c 3 min, and 46 cycles at 94°C 5 s, 60°C 30 s. Fluorescence measurement was done at 60°C.
As for the kit, the analytical specificity was 100%, diagnostic specificity was 95%, diagnostic sensitivity was 95%, and analytical sensitivity (limit of detection [LOD]) was 100 copy/µL for SARS-CoV-2, 200 copy/µL for Flu A and Flu A/H1N1, 250 copy/µL for Flu B and HCoV-NL63, 300 copy/µL for HRSV, HCoV-229E, and HMPV, 350 copy/µL for HCoV-HKU1, 500 copy/µL for HPiV-1, -2, -3, HAdV and HBoV-1, -2, -3.
2.3 Statistical analysis
SPSS, version 16, was used for statistical analysis. To describe the specimens, we performed descriptive analyses based on age, gender, clinical signs, and underlying disease. Median and IQR were used for continuous variables and number and percentage for categorical variables. We used the chi-square test and the Wilcoxon–Mann–Whitney test to analyze categorical and continuous variables, respectively. The significance threshold was p < 0.05. We compared the SARS-CoV-2-infected and noninfected individuals by the proportion of respiratory coinfections and the number of respiratory viruses found.
3 RESULTS
From January 2021 to March 2022, a total of 549 COVID-19 negative samples were collected based on the primary screening and patients' demographic data were obtained using a questionnaire. Of them, a total of 311 specimens (from individuals with the mean age ± SD: 48.2 ± 21.7 years; range: 1–97 years), based on the availability of data and samples as well as quality and quantity of samples, underwent second PCR, which was made by HiTeq 17 Viro Respiratory pathogen One-Step RT-PCR Kit (Genova, Bonda Faravar). From among these 311 individuals, 161 were female (mean age ± SD: 49.7 ± 22.8 years) and 150 were male (mean age ± SD: 48.8 ± 22.5 years). The onset range for the symptom before sampling was filled in 231 patients by questionnaire, in which it was found to be 1–21 days (mean ± SD: 4.2 ± 3.9 days).
According to the results of PCR, we discovered that 55 (17.6%) cases (35 [63.6%] female and 20 [33.4%] males; mean age ± SD 45.7 ± 18.1; range: 13–82 years) were positive for respiratory viruses panel. Detected viral infections included HCoV-OC43/HKU1: 5.4% (17/311), Flu A: 4.5% (14/311), HCoV-229E/NL63: 2.8% (9/311), HMPV: 1.9% (6/311), HPiV-1, -2, -3: 1.2% (4/311), HRSV: 0.9% (3/311), and HAdV: 0.6% (2/311). The co-infection was detected in four samples (1.2%) among the studied cases. The co-infections included Flu A-HMPV (0.6%, 2/311), OC43/HKU1-Flu A (0.3%, 1/311), and OC43/HKU1-NL63/229E (0.3%, 1/311). The sum of positivity rate of respiratory virus infection in patients with negative SARS-CoV-2 was observed to be 17.6% (55/311). Of 55 positive cases, 26 (47.2%) individuals were identified with coronavirus infection (HCoV-OC43/HKU1 and HCoV-229E/NL63), which rate them as predominant infections followed by Flu A virus (25.2%, 14/55).
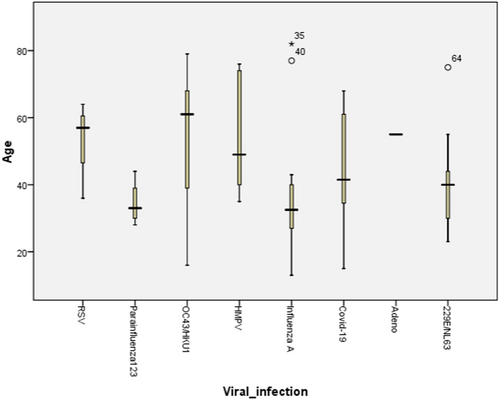
Statistical analyses showed no significant difference between the two sexes (p = 0.054; odds ratio: female/male = 0.6; 95% confidence interval: 0.3–1), age (p = 0.1) (Figure 1), symptom onset before sampling (p = 0.3), and all of the clinical symptoms except for sore throat (0.028), headache (p = 0.016), and body pain (p = 0.0001). Table 1 shows the major clinical symptoms in PCR-positive cases infected by respiratory viruses. As for the age, cases with HPiV infection were the youngest (mean age ± SD: 34.5 ± 6.8) and the patient with HAdV infection was the oldest (55.0 ± 0.0), although the difference was not statistically significant (p = 0.1). Also, considering the symptom onset before sampling, HRSV infection had the lowest value (mean ± SD: 1.5 ± 0.7 days) and the HMPV the longest (mean ± SD: 6.4 ± 3.7 days), but the difference observed was not statistically significant (p = 0.4). By age group, the majority of cases were in 60 years and older group (36.3%) and the 21- to 40-year group (32.4%) came next. Interestingly, viral infection was identified more in the 21- to 40-year group (41.7%) and next in 60 years and older group (28.3%). By the age-specific analysis, it was revealed that human coronaviruses (OC43/HKU1 and 229E/NL63) are the first-ranked pathogens among all age groups; however, Flu A was the dominant pathogen in the 21- to 40-year age group, and two viruses (Flu A and Coronaviruses) were not observed to have statistically significant differences (p = 0.1). Considering the sampling season, most of the samples were collected in spring (43.0%) and interestingly this season included the most positive cases (41.7%), although the difference was not found to be significant (p = 0.8). Moreover, HPiV and Flu A were detected more in autumn, human coronaviruses (OC43/HKU1 and 229E/NL63) in spring, and HRSV and HMPV were detected at a similar rate in all seasons.
| Viral infection | N | Major clinical symptoms | p Value |
|---|---|---|---|
| HCoV-OC43/HKU1 | 17 | Fever (59%), body pain (59%), cough (53%) | 0.346 |
| Influenza A virus | 14 | Fever (57%), body pain (57%), chill (50%), cough (43%) | 0.133 |
| HCoV-229E/NL63 | 9 | Sore throat (78%), malaise (56%), body pain (56%), rhinorrhea (56%) | 0.548 |
| Human metapneumovirus (HMPV) | 6 | Fever (84%), cough (50%), headache (50%) | 0.279 |
| Parainfluenza virus | 4 | Body pain (75%), fever (50%), rhinorrhea (50%) | 0.521 |
| Respiratory syncytial virus (RSV) | 3 | Body pain (100%), fever (67%), chill (67%), malaise (67%) | 0.384 |
| Adenovirus | 2 | Malaise (100%), fever (50%), cough (50%), body pain (50%) | 0.612 |
| Total | 55 | Fever (59%), body pain (52%), cough (44%), malaise (40%) | 0.534 |
4 DISCUSSION
Annually, lower respiratory tract infections account for approximately 4 million mortalities with over 500,000 of them caused by influenza viruses. Although bacterial coinfection post influenza is associated with 25% of their related deaths, fungal coinfections are recognized as equally very important. Co-infection should be considered not only for influenza infection but also for other viral respiratory tract infections, such as SARS-CoV-2, HPiV, human cytomegalovirus, HRSV, human rhinovirus, and HAdV.8
Considering the importance of respiratory viral infections, the present study showed the prevalence of 17.6% (55/311) of various viral respiratory agents in patients diagnosed negative for SARS-CoV-2 infection. In this regard, we found hCoV-OC43/HKU1, Flu A, HCoV-229E/NL63, HMPV, HPiV-1, -2, -3, HRSV, and HAdV of PCR-positive patients, respectively. Also, the co-infection was detected in 1.2% of our studied cases.
In a primary study conducted in Iran, using the HiTeq 17 Viro Respiratory pathogen One-Step RT-PCR Kit (Genova, Bonda Faravar, Bioluence, Tehran, Iran), from December 2020 to March 2021, a total of 197 cases were studied: 106 of these cases were reported as SARS-CoV-2 negative and 91 were identified as positive. They reported the pathogens and the relevant percentages as HMPV (7.1%), HCoV-NL63 (4.06%), Flu B (1.52%), HCoV-HKU1 (1.52%), HRSV (2.03%), HPiV (1.01%), HAdV (1.01%), and HBoV (0.5%).7 Interestingly, Veisi et al.7 found Flu B in 1.52% of their cases and no case of Flu A. They also found HMPV, HCoV-NL63, and HRSV by higher prevalence rates, which may be due to high circulation rates of respiratory viruses, especially from 2020–2021 to 2021–2022 during the COVID-19 pandemic waves. The previous studies in Iran before COVID-19 pandemic had shown the prevalence rates of HRSV, Flu, and HMPV as 18.0%,9 10.5%,10 and 8.9%, respectively.11
During the early months of COVID-19 spread in Iran, co-infection was recorded in 22.3% of death cases, especially for Flu A.12 Co-infection of Flu A with SARS-CoV-2 at the same time in Wuhan was reported to be 57.3%.13 The rate of co-infection we found in the present study was 1.2% making us conclude that, by the widespread implementation of control strategies, influenza virus infection may have been restricted in Iran, similar to the conditions reported in Italy,14 France,15 Brazil,16 and Taiwan.17
In a study on 110,058 ARI patients in China, 34.8% were diagnosed with viral infections including influenza virus for 28.5% of total positive detection, HRSV for 16.8%, HRV for 16.7%, HPiV for 13.1%, HAdV for 10.3%, HCoVs for 5.8%, human bocavirus (HBoV) for 4.6%, and HMPV for 4.1% of cases.1 In another study by Ntagereka et al.,18 a multiplex RT-PCR assay was designed for common acute respiratory infection prevalence in 1352 individuals presenting flu-like illness. They found that Flu A accounted for 5.6%, Flu B for 0.9%, HPiV 1–4 for 0.7%, HMPV for 0.4%, human coronaviruses (ОС43, Е229, NL63, and HKUI) for 0.2%, HRV for 0.2%, HAdV for 0.2%, and HBoV for 0.07% of total cases. Differences may be due to different geographical regions and age groups. As for the age groups, our study showed slightly higher infection rate in 60 years and older age group compared to that in Zhong et al. (28.3% vs. 25.1%) and adult groups (32.4% vs 26.9%). However, the difference in results may be because of the limited spectrum of eight respiratory viruses in the study by Li et al.1 or our small sample size. Meanwhile, regional differences should not be neglected. The HAdV prevalence in our study and others were similar except for Li et al.'s study, which may be due to the duration of their study as well as the sample size.
Li et al.'s1 study reported the rate of infection in children (<5 years) as 46.9%, 26.9% in school-age children (5–17 years), 26.9% in adults (18–59 years), and 25.1% in older people (>60 years). In another study by Ntagereka et al.,18 this rate was 89.6% in adults. In an age-specific analysis of Li et al.'s1 study, it was revealed that HRSV (25.7%), HRV (17.4%), HPiV (15.8%), influenza virus (14.2%), and HAdV (10.7%) were the most prevalent pathogens among children. In addition, Qing and Huihui studied 8202 children (<6 years old) in China in 2020. They reported HAdV in 0.2%, Flu A in 0.3%, HPiV in 3.2%, and HRSV in 9.3% of the cases.19 In the present study, we showed that a majority of cases were human coronaviruses (OC43/HKU1 and 229E/NL63); however, Li et al. did not examine human coronaviruses. It seems that human coronaviruses are predominant in our population, followed by influenza A virus. Differences in geographical regions and sampling years maybe other reasons to explain the differences between studies. Our studied specimens were collected from March 2021 to July 2022 in Iran, during which the fourth to sixth waves of SARS-CoV-2 were recorded. The study by Li et al. was carried out from 2009 to 2019 in China, which makes the two studies different in terms of both the region and the SARS-CoV-2 pandemic. Climatic condition is another factor that could affect the results and may contribute to different infection rates. The Flu A and HPiV infection rates in our study are in accordance with those reported in Ntagereka et al., which may be due to rather regional and climate similarities.
Moreover, according to the study by Li et al.,1 influenza A was found during autumn and winter, just similar to our findings; nevertheless, they reported a higher rate of HPiV in summer, which is different from our findings showing more cases in autumn. In spite of the Li et al.1 study, we found HRSV and HMPV at the same rate in all seasons. Also, for the HCoVs prevalence, they reported summer for higher rate of diagnosed cases, yet we found them to be more prevalent in spring. However, although our rates were not found to be significant, limited number of cases can explain it.
In one study by Avolio et al.,20 prevalence of HAdV in 2020–2021 was reported to be similar to that in our study, although they collected the cases in November and we did it in February. A limited number of cases may have affected on finding. However, HAdV could be found in all seasons and it is not season-based.21 In this study, prevalence of coronaviruses in 2020–2021 showed that they are common in winter similar to the results we found in our study. The HPiV is commonly found in November which is similar to other reports in the literature.20, 22 Our study HMPV cases were identified in winter which is not seen by seasonal spreading; however, in some cases, it was reported to happen more in summer.23, 24
Similar to Ntagereka et al., we did not find differences in the clinical behavior and viruses' infection except for sore throat (0.028), headache (p = 0.016), and body pain (p = 0.0001), either. In the present study, clinical symptoms in the Flu A positive patients were fever (57%), body pain (57%), chill (50%), and cough (43%); with regard to fever and cough, our findings are similar to those by Ntagereka et al. and another study in Costa Rica.25
Moreover, the HiTeq 17 Viro Respiratory pathogen One-Step RT-PCR Kit (GeneovA), made in 2021 in Iran, was used in the present study; other studies have used this kit considering its appropriate sensitivity and specificity to identify various viral respiratory pathogens, including SARS-CoV-2, Flu A, Flu B, Flu A/H1N1, HCoV-NL63, HCoV-229E, HCoV-HKU1, HCoV-229E, HCoV-OC43, HPiV-1, -2, -3, HAdV, HBoV-1, -2, -3, HMPV, and HRSV.7, 26
Furthermore, many countries have used nonpharmacological interventions, which are part of Complementary and Alternative Medicine, to control the spread of viruses at different times. The Iranian government implemented various policies to control the epidemic since the pandemic started. The Nowruz lockdown was one of the strictest measures, but it caused economic problems, which made the government adopt softer interventions with fewer restrictions. This approach led to a surge in cases and deaths. These interventions may have affected the spread of all respiratory viruses, including SARS-CoV-2.27
Mathematical epidemic models are powerful tools that allow healthcare policymakers to predict the effects of virus mutation, vaccination, quarantine, and lockdown on the number of infections and deaths, and to make effective decisions.27
It is important to note that COVID-19 vaccination reached almost 150,078,501 doses in Iran by the time of our study sampling, according to the COVID-19 situation updates for Week 27 (July 3–9, 2022).28 The Influenza vaccine was also available in all parts of the country before autumn, as in previous years.29
Furthermore, studies have shown how temperature and humidity influence the stability and transmission of respiratory viruses. Recent research emphasizes the role of environmental factors, especially temperature and humidity, in shaping the host's immune responses to viral infections in the respiratory tract.30
The present study has some limitations that should also be taken into consideration before any generalization. The limited sample size was a limitation which was due to the costs of the primary and secondary screenings. Finally, although we did not run confirmatory test for the secondary screening results in addition to the first-round screening, we made sure to use appropriate controls in each reason. Multiplex Real-time PCR is very sensitive, so it can sometimes find traces of viruses that indicate past infections, not just current ones. Also, when more than one virus is found, it is important to think about both “co-infection” and “coexistence” as possible scenarios. Also, we lack data on other relevant information, such as medical conditions, that could alter the results. The history of the family or cohabitants is also important. However, we were unable to analyze or present these cases due to missing data.
In conclusion, with regard to the worldwide circulation of respiratory viruses during the COVID-19 pandemic, their identification, prevalence investigation, and coinfection rate in each geographical region, or setting, could help experts in isolation and maintenance of patients. The utilization of panel of respiratory viruses in patients with respiratory infection referred to clinics, as an appropriate tool, is suggested for infection control and diagnosis in primary screening due to the importance of other respiratory viruses in addition to SARS-CoV-2 detection. Widespread improvement of laboratories to updated diagnostic tools, such as Next-Generation Sequencing and Real-time PCR systems, while performing COVID-19 more accurate diagnostic measures, could allow us to diagnose other respiratory viruses. In our study, we found that 17.6% of our samples were positive for various respiratory viruses and 1.2% were identified by co-infection. Coronaviruses were still predominant viral infections identified in the present study (47.2%), followed by the Flu A virus (25.4%). However, the limited sample size may have negatively impacted our results. Multicentric studies making use of greater sample size are recommended to obtain more comprehensive results.
AUTHOR CONTRIBUTIONS
Reyhaneh Sadeh-Tehrani: Software; formal analysis; data curation; investigation. Hanieh Mohammadjafari: Methodology; visualization; formal analysis; investigation. Sheida Alizadeh: Methodology; validation; visualization; data curation; supervision. Maryam Naseroleslami: Conceptualization; writing—original draft; formal analysis; data curation. Mohammad Hadi Karbalaie Niya: Conceptualization; writing—review and editing; methodology; project administration; resources; supervision.
ACKNOWLEDGMENTS
The authors would like to appreciate the sincere help provided by Mehr Hospital personnel, affiliated to Iran University of Medical Sciences, Tehran, Iran. This article is based on a dissertation by two MSc students, Hanieh Mohammadjafari and Reyhaneh Sadeh Tehrani (code: IR.IAU.PS.REC.1401.343 and IR.IAU.PS.REC.1401.342) which were financially supported by Tehran Medical Sciences, Islamic Azad University, Tehran, Iran.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
TRANSPARENCY STATEMENT
The lead author Mohammad Hadi Karbalaie Niya affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author, [M.H. K.N].