Pyroptosis-related gene signatures are associated with prognosis and tumor microenvironment infiltration in head and neck cancer
Abstract
Background and Aims
Recent studies have highlighted the biological significance of pyroptosis in cancer development. Nevertheless, it is still uncertain if pyroptosis also plays a part in immune modulation and the creation of the tumor microenvironment (TME).
Methods
The pyroptosis regulatory genes (PRGs) were comprehensively assessed in 1938 head and neck cancer samples, and systematically correlated these modification patterns with the infiltration characteristics of TME cells. The unsupervised consensus analysis method was used to identify specific pyroptosis clusters. The single-sample gene set enrichment analysis and CIBERSOFT algorithms were used to evaluate the infiltration levels of various immune cell subsets. A principal component analysis algorithm was used to construct the pyrolysis potential index (PPI) to quantify the pyrolysis regulation patterns in head and neck squamous cell carcinoma (HNSC).
Results
Pyrophosphate regulatory genes (PRGs) are often upregulated in tumors due to mutations. PRGs relate to various clinical outcomes and pathways. Molecular subtyping identified pyroptosis patterns, which align with three tumor immunophenotypes: immune-inflamed, immune-excluded, and immune-desert. The PPI measures pyrolysis roles, showing higher PPI in tumor samples linked to subtypes and clinical characteristics. Lower PPI correlates with longer survival, increased immune activity, more tumor mutations, high PD-L1 expression, and mutations in significant genes like PIK3CA. Such patients also experience enhanced immune responses in immunotherapy trials.
Conclusion
We conducted a comprehensive examination of pyroptosis in HNSC and developed a PPI indicator that shows a strong correlation with the variety and intricacy of the TME.
1 INTRODUCTION
Human head and neck squamous cell carcinoma (HNSC) is a prevalent form of cancer, affecting more than 500,000 individuals worldwide annually.1-3 Even with surgical intervention, radiation treatment, and chemotherapy, approximately 50% of patients succumb to the illness. Stage III/IV tumors are the main subgroup of HNSC.4 Patients diagnosed with Stage III/IV have a bleak outlook, as indicated by a 40% recurrence-free survival rate and a 60% overall survival (OS) rate over 5 years.5, 6 Hence, there is a requirement for new therapeutic approaches, either through single-drug treatment or a combination of therapies, to enhance the outlook for primary advanced HNSC. Considering the limitations of HNSC treatment, new treatment targets are crucial to improve the clinical efficacy of HNSC. Therefore, there is an urgent need for new reliable prognostic models for more feasible targeted therapies. According to recent research, it has been found that genes associated with pyroptosis (PRGs) are frequently disrupted in cases of head and neck cancer.7, 8 Nevertheless, as far as we know, there has been no systematic analysis conducted on the carcinogenic impacts of PRGs in head and neck cancer.
Regulated cell death, a pathway of programmed cell death, has a significant impact on the development, maintenance of balance, and disease progression in organisms. Pyroptosis, a recently identified form of programmed cell death, is involved in the pathogenesis of several conditions, such as autoimmune disorders, diseases related to immunodeficiency, neurodegenerative disorders, ischemia–reperfusion injuries, and cancer. CD4+ T lymphocytes (immune cells of HIV infection) exhibit resistance to cellular scorching,9 a process implicated in the development of acute liver injury and acute lung injury.10, 11 Classical cellular scorching of NLRP1 inflammatory vesicles12 is a significant process that plays a crucial role in the neurodegeneration observed in Alzheimer's disease. With the increasing understanding of pyroptosis, its complex biological function has been uncovered. Drug resistance has also been shown to be linked with pyroptosis and PRGs. According to a previous report by Guo et al.,13 the regulation of GW4064 was found to trigger pyroptosis in colorectal cancer cells, leading to enhanced chemosensitivity both in vivo and in vitro. In a recent study, it was discovered that BIX-01294 can boost the effectiveness of chemotherapy in gastric cancer through the stimulation of GSDME-mediated pyroptosis.14 Hence, pyroptosis might have a significant impact on the progression and management of cancer. Exploring the systematic examination of pyroptosis and its disruption in HNSC may prove beneficial for clinical intervention.
Studying the correlation between cell pyroptosis and disease could offer fresh perspectives on the clinical management of the condition. Pyroptosis has been found to have a significant impact on tumor development and antitumor mechanisms, according to the current research. Nevertheless, the examination of its particular role in head and neck cancer remains unexplored. Hence, an organized investigation was carried out to ascertain the levels of expression of PRGs in tissues of head and neck tumors. In addition, the predictive significance and the association between pyroptosis and the immune microenvironment of tumors were investigated. Furthermore, a pyroptotic potential index (PPI) was developed to measure the pyroptosis alteration patterns of individual tumors and forecast the immune therapy response of patients.
2 MATERIAL AND METHODS
2.1 Source of data and pre-processing
Seven publicly accessible datasets (GSE27020,15 GSE31056,16 GSE30784,17 GSE39366,18 GSE41613,19 GSE65858 [LHNG cohort],20 and TCGA-HNSC cohort) were utilized to gather messenger RNAs and clinical information. Data of TCGA-HNSC datasets obtained from The Cancer Genome Atlas (TCGA) website were downloaded, specifically RNA sequencing (RNA-seq; fragments per kilobase million value) data. The R package GEOquery.21 was used to download the microarray data for GSE27020, GSE31056, GSE30784, GSE39366, GSE41613, and GSE65858 from Gene Expression Omnibus (GEO). To remove nonbiological technical biases, the sva package utilized the “ComBat” algorithm.22 The OS data were available for TCGA-HNSC, GSE27020, GSE31056, GSE41613, and LHNG cohorts. Data on somatic mutation and copy number variation (CNV) were obtained from the TCGA database. To depict the CNV pattern of 33 genes involved in pyroptosis on human chromosomes, the R package “Rcircos” was utilized. The quantification of tumor mutation burden (TMB) involved calculating the number of somatic alterations per megabase within the genome, as stated by Hellmann et al. in 2018.23
2.2 Unsupervised clustering analysis using a consensus algorithm
To identify differentially expressed PRGs, a collection of 33 PRGs was extracted from six integrated GEO datasets. Pyroptosis modification patterns were identified using unsupervised clustering analysis, which was based on the 33 genes regulating pyroptosis. This analysis classified patients into three distinct clusters. To ensure classification stability, the “Pam” method underwent unsupervised clustering analysis using the “ConsensuClusterPlus” R package,24 which was repeated 1000 times.
2.3 Functional enrichment analysis and gene set variation analysis
To examine the diversity in biological processes among various pyroptosis modification patterns, we utilized the gene set variation analysis (GSVA)25 technique with the R package “GSVA.” For GSVA analysis, the MSigDB database was used to download the gene sets labeled as “c2.cp.kegg.v6.2.-symbols.” The R package “clusterProfiler” was used to perform KEGG annotation for genes relevant to pyroptosis, with a threshold of FDR < 0.05.26
2.4 Assessment of the infiltration of cells in the tumor microenvironment
Quantification of the infiltration levels of various immune cells in HNSC was conducted through a single-sample gene set enrichment analysis (ssGSEA) as described by Barbie et al. in 2009.27 Recently published studies27, 28 provided gene panels specifically designed for 28 different types of immune cells. The ssGSEA analysis was used to calculate enrichment scores, which represented the relative abundance of tumor microenvironment (TME)-infiltrating cells in each sample.
2.5 Differential expression analysis between distinct phenotypes
Using the “Limma” R package,29 differential expression analysis was conducted to identify differentially expressed genes (DEGs) among different subtypes.
2.6 Principal component analysis dimensionality reduction and establishment of the PPI index
I represent the manifestation of genes related to the m6A phenotype.
2.7 Somatic alteration data
Data on genetic mutations in patients from the TCGA-HNSC group were obtained from the TCGA site (https//www.cancer.gov/tcga/). The calculation of TMB in HNSC was determined by considering the overall count of non-synonymous mutations. HNSC driver genes were analyzed using the ComplexHeatmap package's “oncoplot” function between high and low immune cell infiltration (ICI) scores.32 We identified the 25 most frequently altered genes with the highest mutation rate.
2.8 Prediction of response to chemotherapy/immunotherapy
While immune checkpoint blockade therapies that inhibit T-cell suppressor molecules in cancer treatment have demonstrated remarkable outcomes and the ability to enhance the advancement of advanced cancers, they may not be appropriate for every patient.33, 34 The therapeutic benefit of the PPI score was further analyzed in two immunotherapy cohorts: the IMvigor 2100 cohort, which involved the intervention of advanced urothelial cancer with the anti-PD-L1 antibody atezolizumab,35 and the GSE78220 cohort from GEO, which focused on the treatment of metastatic melanoma with the anti-PD-1 antibody pembrolizumab.36
To forecast clinical responses to immune checkpoints, the TIDE algorithm37 and Subclass mapping38 were employed for the estimation of tumor immune dysfunction and exclusion. Predictions of the response to chemotherapy were also made for every sample using the most extensive pharmacogenomic database accessible to the public (Genomics of Drug Sensitivity in Cancer [GDSC], https//www.cancerrxgene.org/).39 Table S1 listed a total of 149 medications with the capability to address cancer. The R software's “pRRophetic” package was utilized for making predictions. The package utilized ridge regression to calculate the IC50 values of the samples and evaluated the prediction accuracy through 10-fold cross-validation using the GDSC training set.40
3 RESULTS
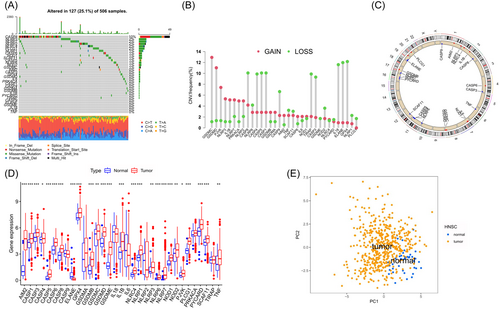
3.1 The genetic variation of PRGs in head and neck cancer is reflected in the landscape
In this study, a total of 33 PRGs were ultimately discovered. Out of the 506 samples, a total of 127 exhibited mutations in m6A regulatory genes, accounting for a prevalence rate of 25.10%. In HNSC samples (Figure 1A), CASP8 displayed the highest mutation frequency, with NIRP3 following closely. Notably, TNF, PJVK, and GSDME, which are involved in prognosis regulation, exhibited no mutations. In the examination of CNV modification occurrence, a dominant CNV modification was observed in 33 regulatory genes, with the majority of genes being linked to copy number amplification. However, cysteinyl aspartate protease (caspase or CASP) family genes like CASP8, CASP4, and CASP1 exhibited a widespread occurrence of CNV deletion (Figure 1B). Figure 1C displays the position of CNV modification of PRGs on chromosomes. Figure 1D clearly showed that HNSC samples were completely differentiated from normal samples based on the expression of the 33 PRGs. An abnormal elevation of most PRGs was observed in tumor samples when compared to normal samples, except for CASP9, ELANE, IL18, and IL6 (Figure 1E). To assess the reciprocal control among the PRGs, a Spearman correlation analysis was conducted (see Figure S1). Other PRGs exhibited a notable and positive correlation with CASP5. In general, the findings indicated that there is significant diversity in the genetic and expression patterns of PRGs between normal and HNSC samples, suggesting that the expression of PRGs plays a crucial part in the onset and progression of HNSC.
3.2 Identification of pyroptosis-related molecular subtypes
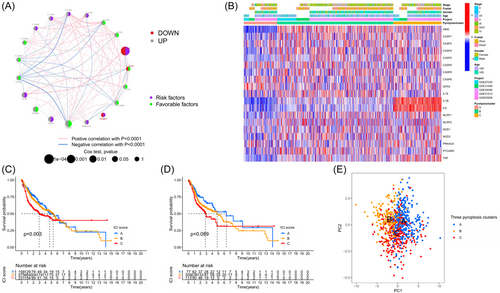
In the meta-GEO cohort, we deemed six GEO datasets (LHNG cohort/GSE65858, GSE41613, GSE39366, GSE31056, GSE30784, and GSE27020) as suitable, as they had clinical data available. The prognostic values of the 33 PRGs in patients with HNSC were revealed using a univariate Cox regression model and the Kruskal–Wallis test (Figure S2A,B; Table S2). The PRG network (Figure 2A) demonstrates the synthesis of the connections, interactions, and prognostic importance of the 33 PRGs in HNSC patients. The discovery indicated that the interaction among PRGs might have a significant impact on the development of pyroptosis alteration patterns and the characterization of TME cell infiltration in distinct tumors. Using the aforementioned assumptions, the consensus cluster analysis was employed to categorize samples into distinct groups with varying pyroptosis alterations, depending on the 33 PRGs' expression. Figures 2B and S3A–C revealed the identification of three separate clusters of modification patterns, consisting of 197 patients in Cluster I, 727 samples in Cluster II, and 286 cases in Cluster III. The three clusters were compared using Kaplan–Meier survival analysis, and the log rank test was employed to determine the significant disparity in survival time. The study revealed that individuals belonging to gene cluster III exhibited a more favorable prognosis, whereas those in gene cluster I had an unfavorable prognosis for OS (based on the log rank test, with a significance level of p < 0.05; as shown in Figure 2C). In addition, the TCGA-HNSC group was confirmed and yielded comparable outcomes (log rank analysis, p < 0.05; Figure 2D). Using PCA, the expression profiles of the 33 PRGs were analyzed to identify three subgroups in the meta-GEO cohort (Figure 2E).
3.3 The infiltration properties of immune cells from the TME in the three clusters associated with pyroptosis
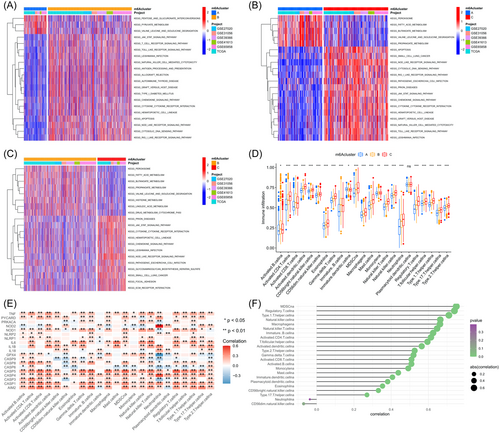
To reveal the biological role of the pyroptosis-related clusters, an analysis of GSVA enrichment was performed. According to GSVA, Cluster A showed significant enrichment in pathways associated with energy metabolisms, such as CITRATE_CYCLE_TCA_CYCLE, PEROXISOME, and FATTY_ACID_METABOLISM. On the other hand, Cluster B exhibited significant enrichment pathways linked to cancer activation and stromal pathways, including the NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY, RIG_I_LIKE_RECEPTOR_SIGNALING_PATHWAY, T_CELL_RECEPTOR_SIGNALING_PATHWAY, and NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY. Interestingly, Cluster C exhibited a significant enrichment in metabolic regulation and signaling pathways related to the stroma (Figure 3A–C).
Using the ssGSEA algorithm, we systematically assessed the TME cell infiltration models and TME signatures in the three ICI clusters. Figure 3D displayed the heatmap of 28 subpopulations of immune infiltrating cells within pyroptosis-associated clusters. Cluster A exhibited a significant presence of B cells that were extremely inexperienced, CD8 T cells, follicular helper T cells, regulatory T cells (Tregs), activated natural killer (NK) cells, monocytes, M0 macrophages, and resting mast cells among the three subtypes. Group B patients exhibited a notably elevated abundance of T cells CD4 memory resting, M1 and M2 macrophages, dendritic cells resting, stromal score, and immune score. Cluster C subjects exhibited a notable rise in the infiltration of plasma cells. In addition, CIBERSORT, an algorithm that employs support vector regression to assess immune cell subpopulations in the TME, was employed to provide further insights into immune infiltration within the three subpopulations. Figure S4 also showed similar findings. Hence, there is speculation that the activation of stroma in Cluster B hinders the immune cells' ability to suppress tumor growth. The additional examination also verified that stromal stimulation in the Cluster B subtype was greatly intensified, and the mechanisms associated with cell death, the pathway of signaling through toll-like receptors, and the pathway of signaling through T cell receptors were demonstrated, providing further validation of our supposition.
Spearman's correlation analyses (Figure 3E) were conducted to investigate the precise association between every type of TME infiltration cell and each PRG. Enhanced ICI showed a significant correlation with the high expression of several PRGs like CASP family members and NOD2, while neutrophil infiltration levels were negatively correlated with the expression of GPX4, CASP3, and CASP6. NLRC4 caught our interest among these PRGs due to its notable association with prognosis and immune infiltration, as depicted in Figure 3F.
3.4 DEGs associated with pyroptosis in head and neck cancer
The “limma” package of the R software was used to determine the transcriptomic variation between different pyroptosis phenotypes. The empirical Bayesian approach (Figure 4A) was used to analyze the 1276 DEGs that were common to the three clusters. Figure 4B,C illustrates that DEGs were concentrated in immune-related Gene Ontology categories, such as T cell stimulation, release of neutrophil contents, immune receptor function, and neutrophil activation in immune response. In addition, they were also found in pathways associated with immune/cancer interactions, including the TNF signaling pathway, the IL-17 signaling pathway, the NF-kappa B signaling pathway, the PI3K-Akt signaling pathway, and the differentiation of Type 17 T helper cells (Th17).

Out of these DEGs, a total of 545 DEGs were recognized as prognostic genes according to Table 1. The prognostic DEGs were then analyzed by the unsupervised clustering method. In the meta-GEO cohort, three genomic clusters, specifically gene Clusters A, B, and C, were discovered. Figure 5A displayed the transcriptome profiles of the DEGs within the three genomic clusters, as depicted by the heat map. In gene cluster C, among the 33 PRGs, AIM2, CASP1, CASP4, CASP8, GSDMD, GSDME, IL1B, IL6, NLRP1, NOD1, PLCG1, and TNF showed significant upregulation. On the other hand, gene cluster A exhibited upregulation of CASP3, CASP6, CASP9, GPX4, GSDMB, and PRKACA. Aberrant expression of GSDMA, GSDMC, IL18, NOD2, PYCARD, and SCAF11 was observed in gene Cluster B (Figure 5B). Next, the examination focused on the particular association between every type of TME infiltration cell and each PRG. Gene Cluster A exhibited elevated levels of stimulated B cell, eosinophil, stimulated CD8 T cell, and regulatory T cell. Figure 5C showed that Gene cluster C exhibited enrichment of activated CD4 T cell, activated dendritic cell, CD56 bright NK cell, CD56 dim NK cell, gamma delta T cell, immature B cell, MDSC, macrophage, mast cell, monocyte, NK T cell, NK cell, regulatory T cell, Th1 cell, Th17 cell, and Th2.
| ID | HR | HR.95L | HR.95H | p Value |
|---|---|---|---|---|
| XYLT2 | 1.29691 | 1.076004 | 1.56317 | 0.006355 |
| LARP6 | 1.266636 | 1.125973 | 1.424873 | 8.31E−05 |
| NOX4 | 1.139376 | 1.026343 | 1.264857 | 0.014376 |
| COL10A1 | 1.062942 | 1.008538 | 1.120282 | 0.02278 |
| SULF1 | 1.077868 | 1.010165 | 1.150109 | 0.02348 |
| ACVR1 | 1.159116 | 1.002066 | 1.34078 | 0.046839 |
| FOXF2 | 1.14116 | 1.023744 | 1.272041 | 0.017146 |
| EGLN3 | 1.086472 | 1.002792 | 1.177135 | 0.042544 |
| P4HA1 | 1.476689 | 1.304708 | 1.67134 | 6.83E−10 |
| STC2 | 1.271687 | 1.160355 | 1.3937 | 2.72E−07 |
| HOXC6 | 1.113041 | 1.001791 | 1.236646 | 0.046232 |
| GLG1 | 1.256676 | 1.03427 | 1.526907 | 0.021503 |
| CDH11 | 1.098933 | 1.018988 | 1.185149 | 0.014362 |
| TGM1 | 0.941318 | 0.895893 | 0.989046 | 0.016556 |
| LAMB3 | 1.125807 | 1.030175 | 1.230318 | 0.008888 |
| APP | 1.25389 | 1.096859 | 1.433403 | 0.000919 |
| ARL4C | 1.146228 | 1.018068 | 1.29052 | 0.024072 |
| NID1 | 1.164153 | 1.061618 | 1.276592 | 0.001233 |
| SERPINE2 | 1.127064 | 1.044853 | 1.215743 | 0.001966 |
| DKK3 | 1.181 | 1.068068 | 1.305873 | 0.001178 |
| MAGEH1 | 1.139605 | 1.004257 | 1.293195 | 0.042784 |
| MMP13 | 1.051916 | 1.012552 | 1.09281 | 0.009295 |
| LHFPL2 | 1.166791 | 1.011219 | 1.346297 | 0.03462 |
| GNA12 | 1.428293 | 1.209139 | 1.687169 | 2.73E−05 |
| PTK7 | 1.193968 | 1.039851 | 1.370925 | 0.011932 |
| KLK12 | 0.931325 | 0.888362 | 0.976367 | 0.003152 |
| FARP1 | 1.14206 | 1.002505 | 1.301041 | 0.045761 |
| LRP12 | 1.226166 | 1.089555 | 1.379907 | 0.000717 |
| TMEM184B | 1.330451 | 1.11395 | 1.58903 | 0.001628 |
| TPST1 | 1.205569 | 1.078002 | 1.348231 | 0.001052 |
| CSTB | 0.861523 | 0.785539 | 0.944856 | 0.001556 |
| TRAM2 | 1.161685 | 1.011715 | 1.333886 | 0.033577 |
| MAP1B | 1.130082 | 1.027339 | 1.2431 | 0.011918 |
| CRNN | 0.965201 | 0.934299 | 0.997124 | 0.032891 |
| NEDD9 | 1.212145 | 1.081247 | 1.35889 | 0.000968 |
| DSG2 | 1.113117 | 1.024583 | 1.209302 | 0.011268 |
| HSP90B1 | 1.360787 | 1.118377 | 1.65574 | 0.002086 |
| ETS1 | 1.147718 | 1.01515 | 1.297599 | 0.027802 |
| TGM3 | 0.947095 | 0.914268 | 0.981102 | 0.002528 |
| DPY19L1 | 1.180051 | 1.021605 | 1.363073 | 0.024416 |
| INHBB | 1.18258 | 1.087874 | 1.285531 | 8.23E−05 |
| NFE2L1 | 1.241567 | 1.049791 | 1.468376 | 0.011485 |
| PPL | 0.881535 | 0.819119 | 0.948707 | 0.000765 |
| INHBA | 1.184569 | 1.102599 | 1.272634 | 3.67E−06 |
| MMP2 | 1.111267 | 1.015929 | 1.215552 | 0.021151 |
| RAB25 | 0.868311 | 0.806203 | 0.935204 | 0.000192 |
| PLAUR | 1.228336 | 1.099381 | 1.372416 | 0.000279 |
| LITAF | 1.186823 | 1.00714 | 1.398562 | 0.040864 |
| MSC | 1.200947 | 1.094987 | 1.31716 | 0.000102 |
| SOAT1 | 1.276423 | 1.096193 | 1.486287 | 0.001675 |
| ATP2B4 | 0.832925 | 0.706825 | 0.981523 | 0.029063 |
| CHPF | 1.269965 | 1.110793 | 1.451945 | 0.000469 |
| PTPRR | 1.253903 | 1.026589 | 1.531549 | 0.026616 |
| CSTA | 0.853663 | 0.797013 | 0.91434 | 6.30E−06 |
| PITX1 | 0.802528 | 0.73286 | 0.878818 | 2.05E−06 |
| SLC16A6 | 0.870687 | 0.76798 | 0.98713 | 0.030601 |
| ALS2CL | 0.860694 | 0.756226 | 0.979593 | 0.02307 |
| MICAL2 | 1.219536 | 1.080396 | 1.376596 | 0.001323 |
| MSN | 1.173952 | 1.018028 | 1.353757 | 0.027405 |
| BCAT1 | 1.121793 | 1.030633 | 1.221017 | 0.007867 |
| EVPL | 0.85879 | 0.786391 | 0.937854 | 0.000704 |
| GLTP | 0.819513 | 0.741 | 0.906345 | 0.000107 |
| TGFBI | 1.120726 | 1.045805 | 1.201016 | 0.001244 |
| ANXA5 | 1.535987 | 1.298945 | 1.816288 | 5.21E−07 |
| FUT6 | 0.843177 | 0.767389 | 0.92645 | 0.000386 |
| DENND2D | 0.726704 | 0.626423 | 0.843038 | 2.51E−05 |
| AXL | 1.203911 | 1.078942 | 1.343355 | 0.000904 |
| ZNF639 | 1.259112 | 1.06078 | 1.494527 | 0.008422 |
| CYB5R3 | 1.260687 | 1.017797 | 1.561539 | 0.033878 |
| PDIA6 | 1.221262 | 1.03442 | 1.441853 | 0.018304 |
| CNIH3 | 1.209005 | 1.019001 | 1.434439 | 0.029575 |
| APLP2 | 1.266795 | 1.075523 | 1.492083 | 0.004629 |
| MAL | 0.960304 | 0.923745 | 0.99831 | 0.040817 |
| CALML5 | 0.877656 | 0.840766 | 0.916165 | 2.58E−09 |
| SCEL | 0.926024 | 0.876468 | 0.978382 | 0.006166 |
| GABARAPL1 | 1.193956 | 1.0504 | 1.35713 | 0.006682 |
| NRSN2 | 1.23226 | 1.062689 | 1.42889 | 0.005694 |
| DDOST | 1.349804 | 1.071605 | 1.700226 | 0.010858 |
| SPINK5 | 0.887729 | 0.845668 | 0.931881 | 1.52E−06 |
| PTDSS1 | 1.277505 | 1.080392 | 1.51058 | 0.004179 |
| NDRG1 | 1.11878 | 1.012763 | 1.235895 | 0.027131 |
| TM7SF2 | 0.888298 | 0.813305 | 0.970206 | 0.008486 |
| FUT3 | 0.882012 | 0.81581 | 0.953587 | 0.001612 |
| CTSZ | 1.141007 | 1.011762 | 1.286763 | 0.031508 |
| TWSG1 | 1.204773 | 1.040692 | 1.394723 | 0.012634 |
| TPM1 | 1.181625 | 1.076333 | 1.297218 | 0.000457 |
| CLSTN2 | 1.127845 | 1.008545 | 1.261257 | 0.034934 |
| SNX10 | 1.16279 | 1.043032 | 1.296298 | 0.006534 |
| ERF | 1.227076 | 1.02442 | 1.469822 | 0.026288 |
| OLR1 | 1.186432 | 1.098566 | 1.281325 | 1.33E−05 |
| ZNF281 | 1.407943 | 1.20498 | 1.645093 | 1.65E−05 |
| TOR1A | 1.654326 | 1.301684 | 2.102503 | 3.86E−05 |
| RASAL1 | 0.866299 | 0.762803 | 0.983837 | 0.027037 |
| SCG5 | 1.232138 | 1.148495 | 1.321872 | 5.88E−09 |
| ARPC1B | 1.163229 | 1.001602 | 1.350937 | 0.047598 |
| TMED2 | 1.410183 | 1.151511 | 1.726962 | 0.000886 |
| CYP2C18 | 0.875504 | 0.819944 | 0.934829 | 7.05E−05 |
| EFEMP1 | 1.10013 | 1.001424 | 1.208565 | 0.046632 |
| LY6G6C | 0.927623 | 0.874811 | 0.983623 | 0.012002 |
| STX2 | 1.35379 | 1.137977 | 1.610531 | 0.000629 |
| LASP1 | 1.376742 | 1.118076 | 1.695251 | 0.002603 |
| NMU | 0.901952 | 0.844897 | 0.96286 | 0.001967 |
| HOPX | 0.873536 | 0.808728 | 0.943537 | 0.000587 |
| ST3GAL2 | 1.22066 | 1.010487 | 1.474548 | 0.038624 |
| JAM3 | 1.145645 | 1.005789 | 1.304948 | 0.040672 |
| MT1F | 1.298073 | 1.159978 | 1.452607 | 5.47E−06 |
| PRKAB2 | 1.175324 | 1.011677 | 1.365442 | 0.03471 |
| TNFRSF12A | 1.240456 | 1.128225 | 1.363851 | 8.45E−06 |
| SCG2 | 1.195768 | 1.110423 | 1.287672 | 2.22E−06 |
| ALDH1B1 | 1.244187 | 1.108113 | 1.39697 | 0.000218 |
| ALOX12 | 0.926048 | 0.863976 | 0.992579 | 0.029978 |
| FAM135A | 0.812141 | 0.706951 | 0.932984 | 0.003281 |
| SIRT7 | 0.786733 | 0.630071 | 0.982347 | 0.034246 |
| TREM1 | 1.151536 | 1.063599 | 1.246743 | 0.000499 |
| NCF2 | 1.177597 | 1.051464 | 1.318861 | 0.004682 |
| NLRX1 | 0.813027 | 0.700135 | 0.944122 | 0.006651 |
| TTC9 | 0.841095 | 0.761679 | 0.928792 | 0.000627 |
| THBS1 | 1.31116 | 1.213337 | 1.41687 | 7.49E−12 |
| SPRR3 | 0.932931 | 0.902859 | 0.964004 | 3.28E−05 |
| RAPGEFL1 | 0.855302 | 0.786697 | 0.929891 | 0.000248 |
| ATP6V1C1 | 1.442359 | 1.176376 | 1.768483 | 0.000429 |
| ABLIM1 | 0.787821 | 0.701411 | 0.884876 | 5.74E−05 |
| MPP1 | 1.226327 | 1.068634 | 1.407291 | 0.00367 |
| KLF10 | 1.213793 | 1.060158 | 1.389691 | 0.005016 |
| PIK3IP1 | 0.85421 | 0.747616 | 0.976001 | 0.020495 |
| ALDH18A1 | 1.222674 | 1.021979 | 1.462781 | 0.027977 |
| LLGL2 | 0.745525 | 0.639405 | 0.869256 | 0.000178 |
| SLC39A8 | 1.136591 | 1.002131 | 1.289093 | 0.04625 |
| KLK10 | 0.944836 | 0.893685 | 0.998913 | 0.04569 |
| SLC25A10 | 0.745121 | 0.631843 | 0.878707 | 0.000471 |
| CD63 | 1.514009 | 1.260224 | 1.8189 | 9.39E−06 |
| TIAM1 | 0.787827 | 0.687622 | 0.902635 | 0.000591 |
| NDFIP1 | 1.517489 | 1.195944 | 1.925486 | 0.000597 |
| PSCA | 0.924598 | 0.867642 | 0.985293 | 0.015662 |
| KLK11 | 0.92953 | 0.879525 | 0.982379 | 0.009595 |
| BSPRY | 0.838713 | 0.771977 | 0.911218 | 3.21E−05 |
| TINAGL1 | 1.21401 | 1.101211 | 1.338363 | 9.71E−05 |
| GFPT2 | 1.178785 | 1.075027 | 1.292557 | 0.000467 |
| PANX1 | 1.146005 | 1.00944 | 1.301045 | 0.035283 |
| SFXN3 | 1.351727 | 1.147802 | 1.591883 | 0.000304 |
| LYPD3 | 0.861532 | 0.804805 | 0.922257 | 1.80E−05 |
| ERBB3 | 0.768161 | 0.670993 | 0.879399 | 0.000132 |
| KBTBD2 | 1.283048 | 1.050985 | 1.566352 | 0.014346 |
| LGALS1 | 1.23354 | 1.120066 | 1.35851 | 2.02E−05 |
| ZNF750 | 0.87818 | 0.821586 | 0.938672 | 0.000132 |
| FHL2 | 1.275099 | 1.130784 | 1.437832 | 7.32E−05 |
| DBI | 0.76199 | 0.645226 | 0.899884 | 0.00136 |
| OVOL1 | 0.893804 | 0.812772 | 0.982914 | 0.020592 |
| HSPA4L | 0.859612 | 0.776346 | 0.951809 | 0.003613 |
| EPHX2 | 0.858396 | 0.776478 | 0.948956 | 0.002847 |
| MGAT1 | 1.371482 | 1.110695 | 1.6935 | 0.003329 |
| DBNDD1 | 0.882103 | 0.78301 | 0.993737 | 0.039084 |
| SCNN1A | 0.87176 | 0.813248 | 0.934481 | 0.000108 |
| ITGA3 | 1.1501 | 1.041548 | 1.269966 | 0.005697 |
| CYP2C9 | 0.799505 | 0.669947 | 0.954118 | 0.013114 |
| NUDCD3 | 1.413573 | 1.092916 | 1.828308 | 0.008368 |
| GDPD5 | 1.218913 | 1.033698 | 1.437314 | 0.018568 |
| CTSB | 1.174501 | 1.026233 | 1.344191 | 0.019489 |
| SH2B3 | 1.149417 | 1.006717 | 1.312344 | 0.039499 |
| TSPO | 0.705501 | 0.578377 | 0.860565 | 0.000579 |
| CES3 | 0.825806 | 0.694592 | 0.981808 | 0.030165 |
| SIL1 | 1.535338 | 1.250923 | 1.884419 | 4.10E−05 |
| FNDC3A | 1.242389 | 1.048914 | 1.471552 | 0.011975 |
| HDAC9 | 1.413298 | 1.174284 | 1.700961 | 0.000253 |
| GIPC1 | 0.785315 | 0.635383 | 0.970626 | 0.025366 |
| CANX | 1.348199 | 1.103924 | 1.646527 | 0.003396 |
| GLS | 1.209414 | 1.047557 | 1.396278 | 0.009493 |
| HOXA1 | 1.330354 | 1.161726 | 1.523459 | 3.66E−05 |
| RGS19 | 1.27153 | 1.053432 | 1.534783 | 0.012344 |
| SAMD4A | 1.278198 | 1.141071 | 1.431805 | 2.24E−05 |
| DUOX1 | 0.881145 | 0.801205 | 0.969061 | 0.009117 |
| PRSS12 | 0.807898 | 0.738412 | 0.883923 | 3.34E−06 |
| ARHGEF7 | 1.290341 | 1.010404 | 1.647836 | 0.04106 |
| KTN1 | 1.286827 | 1.095467 | 1.511616 | 0.002141 |
| ENSA | 0.733579 | 0.579666 | 0.928359 | 0.009918 |
| NTAN1 | 1.278397 | 1.011764 | 1.615297 | 0.039594 |
| NTRK2 | 0.930127 | 0.874504 | 0.989289 | 0.021321 |
| EPN3 | 0.838107 | 0.742247 | 0.946347 | 0.004375 |
| LCN2 | 0.931653 | 0.887539 | 0.97796 | 0.00423 |
| MAPK13 | 0.864323 | 0.77396 | 0.965237 | 0.009655 |
| CRYM | 0.888664 | 0.800426 | 0.986629 | 0.02695 |
| PSD3 | 0.876018 | 0.779679 | 0.984261 | 0.025957 |
| ULBP1 | 1.188275 | 1.045271 | 1.350843 | 0.008371 |
| ICAM1 | 1.101498 | 1.003607 | 1.208937 | 0.041772 |
| NRIP1 | 1.137867 | 1.014004 | 1.27686 | 0.028058 |
| CAMK2N1 | 1.329294 | 1.214886 | 1.454477 | 5.68E−10 |
| MRAS | 1.154194 | 1.022469 | 1.30289 | 0.020377 |
| POMT2 | 1.356344 | 1.08254 | 1.699399 | 0.008065 |
| SLC25A32 | 1.344068 | 1.118813 | 1.614675 | 0.00158 |
| CEACAM1 | 0.90788 | 0.839326 | 0.982034 | 0.015842 |
| PLCD1 | 0.789141 | 0.684276 | 0.910077 | 0.001133 |
| FSTL3 | 1.21902 | 1.119181 | 1.327765 | 5.56E−06 |
| ADA | 1.451826 | 1.277885 | 1.649443 | 1.03E−08 |
| RNF14 | 1.394559 | 1.092437 | 1.780236 | 0.007593 |
| BCAR3 | 1.29701 | 1.167982 | 1.44029 | 1.15E−06 |
| RABGGTB | 1.259644 | 1.013218 | 1.566003 | 0.037692 |
| EMP3 | 1.14563 | 1.020103 | 1.286602 | 0.021669 |
| SASH1 | 0.87454 | 0.786825 | 0.972034 | 0.01292 |
| SPRR1A | 0.929043 | 0.891527 | 0.968138 | 0.000466 |
| TRIM32 | 1.516757 | 1.292418 | 1.780037 | 3.38E−07 |
| CEBPB | 1.275988 | 1.095249 | 1.486553 | 0.001763 |
| RAB31 | 1.162093 | 1.027721 | 1.314034 | 0.01657 |
| PAX9 | 0.883506 | 0.81241 | 0.960825 | 0.003808 |
| CYP2C19 | 0.374608 | 0.157314 | 0.892044 | 0.026553 |
| ATG12 | 1.377945 | 1.076558 | 1.763707 | 0.010905 |
| IVL | 0.929627 | 0.881849 | 0.979994 | 0.006715 |
| TNPO1 | 1.297919 | 1.060612 | 1.588324 | 0.011368 |
| WNT4 | 0.876398 | 0.79441 | 0.966848 | 0.00847 |
| ILK | 1.273212 | 1.069984 | 1.515041 | 0.006482 |
| IL21R | 0.808876 | 0.703571 | 0.929941 | 0.002876 |
| SLC20A1 | 1.30057 | 1.126854 | 1.501066 | 0.000327 |
| CD300C | 1.213318 | 1.035551 | 1.421601 | 0.01675 |
| B4GALT3 | 1.729395 | 1.369255 | 2.18426 | 4.27E−06 |
| SPRR1B | 0.946396 | 0.904443 | 0.990295 | 0.017241 |
| CAV1 | 1.11067 | 1.024164 | 1.204484 | 0.011178 |
| CHST11 | 1.193421 | 1.064028 | 1.338549 | 0.002529 |
| IL2RB | 0.884235 | 0.808694 | 0.966833 | 0.006929 |
| PTPRE | 1.221373 | 1.040914 | 1.433116 | 0.014224 |
| DYSF | 1.170588 | 1.046729 | 1.309103 | 0.005774 |
| DLX5 | 0.889593 | 0.811969 | 0.974637 | 0.012023 |
| STAP2 | 0.745028 | 0.649938 | 0.85403 | 2.39E−05 |
| PTPN13 | 0.793361 | 0.716558 | 0.878396 | 8.36E−06 |
| FUT2 | 0.786835 | 0.709738 | 0.872307 | 5.20E−06 |
| PXN | 1.392276 | 1.20364 | 1.610476 | 8.38E−06 |
| CDS1 | 0.857526 | 0.756304 | 0.972296 | 0.016469 |
| MTMR6 | 1.301271 | 1.059251 | 1.598588 | 0.012134 |
| PEA15 | 1.351756 | 1.08901 | 1.677895 | 0.006272 |
| CFL1 | 1.25947 | 1.025175 | 1.547312 | 0.028041 |
| PGAP1 | 0.798501 | 0.688303 | 0.926342 | 0.00298 |
| ACP2 | 1.220475 | 1.013312 | 1.469989 | 0.03579 |
| HTR7 | 1.147172 | 1.031092 | 1.27632 | 0.011653 |
| IGFBP2 | 0.923603 | 0.869325 | 0.98127 | 0.010116 |
| DSE | 1.220392 | 1.073382 | 1.387536 | 0.002356 |
| SPCS3 | 1.419239 | 1.172052 | 1.718559 | 0.000336 |
| LAD1 | 0.886266 | 0.788615 | 0.99601 | 0.042654 |
| NOD1 | 1.35881 | 1.07148 | 1.723191 | 0.011421 |
| APOE | 1.085117 | 1.008283 | 1.167807 | 0.02925 |
| TGM5 | 0.889898 | 0.81118 | 0.976254 | 0.013567 |
| FLII | 1.295056 | 1.044068 | 1.606381 | 0.018657 |
| TACSTD2 | 0.82502 | 0.729514 | 0.93303 | 0.002182 |
| SERPINA1 | 1.108649 | 1.017782 | 1.207629 | 0.018082 |
| SLC9A3R1 | 0.829647 | 0.741314 | 0.928507 | 0.001148 |
| CEACAM5 | 0.916182 | 0.87293 | 0.961577 | 0.000388 |
| SPTBN2 | 0.835372 | 0.738075 | 0.945495 | 0.004413 |
| BMP7 | 0.92411 | 0.856026 | 0.99761 | 0.043254 |
| GNRHR | 0.715716 | 0.547437 | 0.935722 | 0.014454 |
| VASP | 1.240091 | 1.014248 | 1.516224 | 0.035915 |
| FGFR2 | 0.822351 | 0.732041 | 0.923801 | 0.000983 |
| APPL2 | 0.805459 | 0.6805 | 0.953363 | 0.011896 |
| PVR | 1.495225 | 1.242913 | 1.798756 | 1.99E−05 |
| POPDC3 | 1.134423 | 1.064587 | 1.20884 | 1.00E−04 |
| PRUNE2 | 1.127257 | 1.03603 | 1.226518 | 0.005402 |
| DUOX2 | 0.887911 | 0.822443 | 0.95859 | 0.002349 |
| ACO1 | 1.241225 | 1.042726 | 1.47751 | 0.015076 |
| RNF121 | 1.299652 | 1.096719 | 1.540135 | 0.00248 |
| TRIM29 | 0.819545 | 0.74933 | 0.89634 | 1.33E−05 |
| NTF3 | 0.774544 | 0.654953 | 0.915971 | 0.002829 |
| DSG1 | 0.946231 | 0.909674 | 0.984258 | 0.005973 |
| VEGFC | 1.184812 | 1.104304 | 1.27119 | 2.32E−06 |
| ARHGAP29 | 1.309174 | 1.138061 | 1.506014 | 0.000164 |
| PLXNA2 | 0.808917 | 0.698084 | 0.937347 | 0.004794 |
| FADS3 | 1.316445 | 1.181131 | 1.467262 | 6.76E−07 |
| ECHDC2 | 0.793552 | 0.707792 | 0.889704 | 7.41E−05 |
| STK10 | 1.214303 | 1.011303 | 1.458053 | 0.03749 |
| ERBB2 | 0.730551 | 0.618231 | 0.863277 | 0.000228 |
| SMAD6 | 0.786044 | 0.633583 | 0.975192 | 0.028647 |
| SNAP23 | 1.283683 | 1.038469 | 1.586799 | 0.020945 |
| MPZL2 | 0.862386 | 0.775985 | 0.958407 | 0.005983 |
| SACS | 1.238147 | 1.066606 | 1.437277 | 0.004994 |
| HMGN3 | 0.806495 | 0.685501 | 0.948845 | 0.009511 |
| FPR1 | 1.115472 | 1.017943 | 1.222346 | 0.019236 |
| GNPDA1 | 1.448034 | 1.152205 | 1.819817 | 0.001498 |
| ACTN1 | 1.34765 | 1.203247 | 1.509383 | 2.47E−07 |
| SGPP1 | 1.206766 | 1.052015 | 1.384281 | 0.007271 |
| FLG | 0.931548 | 0.873764 | 0.993153 | 0.029988 |
| APOC1 | 1.071971 | 1.005067 | 1.143327 | 0.034542 |
| TWISTNB | 1.286094 | 1.048884 | 1.576951 | 0.015573 |
| LTBP4 | 0.869115 | 0.762762 | 0.990296 | 0.035171 |
| MRPL15 | 1.287125 | 1.073292 | 1.543561 | 0.006469 |
| CYP4F12 | 0.791949 | 0.718075 | 0.873424 | 3.03E−06 |
| MSMB | 0.93112 | 0.883321 | 0.981506 | 0.007949 |
| ALOX12B | 0.912794 | 0.863835 | 0.964529 | 0.001179 |
| NT5E | 1.14313 | 1.058409 | 1.234632 | 0.000662 |
| DLG3 | 0.799638 | 0.679232 | 0.941389 | 0.007246 |
| DOCK2 | 0.880404 | 0.785188 | 0.987166 | 0.029172 |
| CBR3 | 0.858993 | 0.777351 | 0.94921 | 0.002855 |
| ZMAT5 | 0.789994 | 0.64945 | 0.960951 | 0.01835 |
| ACTB | 1.378478 | 1.112325 | 1.708315 | 0.003362 |
| LRRC8D | 0.860994 | 0.745888 | 0.993864 | 0.040951 |
| CXADR | 0.874655 | 0.792088 | 0.965828 | 0.008115 |
| PLEKHH3 | 0.695225 | 0.55854 | 0.86536 | 0.001135 |
| SERPINI2 | 0.652481 | 0.44222 | 0.962715 | 0.031443 |
| VSIG4 | 1.134556 | 1.045978 | 1.230636 | 0.002336 |
| PNMA1 | 1.362288 | 1.171819 | 1.583716 | 5.74E−05 |
| ATP10B | 0.814037 | 0.735359 | 0.901133 | 7.27E−05 |
| PSEN2 | 1.411232 | 1.180407 | 1.687193 | 0.000157 |
| DSP | 0.874386 | 0.80356 | 0.951456 | 0.001842 |
| TAF7 | 1.242086 | 1.011634 | 1.525036 | 0.038412 |
| GCHFR | 0.844993 | 0.758276 | 0.941628 | 0.002299 |
| MRPL22 | 1.330421 | 1.050505 | 1.684923 | 0.017848 |
| PELI1 | 0.848755 | 0.743583 | 0.968803 | 0.015119 |
| BTG3 | 0.75541 | 0.649228 | 0.878957 | 0.000284 |
| CALML3 | 0.866027 | 0.807416 | 0.928892 | 5.75E−05 |
| GRB7 | 0.838471 | 0.740252 | 0.949722 | 0.00558 |
| UFM1 | 1.568464 | 1.268364 | 1.939569 | 3.27E−05 |
| YKT6 | 1.326238 | 1.11229 | 1.581337 | 0.001657 |
| MS4A4A | 1.104977 | 1.00836 | 1.210852 | 0.032493 |
| FCF1 | 1.394169 | 1.144442 | 1.698388 | 0.000968 |
| CYP3A5 | 0.903708 | 0.825195 | 0.989692 | 0.029006 |
| POF1B | 0.871354 | 0.815019 | 0.931582 | 5.39E−05 |
| MAP4K4 | 1.357555 | 1.137793 | 1.619764 | 0.000692 |
| DCUN1D4 | 1.381508 | 1.122628 | 1.700086 | 0.002269 |
| HOOK1 | 0.800381 | 0.717044 | 0.893403 | 7.21E−05 |
| INPP4B | 1.264711 | 1.124329 | 1.42262 | 9.15E−05 |
| ITGB3 | 1.342908 | 1.110412 | 1.624085 | 0.002368 |
| TOR1AIP2 | 1.46059 | 1.119275 | 1.905985 | 0.005275 |
| ELK3 | 1.243723 | 1.060664 | 1.458377 | 0.007253 |
| JMJD6 | 1.839798 | 1.385036 | 2.443876 | 2.57E−05 |
| ANGEL1 | 1.462939 | 1.179326 | 1.814758 | 0.00054 |
| POLR1E | 1.2169 | 1.011817 | 1.46355 | 0.037095 |
| CD177 | 0.905363 | 0.844974 | 0.970068 | 0.004761 |
| ADAMTS6 | 1.280755 | 1.054227 | 1.55596 | 0.012713 |
| PLA2G3 | 0.876249 | 0.811239 | 0.946468 | 0.000783 |
| KRT24 | 0.925329 | 0.883956 | 0.968638 | 0.000883 |
| TGFBR3 | 0.869409 | 0.792133 | 0.954223 | 0.003213 |
| PTPRC | 0.905981 | 0.82642 | 0.993201 | 0.035253 |
| ZNF787 | 1.259758 | 1.001004 | 1.5854 | 0.049009 |
| TFPI2 | 1.13683 | 1.07384 | 1.203514 | 1.04E−05 |
| FHOD1 | 1.318062 | 1.117717 | 1.554317 | 0.001027 |
| HIBCH | 0.791832 | 0.668322 | 0.938169 | 0.006983 |
| RIPK2 | 1.30418 | 1.095725 | 1.552294 | 0.002801 |
| CD28 | 0.793107 | 0.670969 | 0.937478 | 0.006595 |
| IL10RA | 0.895269 | 0.809 | 0.990737 | 0.032356 |
| PRPSAP1 | 0.7911 | 0.658708 | 0.950102 | 0.012151 |
| HK3 | 1.148428 | 1.029546 | 1.281036 | 0.013056 |
| TGFBR2 | 1.157698 | 1.023999 | 1.308853 | 0.01935 |
| ARMC1 | 1.265347 | 1.038826 | 1.541263 | 0.019364 |
| EYA2 | 0.93329 | 0.874759 | 0.995737 | 0.036686 |
| DSC2 | 0.895156 | 0.830036 | 0.965384 | 0.004051 |
| TMOD1 | 1.0859 | 1.008653 | 1.169063 | 0.028612 |
| CEACAM6 | 0.925723 | 0.881357 | 0.972323 | 0.002069 |
| SCNN1B | 0.848823 | 0.785319 | 0.917463 | 3.61E−05 |
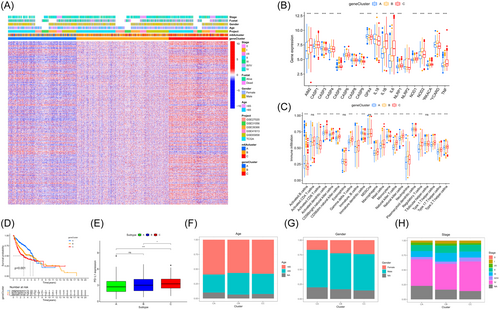
According to survival analysis, patients with gene cluster C exhibited a more favorable prognosis, whereas those with gene cluster A showed an unfavorable prognosis for OS (log rank test, p < 0.001; Figure 5D). Furthermore, it was noted that individuals with a progressed clinical phase were characterized by the gene Cluster B subgroup, while those exhibiting decreased PD-L1 expression were predominantly found in gene Cluster B and C subgroups (Figure 5E). The results also indicated that stratifications exhibited distinct clinicopathologic characteristics and were identified as gene Clusters A, B, and C (Figure 5F,G,I). The regulatory genes showed notable variations among the three subgroups of PRGs signature, aligning with the anticipated outcomes of the pyroptosis modification pattern.
3.5 Establishment of the PPI index and exploration of its clinical relevance
While our results validate the involvement of pyroptosis in prognosis and regulation of immune invasion, it is important to note that these analyses were solely conducted on patient populations and did not provide an accurate prediction of the alteration of pyroptosis in an individual tumor. Hence, an indicator of future outcomes score was formulated and designated as the PPI index relying on the PCA algorithm. The meta-GEO cohort patients were divided into two categories based on their PPI scores, using the most accurate threshold values. Figure 6A illustrates the allocation of patients among the three gene clusters. Figure 6B also assessed the correlation between established immune signatures and the PPI score to provide a clearer depiction of the PRG signature's characteristics. In Figure 6C, pyroptosis Cluster A exhibited a greater PPI compared to Cluster C and Cluster B, while in Figure 6D, gene Cluster A displayed a higher PPI than Cluster C and Cluster B. Spearman's correlation analysis was utilized to investigate the association between identified biological traits and PPI scores. The scatter diagram indicated a significant positive correlation between PPI scores and cell cycle regulators (Figure 6E) as well as the WNT pathway (Figure 6F) while displaying a negative correlation with mismatch repair (Figure 6G) and angiogenesis (Figure 6H).
Following the assessment of the predictive significance of PPI scores, the meta-GEO cohort was examined to analyze immune activity and tolerance conditions. In Figure 6I, the Wilcoxon test indicated that the high PPI group exhibited significant overexpression of the majority of genes related to immune checkpoints and immune activity. Gene set enrichment analysis (GSEA) revealed that GLYCEROPHOSPHOLIPID_METABOLISM, FATTY_ACID_METABOLISM, and the ERBB_SIGNALING_PATHWAY were significantly enriched in the higher PPI score group, whereas ECM_RECEPTOR_INTERACTION, the JAK_STAT_SIGNALING_PATHWAY, TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY, NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY, and the B_CELL_RECEPTOR_SIGNALING_PATHWAY were enriched in the lower PPI group (Figure 6J).
According to the Kaplan–Meier plotter, patients in the lower PPI score group exhibited significantly reduced OS rates compared to those in the higher PPI score group in the meta-GEO cohort (log rank test, p < 0.05; Figure 7A), as well as in GSE31056 (log rank test, p < 0.05; Figure 7B), GSE41613 (log rank test, p < 0.05; Figure 7C), GSE65858 (log rank test, p < 0.05; Figure 7D), and TCGA HNSC (log rank test, p < 0.05; Figure 7E) cohorts.

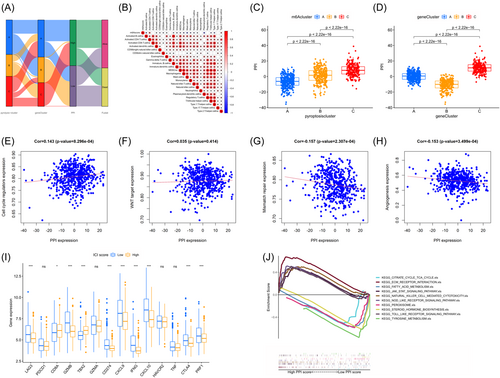
3.6 Relationship between PPI scores and somatic mutations
Increasing evidence suggests a link between genetic mutations in the tumor genome and the response to immunotherapy. Apart from the expression of PD-L1, TMB serves as a separate biomarker for the response to ICI.41, 42 The presence of genetic mutations in tumors is linked to the effectiveness of immunotherapy in various types of tumors and treatment methods, such as CPIs and cellular-based therapy.42, 43 This is a metric for genetic changes in a tumor and is a significant genomic indicator that is strongly linked to immunotherapy and predicting survival.44, 45 To understand the genetic impact of each PPI subgroup, the relationship between TMB and PPI scores was investigated, given the significant clinical significance of TMB. According to Figure 8A, individuals belonging to the high PPI score subgroup exhibited a notably greater TMB compared to those in the low PPI score subgroup (Wilcoxon test p < 0.05). Furthermore, the correlation analysis revealed a significant and positive correlation between PPI scores and TMB (Spearman coefficient R = 0.35, p < 0.05; Figure 8B). According to the findings in Figure 8C, patients with a low TMB exhibited a superior PPI compared to individuals with a high TMB (log rank test, p < 0.05). The analysis of survival, stratified by TMB status, revealed an association between predictions made using PPI scores. Both the high and low TMB subgroups showed significant differences in survival (log rank test, p < 0.05 for high TMB and high PPI scores (HH) compared to high TMB and low PPI scores (HL), and low TMB and high PPI scores (LH) compared to low TMB and low PPI scores (LL); Figure 8D). In general, these discoveries indicate that the PPI score could be unrelated to TMB and a reliable gauge of possible factors that can determine the reaction to immunotherapy.
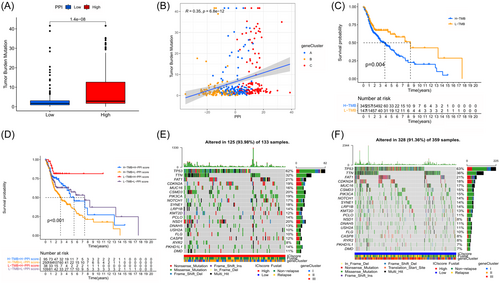
Moreover, maftools32 were utilized to analyze significantly mutated genes in HNSC samples, comparing the low and high PPI score subgroups. Using a chi-square test, Figure 8E and Table 2 helped identify the 25 driver genes that had the most frequent alterations. Significant differences were observed between the low and high PPI score subgroups for NSD1, PTPRZ1, GRM1, ELMO1, FLNA, VPS13C, CUX2, and PKD1L1 among these genes. The discovery of these findings could offer fresh perspectives on the processes of tumor PPI composition and gene mutation in immune checkpoint blockade treatment.
| Gene | H-wild | H-mutation | L-wild | L-mutation | p Value |
|---|---|---|---|---|---|
| NSD1 | 106 (79.7%) | 27 (20.3%) | 331 (92.2%) | 28 (7.8%) | 0.000179 |
| PTPRZ1 | 125 (93.98%) | 8 (6.02%) | 354 (98.61%) | 5 (1.39%) | 0.011649 |
| GRM1 | 122 (91.73%) | 11 (8.27%) | 349 (97.21%) | 10 (2.79%) | 0.015432 |
| ELMO1 | 126 (94.74%) | 7 (5.26%) | 355 (98.89%) | 4 (1.11%) | 0.015467 |
| FLNA | 123 (92.48%) | 10 (7.52%) | 350 (97.49%) | 9 (2.51%) | 0.021506 |
| VPS13C | 122 (91.73%) | 11 (8.27%) | 348 (96.94%) | 11 (3.06%) | 0.025343 |
| CUX2 | 126 (94.74%) | 7 (5.26%) | 354 (98.61%) | 5 (1.39%) | 0.032137 |
| PKD1L1 | 126 (94.74%) | 7 (5.26%) | 354 (98.61%) | 5 (1.39%) | 0.032137 |
| PCDHA6 | 126 (94.74%) | 7 (5.26%) | 354 (98.61%) | 5 (1.39%) | 0.032137 |
| ASXL3 | 131 (98.5%) | 2 (1.5%) | 334 (93.04%) | 25 (6.96%) | 0.032441 |
| PKHD1L1 | 115 (86.47%) | 18 (13.53%) | 334 (93.04%) | 25 (6.96%) | 0.034684 |
| ERICH3 | 123 (92.48%) | 10 (7.52%) | 349 (97.21%) | 10 (2.79%) | 0.035363 |
| GRIN2A | 123 (92.48%) | 10 (7.52%) | 349 (97.21%) | 10 (2.79%) | 0.035363 |
| FAT4 | 120 (90.23%) | 13 (9.77%) | 343 (95.54%) | 16 (4.46%) | 0.044565 |
| EPHA2 | 132 (99.25%) | 1 (0.75%) | 340 (94.71%) | 19 (5.29%) | 0.044637 |
3.7 PPI score for response to immunotherapy
We assessed the correlation between clinical characteristics and established PPI scores. The advanced TNM stage (Figure 9A,B) was correlated with high PPI scores. The analysis of survival, stratified by PPI scores, indicated a correlation with survival outcomes (Figure 9C,D). Furthermore, there was a positive correlation between higher PPI scores and objective response to therapy in the meta-GEO cohort, as depicted in Figure 9E,F. In the meantime, stratified survival analysis indicated a correlation between the PPI and the outcome success of the primary therapy. Among patients with partial/complete response, high PPI scores were related to worse survival, similar results were obtained among patients with stable/progressive disease (Figure 9G,H).
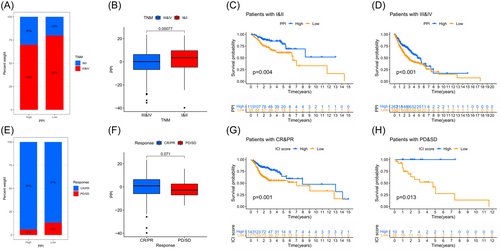
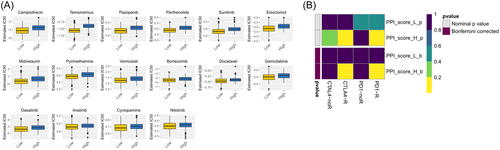
Despite not being approved as a conventional treatment for HNSC, immunological checkpoint medications were assessed using the submap algorithm to estimate the probability of immunotherapy response in the meta-GEO cohort. The findings revealed that individuals with high PPI scores were more likely to exhibit a favorable response to immunotherapy compared to those with low PPI scores (p < 2.2e−16). Furthermore, immunotherapy yielded more favorable outcomes in individuals with high PPI scores as per the TIDE algorithm (p < 0.05). The findings indicated that individuals with elevated PPI scores exhibited strong responsiveness to programmed cell death 1 (PD1) immunotherapy (p < 0.05 after Bonferroni correction) (Figure 10A). Treating patients with HNSC often involves the use of anticancer medication as a fundamental therapeutic approach. The purpose of the PPI model was to evaluate the reaction to antitumor medications in two molecular subcategories using the GDSC cell line dataset. The accuracy of the method's prediction was validated using 10-fold cross-validation, while the IC50 was utilized to estimate the sensitivity of the response. Out of the 130 categories of anticancer medication reactions, 74 drugs exhibited variances among the two PPI clusters as indicated in Table 3. The findings indicated that elevated PPI scores exhibited greater responsiveness to 68 different types of medications in comparison to lower PPI scores. Figure 10B showed that temsirolimus, camptothecin, pazopanib, parthenolide, and docetaxel exhibited significant promise in treating HNSC (p < 0.05). Specifically, camptothecin and docetaxel have been extensively utilized as primary chemotherapy in clinical settings.
| Drug | p Value |
|---|---|
| CEP.701 | 5.71E−42 |
| Temsirolimus | 3.14E−39 |
| NU.7441 | 1.28E−38 |
| AZD.2281 | 3.90E−32 |
| WO2009093972 | 6.70E−29 |
| GSK269962A | 6.09E−27 |
| Axitinib | 3.60E−24 |
| AZ628 | 1.57E−22 |
| BX.795 | 3.44E−21 |
| AZD7762 | 8.60E−21 |
| Pazopanib | 2.01E−20 |
| AZD8055 | 4.31E−20 |
| TW.37 | 6.08E−19 |
| Parthenolide | 7.90E−19 |
| IPA.3 | 1.06E−18 |
| Cytarabine | 1.04E−17 |
| GDC.0449 | 1.59E−16 |
| PLX4720 | 4.32E−16 |
| VX.702 | 4.90E−16 |
| PD.173074 | 5.16E−15 |
| NVP.BEZ235 | 1.61E−14 |
| Camptothecin | 1.75E−13 |
| Sunitinib | 2.01E−13 |
| AP.24534 | 2.51E−13 |
| Elesclomol | 6.55E−13 |
| NVP.TAE684 | 7.17E−12 |
| JNJ.26854165 | 1.26E−11 |
| AG.014699 | 2.96E−11 |
| DMOG | 5.73E−11 |
| Midostaurin | 8.20E−11 |
| Pyrimethamine | 4.23E−10 |
| LFM.A13 | 2.50E−09 |
| AZD6482 | 5.36E−09 |
| Vorinostat | 1.17E−08 |
| ABT.888 | 1.99E−08 |
| CGP.60474 | 2.71E−08 |
| Bortezomib | 1.09E−07 |
| Docetaxel | 2.20E−07 |
| SB.216763 | 2.86E−07 |
| PAC.1 | 3.07E−07 |
| AMG.706 | 3.26E−07 |
| BMS.754807 | 1.39E−06 |
| OSI.906 | 1.69E−06 |
| Gemcitabine | 1.74E−06 |
| JNK.Inhibitor.VIII | 1.79E−06 |
| CHIR.99021 | 1.80E−06 |
| Dasatinib | 3.92E−06 |
| WH.4.023 | 8.50E−06 |
| MS.275 | 3.96E−05 |
| ZM.447439 | 4.32E−05 |
| XMD8.85 | 0.000107 |
| Imatinib | 0.00011 |
| BI.D1870 | 0.000121 |
| BMS.509744 | 0.000199 |
| Z.LLNle.CHO | 0.000209 |
| CMK | 0.000213 |
| PF.02341066 | 0.000248 |
| GDC0941 | 0.00041 |
| Cyclopamine | 0.000563 |
| BAY.61.3606 | 0.001202 |
| Nilotinib | 0.001397 |
| AS601245 | 0.001475 |
| KU.55933 | 0.002407 |
| SL.0101.1 | 0.002917 |
| Vinblastine | 0.003376 |
| PHA.665752 | 0.003836 |
| SB590885 | 0.004598 |
| Bexarotene | 0.004692 |
| Cisplatin | 0.011966 |
| A.770041 | 0.016861 |
| RO.3306 | 0.017973 |
| X681640 | 0.021013 |
| Methotrexate | 0.024357 |
| Obatoclax.Mesylate | 0.029282 |
4 DISCUSSION
Pyroptosis, alternatively referred to as cellular inflammatory necrosis, is initiated by the destruction of cell membranes resulting from the functioning of the majority of CASP family genes.46 Cells undergoing pyroptosis display cellular enlargement and multiple bubble-shaped protrusions. The activation of CASP-1/4/5/11 is necessary for its dependency. The GSDMD-N protein creates minuscule openings measuring 1.1–2.4 nm across the cellular membrane.47 Pyroptosis, serving as a type of programmed cellular demise, functions as the principal mechanism against infections. In addition, pyroptosis has demonstrated significant involvement in the development of tumors.48, 49 Pyroptosis, a process linked to tumorigenesis, invasion, and metastasis, involves the presence of inflamed vesicles, gastrin, and proinflammatory cytokines as crucial elements. Furthermore, pyroptosis significantly contributes to the innate immune response against intracellular pathogens and is also implicated in the development of fatal infectious shock. Although cellular scorch death plays an important role in tumor development and antitumor processes, its specific role in HNSC has not been adequately explored. Consequently, we carried out a comprehensive investigation to assess the levels of gene expression associated with scorch death in both normal and HNSC tissues. In addition, the predictive significance of these genes was investigated, along with examining the association between scorch-induced mortality and the immune microenvironment of the tumor.
Initially, the expression levels of 33 presently recognized PRGs in HNSC and normal tissues were assessed, revealing significant differential expression in the majority of them. A meta-study was then conducted to analyze 1341 HNSC samples. The specimens were categorized into three separate subcategories, distinguished by varying immune characteristics, that were associated with diverse antitumor immune responses. An immune-inflamed phenotype was observed in Pyroptosis-C1, which was identified by the presence of immune activation and infiltration of tumor-infiltrating lymphocytes. An immune-excluded phenotype was observed in Pyroptosis-C2, which was identified by the existence of immune cells and stroma, along with the activation of EMT, TGF-β, and Wnt signaling pathways. The immune-desert phenotype was associated with an immunosuppressive TME in Pyroptosis-C3.
In the meantime, we conducted a thorough examination of the PRGs' expression and devised a technique for measuring the PPI index in HNSC. The results indicate that the PPI score serves as a reliable prognostic biomarker and predictor in evaluating the response to immunotherapy. The alteration of pyroptosis in the TME of HNSC has been documented to enhance immune suppression, which is linked to the survival and advancement of tumors.50 Nevertheless, it is important to take into account molecular subcategories when implementing pyroptosis regulation as a therapeutic approach. We hypothesized that patients with a high PPI score may experience a favorable prognosis due to the presence of an antitumor immune response, suggesting potential benefits from immunotherapy. A negative outcome was linked to the immune-cold characteristic in the group with a low PPI score. Our previous findings are consistent with the high sensitivity of immunotherapy to high PPI scores, as predicted by TIDE and submap. While the precise manner in which pyroptosis controls the growth and proliferation of tumor cells is still unknown, the correlation we witnessed between PPI and key characteristics of cancer has the potential to enhance our comprehension of the function of pyroptosis. In the majority of cancers, GSEA analysis revealed a strong correlation between the extent of pyroptosis and tumor-associated characteristics. In cancer, pyroptosis genes can have both oncogenic and tumor-suppressive functions, while protein–protein interaction serves as a safeguarding element in HNSC. Hence, the integration of the PPI subgroup and immune-related gene expression profiles may offer a new strategy for creating personalized treatment plans tailored to each patient.
To summarize, the results of this research not only enhanced our understanding of the process involved in pyroptosis in the HNSC TME but also introduced a new possible prognostic biomarker, the PPI score, to assist in precision immunotherapy.
AUTHOR CONTRIBUTIONS
Yan Long: Supervision; visualization; writing—original draft. Yadong Wu: Conceptualization; software; supervision. Juxiang Peng: Conceptualization; investigation; methodology. Jukun Song: Conceptualization; supervision; writing—original draft; writing—review and editing. Na Li: Visualization; writing—review and editing.
ACKNOWLEDGMENTS
The work was supported by the Guizhou Provincial People's Hospital Youth Fund GZSYQN[2016]09.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
Open Research
DATA AVAILABILITY STATEMENT
Data availability could be obtained from TCGA and GEO websites.