HBV RNA Profiles in Patients With Chronic Hepatitis B Under Different Disease Phases and Antiviral Therapy
Abstract
Background and Aims
Large-scale comprehensive studies on HBV RNA in chronic hepatitis B are lacking. We aimed to study the HBV RNA profile and its correlation with other viral markers in patients with chronic hepatitis B who are treatment-naïve and patients receiving nucleos(t)ide analogues (NA).
Approach and Results
Biomarkers, including HBV RNA and hepatitis B core-related antigen (HBcrAg), were measured in 388 patients. Of these, 246 were treatment-naïve and were categorized into HBeAg-positive chronic infection (n = 41), HBeAg-positive chronic hepatitis (n = 81), HBeAg-negative chronic infection (n = 39), HBeAg-negative chronic hepatitis (n = 66), and HBsAg seroclearance (n = 19). These biomarkers were also measured in 142 patients who were NA-treated receiving tenofovir or entecavir at baseline, week 48, and week 96. The pattern of serum HBV RNA levels mirrored HBV DNA (1-2 logs higher than HBV RNA) and HBcrAg in patients who were treatment-naïve. HBV RNA correlated best with HBcrAg (r = 0.84) and to a lesser extent with HBV DNA (r = 0.737) (both P < 0.001). In patients with HBsAg seroclearance, 15.8% and 15.8% had detectable serum HBV RNA and HBcrAg, respectively. NA treatment reduced serum HBV RNA by 1.46 logs and 1.77 logs at weeks 48 and 96, respectively. At week 96 of NA therapy, only 19.1% patients who were tenofovir-treated and 25.7% patients who were entecavir-treated had unquantifiable HBV RNA (P > 0.05). In patients who were treated and had undetectable HBV DNA, 77.5% and 30% had quantifiable HBV RNA and HBcrAg, respectively.
Conclusions
HBV RNA showed distinct and corresponding profiles in patients with HBV in different disease phases. HBV RNA and HBcrAg could be used to monitor residual transcriptional activities in patients with HBsAg seroclearance. NA led to reduction of serum HBV RNA. Monitoring of viral activities can still be achieved in patients with undetectable HBV DNA by serum HBV RNA.
Abbreviations
-
- ALT
-
- alanine aminotransferase
-
- anti-HBs
-
- serum antibody to HBsAg
-
- AUROC
-
- area under the receiver operating characteristics curve
-
- cccDNA
-
- covalently closed circular DNA
-
- CHB
-
- chronic hepatitis B
-
- CpAM
-
- core protein allosteric modulator
-
- ETV
-
- entecavir
-
- HBcrAg
-
- hepatitis B core-related antigen
-
- IQR
-
- interquartile range
-
- LLOD
-
- lower limit of detection
-
- LLOQ
-
- lower limit of quantification
-
- NA
-
- nucleos(t)ide analogue
-
- pgRNA
-
- pregenomic RNA
-
- qHBeAg
-
- quantitative HBeAg
-
- qHBsAg
-
- quantitative HBsAg
-
- TAF
-
- tenofovir alafenamide
-
- TDF
-
- tenofovir disoproxil fumarate
Chronic hepatitis B (CHB) infection affects 292 million individuals globally and can lead to considerable morbidity and mortality in 15%-40% of the infected persons.(1, 2) Remarkable advances in understanding the life cycle of HBV has led to the identification of serum HBV biomarkers, which reflect the status of HBV in the human body. The most well-known and widely utilized biomarker is serum HBV DNA, which is produced from reverse transcription of the pregenomic RNA (pgRNA), a process that takes place inside the nucleocapsid. Serum HBV DNA quantification has been used to assess disease phase, predict liver-related complications, and determine the need for antiviral therapy. With the availability of three potent first-line nucleos(t)ide analogues (NAs), serum HBV DNA rapidly becomes undetectable after commencing therapy, and in most cases, it cannot be used further as a tool to predict disease course.(3) Moreover, NAs only inhibit one of the many steps of the viral life cycle, whereas ongoing transcription, translation, and further downstream events remain unaffected. Hence, there is a need for alternative biomarkers to assess for ongoing viral activity that is not reflected by HBV DNA, especially when it is undetectable.
Ideally, measuring the intrahepatic covalently closed circular DNA (cccDNA) would be the most direct way to assess replication-competent viral reservoir. However, there are limitations, including the need for liver biopsy, together with the lack of a standardized method to quantify cccDNA. Although there has been a recent increase in interest in the use of serum quantitative HBsAg (qHBsAg), this might not be a good alternative, as it is produced not only from cccDNA but also from integrated DNA.(4) Moreover, it is hard to assess the effect of NA on the virus because the level of qHBsAg remains almost static in patients who were NA-treated with CHB for a long duration.(5) Another potential biomarker is serum HBV RNA, which is detectable in the serum as encapsidated virion-containing pgRNA.(6) Its profile in the natural history of CHB has been described in small-scale studies involving Chinese patients.(6, 7) Recently, performance of HBV RNA assays has been improved to approach the standards of the World Health Organization requirements and is 6-fold more sensitive over in-house assays used in studies.(8) A third potential biomarker is hepatitis B core-related antigen (HBcrAg), which consists of 3 related proteins sharing an identical 149 amino acid sequences: HBeAg, hepatitis B core antigen, and a truncated 22 kDa precore protein.(9) HBcrAg has been shown to correlate with disease activity in both patients who were untreated and treated and is a potential surrogate marker for cccDNA.(10-12) Recent studies have also reported the longitudinal profile of both HBV RNA and HBcrAg in patients who were NA-treated and mostly non-Chinese, and it has been shown to correlate with liver histopathology(13) and predict viral rebound after NA cessation.(6, 14-16) Although both markers are surrogates for intrahepatic cccDNA, their coherence with each other has been controversial.(13, 15, 17)
We aimed to examine the profiles of HBV RNA and HBcrAg in a large-scale cohort of treatment-naïve and NA-treated Chinese patients with CHB.
Patients and Methods
Patients
The present study recruited patients with CHB aged ≥18 from the Department of Medicine, Queen Mary Hospital, the University of Hong Kong. All patients were seropositive for HBsAg for at least 6 months. Patients would not be enrolled for the study if they were pregnant or had concomitant hepatitis C or D virus, human immunodeficiency virus infection, Wilson’s disease, autoimmune hepatitis, primary biliary cholangitis, nonalcoholic steatohepatitis, significant alcohol intake (20 g/day for female patients or 30 g/day for male patients), or history of HCC. Patients were followed up every 3-6 months for clinical assessment, blood test monitoring, and ultrasonography.
For the patients who were treatment-naïve, they were enrolled from March 2010 to July 2013, and the results of serum HBsAg and HBcrAg levels were published.(18) Briefly, among the patients included in the original study, 246 patients with available serum samples were recruited. These include 122 patients who were HBeAg positive, 105 patients who were HBeAg negative, and 19 patients with HBsAg seroclearance. For patients who were HBeAg positive, they were classified as either HBeAg-positive chronic hepatitis (n = 81) if serum alanine aminotransferase (ALT) was elevated or HBeAg-positive chronic infection (n = 41) if serum ALT was normal. For patients who were HBeAg-negative, they were classified as either HBeAg-negative chronic hepatitis (n = 66) if serum ALT was elevated or HBeAg-negative chronic infection (n = 39) if serum ALT was normal.(3) The upper limit of normal for ALT was set as 40 U/L according to international guidelines for defining the disease phase.(3) Nineteen patients with HBsAg seroclearance, defined as those with previously documented seropositivity for HBsAg who subsequently underwent sustained HBsAg seroclearance for ≥6 months with or without development of antibodies to HBsAg, were recruited.
The patients who were NA-treated had previously had serum HBcrAg measurement performed, and the results were published.(5, 19) Those with available serum samples, either HBeAg-positive (n = 86) or HBeAg-negative (n = 56), were enrolled when they were started on one of the three first-line oral NAs: entecavir (ETV) 0.5 mg daily, tenofovir disoproxil fumarate (TDF) 300 mg daily, or tenofovir alafenamide (TAF) 25 mg daily. Only patients who were not previously exposed to other NAs or pegylated interferon treatment were recruited. Indications for initiation of oral NA were different between ETV and tenofovir because of differences in local practice in various periods. For ETV, indications included (1) patients who were HBeAg-positive with elevated ALT (defined as >50 U/L at that time) with HBV DNA >20,000 IU/mL, (2) patients who were HBeAg-negative with elevated ALT with HBV DNA >2,000 IU/mL, or (3) patients with cirrhosis with detectable serum HBV DNA. For tenofovir, indications included patients with ALT > 60 U/L for male patients or >38 U/L for female patients together with HBV DNA > 20,000 IU/mL, regardless of HBeAg status. HBV biomarkers were measured on the day of initiation of NA, at week 48 of NA therapy, and at week 96 of NA therapy.
Informed consent in writing was obtained from each patient and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the institutional review committee. The present study was approved by the Institutional Review Board of the University of Hong Kong and the Hospital Authority Hong Kong West Cluster, Hong Kong.
Serum HBV RNA Concentrations
Serum HBV RNA was measured using the m2000 system, which detects amplicons in the HBV X and core targets (Abbott Diagnostics, Abbott Park, IL) with a lower limit of quantification (LLOQ) of 44.6 U/mL (i.e., 1.65 log10 U/mL). The assay linearity was maintained on clinical samples with high concentration up to 7.62 log10 U/mL. This assay was designed for duplexed detection of two targets in the 5′ and 3′ ends of the full-length pgRNA. The X and core target detection has been shown to be comparable (strong linear concordance R2 = 0.96) across the quantitative range of the assay, supporting the idea that the serum RNA detected is primarily full-length pgRNA.(8, 20) Those values below the LLOQ (nonquantifiable) but still detectable are also recorded and arbitrarily defined as 1.65 log10 U/mL for statistical analysis. Undetectable results were regarded as negative (0 log10 U/mL). Every unit of HBV RNA is equivalent to 1 IU of HBV DNA.(8)
Serum HBV DNA Concentrations
Serum HBV DNA levels were measured by a quantitative real-time PCR assay [Cobas Taqman assay (Roche Diagnostics, Branchbrug, NJ)] with a lower limit of detection (LLOD) of 10 IU/mL. The values of HBV DNA were log transformed and expressed in log10 IU/mL.
Serum HBsAg Quantification
Serum qHBsAg levels were measured using the Cobas Taqman assay (Roche Diagnostics, Gmbh, Mannheim, Germany) with an LLOD of 50 mIU/mL (i.e., 1.7 log10 mIU/mL).
Serum HBcrAg Quantification
Serum HBcrAg were measured by the Lumipulse G HBcrAg chemiluminescence Enzyme Immunoassay (Fujirebio, Tokyo, Japan). The linear range of the HBcrAg assay ranged from 1,000 to 10,000,000 unit per milliliter (U/mL), while the LLOD was 100 U/mL. Values above the LLOD (i.e., 100-1,000 U/mL) are also presented for statistical analysis. This same analysis has been verified and used in our studies.(18, 21) The values of HBcrAg were log transformed and were expressed in log10 U/mL.
Serum Antibody to HBsAg
In patients with HBsAg seroclearance, serum antibody to HBsAg (anti-HBs) was measured using Elecsys Anti-HBs II (Cobas e411 analyzer, Roche Diagnostics, GmbH, Mannheim, Germany) with an LLOQ of 10 mIU/mL.
Serum Quantitative HBeAg
Serum quantitative HBeAg (qHBeAg) levels were measured in patients who were HBeAg-positive by an ARCHITECT i2000SR analyzer with a linear range of 0.09-700 Paul-Ehrlich Institute (PEI) U/mL. Samples above 700 PEI U/mL will be diluted. The results were log transformed and were expressed in log10 PEI U/mL.
HBV Genotyping
HBV genotyping was performed using the INNO-LiPA HBV genotyping assay (Innogenetics, Ghent, Belgium).
Statistical Analyses
Continuous variables were expressed in median (interquartile range [IQR]). Comparison of continuous variables was performed using Mann-Whitney U test and Kruskal-Wallis test. Categorical variables were compared using Pearson's χ2 test or Fisher's exact test, as appropriate. Pearson correlation coefficients were analyzed to assess the relationship between serum biomarkers. Receiver operating characteristic (ROC) curves were drawn to determine the performance characteristics of each biomarker for determining the disease phase. The biomarker with the highest area under the ROC (AUROC) value was further examined to identify the Youden's index by the equation (sensitivity + specificity - 1) in determination of the cutoff level. Subgroup analysis would be performed on those with HBsAg seroclearance, and those receiving different first-line oral NAs. All statistical analyses were performed using SPSS version 25 (SPSS, Chicago, IL). A two-sided P value of <0.05 was considered statistically significant.
Results
Treatment-Naïve Patients
Baseline Characteristics
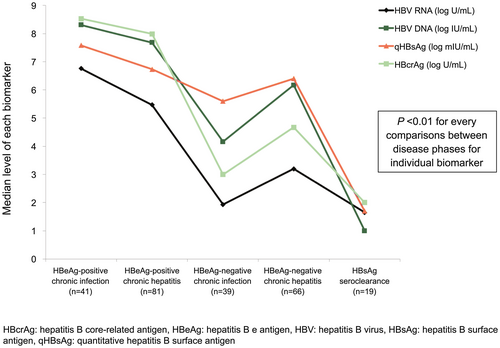
A detailed summary of the baseline characteristics for the 246 patients is shown in Table 1. The predominant genotypes were B (121; 49.2%) and C (125; 50.8%). All four viral biomarkers, including HBV DNA, HBV RNA, qHBsAg, and HBcrAg, were detected in 100% of patients who were HBeAg-positive. For HBeAg-negative chronic hepatitis, HBV RNA, DNA, qHBsAg, and HBcrAg were still detected in 97%, 100%, 100%, and 87.9%, respectively. For HBeAg-negative chronic infection, while both DNA and qHBsAg were still detected in 100%, HBV RNA and HBcrAg were detected in much lower proportion of patients (79.5% and 69.2%, respectively). Serum levels of all four biomarkers were highest in patients with HBeAg-positive chronic infection followed by HBeAg-positive chronic hepatitis, HBeAg-negative chronic hepatitis, and HBeAg-negative chronic infection and were lowest in patients with HBsAg seroclearance (Fig. 1). The median level of HBV DNA was shown to be consistently higher than HBV RNA by 1-2 log10 in both HBeAg-positive and HBeAg-negative phases. No significant differences in HBV RNA levels were observed between genotype B and C patients (P = 0.797). However, when subgroup analysis was performed for different disease phases, genotype B had higher median HBV RNA compared with genotype C in HBeAg-positive chronic hepatitis (6.22 vs. 4.71; P = 0.004) but not in other disease phases. The role of serum HBV biomarkers in differentiating chronic hepatitis versus chronic infection in HBeAg-positive and HBeAg-negative groups by AUROC analysis is shown in the Supporting Figs. S1 and S2.
| HBeAg-Positive Chronic Infection (n = 41) | HBeAg-Positive Chronic Hepatitis (n = 81) | HBeAg-Negative Chronic Infection (n = 39) | HBeAg-Negative Chronic Hepatitis (n = 66) | HBsAg Seroclearance (n = 19) | |
|---|---|---|---|---|---|
| Male | 29 (55.8%) | 67 (63.8%) | 65 (68.4%) | 65 (67%) | 41 (74.5%) |
| Age | 30.4 (28.4-37.9) | 35.1 (28.1-44.6) | 55.9 (46.7-63.3) | 50.4 (44.9-56.3) | 54.9 (49.2-60.7) |
| Albumin | 44 (42-45) | 42 (38-44) | 43 (42-45) | 42 (41-44) | 44 (41-45) |
| Bilirubin | 9 (8-13) | 13 (11-19) | 13 (8-15) | 13 (9-17) | 11 (9-15) |
| ALT | 23 (19-31) | 131 (73-297) | 32 (21-36) | 81 (60-156) | 29 (13-34) |
| Detectable (%) | |||||
| RNA | 41 (100%) | 81 (100%) | 31 (79.5%) | 64 (97%) | 3 (15.8%) |
| DNA | 41 (100%) | 81 (100%) | 39 (100%) | 66 (100%) | 0 (0%) |
| qHBsAg | 41 (100%) | 81 (100%) | 39 (100%) | 66 (100%) | 0 (0%) |
| HBcrAg | 41 (100%) | 81 (100%) | 27 (69.2%) | 58 (87.9%) | 3 (15.8%) |
| HBV RNA (log10 U/mL) | 6.76 (6.54-7.10) | 5.47 (3.97-6.74) | 1.93 (1.65-3.46) | 3.20 (1.65- 4.12) | UD [UD-1.84] |
| HBV DNA (log10 IU/mL) | 8.31 (7.92-8.46) | 7.68 (7.10-8.11) | 4.16 (2.88-5.19) | 6.17 (5.21-7.07) | UD |
| qHBsAg (log10 mIU/mL) | 7.58 (6.93-7.87) | 6.73 (6.19-7.49) | 5.60 (4.76-6.31) | 6.40 (6.04-6.70) | UD |
| HBcrAg (log10 U/mL) | 8.53 (8.31-8.72) | 7.99 (7.01-8.54) | 3.00 (UD-4.15) | 4.67 (3.68-5.68) | UD [UD-3.34] |
| qHBeAg (log10 PEI U/mL) | 2.91 (2.62-3.19) | 1.87 (0.47-2.79) | — | — | — |
Note:
- Median value ( ) refers to interquartile range; [ ] refers to range.
- Abbreviation: UD, undetectable.
Transcriptional Efficiency
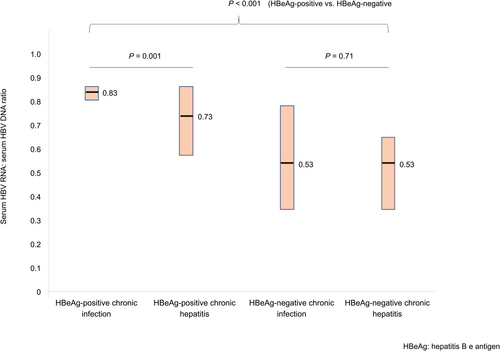
The ratio of serum HBV RNA to HBV DNA was calculated to assess the reverse transcriptional efficiency of pgRNA.(7) The median HBV RNA to DNA ratio was 0.83 (IQR: 0.80-0.85), 0.73 (IQR: 0.58-0.85), 0.53 (IQR: 0.35-0.78), and 0.53 (IQR: 0.36-0.65) for HBeAg-positive chronic infection, HBeAg-positive chronic hepatitis, HBeAg-negative chronic infection, and HBeAg-negative chronic hepatitis, respectively (Fig. 2). The RNA to DNA ratio was significantly higher in patients who were HBeAg-positive than patients who were HBeAg-negative (0.79 vs. 0.53; P < 0.001). Although the ratio was significantly lower in chronic hepatitis phase compared with chronic infection phase for patients who were HBeAg-positive (0.73 vs. 0.83; P = 0.001), the ratio was not significantly different for patients who were HBeAg-negative (0.53 vs. 0.53; P = 0.71) (Fig. 2).
Profile of Viral Biomarkers in Patients With HBsAg Seroclearance
Nineteen patients (73.7% male) with HBsAg seroclearance were recruited. The samples were tested for HBV biomarkers at a median interval of 1.6 (IQR: 0.5-3.6) years after HBsAg seroclearance. Twelve patients (63.1%) developed anti-HBs at median level of 40 mIU/mL (range: 10-1,000 mIU/mL). None of the 19 patients had detectable serum qHBsAg and HBV DNA. Three (15.8%) had detectable HBV RNA (1 patient had HBV RNA level of 1.84 log10 U/mL, and 2 patients had detectable but unquantifiable HBV RNA). Among these 3 patients with detectable serum RNA, only 1 patient developed anti-HBs (44 mIU/mL). On the other hand, 3 out of 19 patients (15.8%) had detectable serum HBcrAg (2.3, 3.0, and 3.34 log10 U/mL), and among these 3 patients, 2 of them also had detectable serum RNA.
Correlation Between Viral Biomarkers
The correlation coefficients between the viral biomarkers are shown in Table 2. Serum HBV RNA correlates best with serum HBcrAg (r = 0.84; P < 0.001) and to a lesser extent with serum HBV DNA (r = 0.737; P < 0.001). The correlation plots for HBV RNA versus HBV DNA are shown in Supporting Figs. S3 and S5.
| HBV DNA | qHBsAg | HBcrAg | qHBeAg | |
|---|---|---|---|---|
| All Treatment-Naïve (n = 246) | ||||
| HBV RNA | 0.737 (P < 0.001) | 0.594 (P < 0.001) | 0.840 (P < 0.001) | |
| HBV DNA | — | 0.825 (P < 0.001) | 0.840 (P < 0.001) | |
| qHBsAg | — | — | 0.669 (P < 0.001) | |
| HBeAg-Positive (n = 122) | ||||
| HBV RNA | 0.419 (P < 0.001) | 0.299 (P < 0.001) | 0.609 (P < 0.001) | 0.679 (P < 0.001) |
| HBV DNA | — | 0.474 (P < 0.001) | 0.532 (P < 0.001) | 0.346 (P < 0.001) |
| qHBsAg | — | — | 0.35 (P < 0.001) | 0.202 (r = 0.025) |
| HBcrAg | — | — | — | 0.686 (P < 0.001) |
| HBeAg-Negative (n = 105) | ||||
| HBV RNA | 0.482 (P < 0.001) | 0.291 (P = 0.007) | 0.566 (P < 0.001) | |
| HBV DNA | — | 0.546 (P < 0.001) | 0.656 (P < 0.001) | |
| qHBsAg | — | — | 0.436 (P < 0.001) | |
When subgroup analysis is performed separately for patients who are HBeAg-positive and patients who are HBeAg-negative, serum HBV RNA still showed good linear correlation with serum HBcrAg (r = 0.609 for HBeAg-positive and r = 0.566 for HBeAg-negative; both P < 0.001). Serum HBV RNA also correlated well with qHBeAg (r = 0.679; P < 0.001) in patients who were HBeAg-positive. The correlation is weak between qHBsAg and HBV RNA (r = 0.29-0.30) and modest between HBV DNA and HBV RNA (r = 0.42-0.48).
Serum ALT correlated weakly with HBV DNA (r = 0.249; P < 0.001), qHBsAg (r = 0.218; P < 0.001), and HBcrAg (r = 0.268; P < 0.001) but not with HBV RNA (r = 0.032; P = 0.616).
Patients Who Were NA-Treated
Baseline Characteristics
Among 142 patients who received first-line oral NA, 37 received TAF, 31 received TDF, and 74 received ETV. The median age is 45.5 (IQR 33.8-56) years old, 52.1% male, with median ALT of 76 U/L (IQR 47-137). The baseline serum HBV RNA, DNA, and HBcrAg were 4.92 (IQR: 3.35-6.84) log10 U/mL, 7.15 (IQR: 5.41-8.04) log10 IU/mL, and 4.66 (2.93-5.49) log10 U/mL, respectively. The summary of the baseline characteristics for the 142 patients according to HBeAg status and NA are shown in Tables 3 and 4.
| All Patients (n = 142) | HBeAg-Positive Chronic Hepatitis (n = 86) | HBeAg-Negative Chronic Hepatitis (n = 56) | |
|---|---|---|---|
| Male | 74 (52.1%) | 53 (61.6%) | 21 (37.5%) |
| Age | 45.5 (33.8-56) | 36.5 (30.8-51) | 56 (46.8-63) |
| ALT | 76 (47-137) | 84 (57-172) | 59 (41-106) |
| Detectable (%) | |||
| RNA | 138 (97.2%) | 86 (100%) | 51 (92.7%) |
| DNA | 142 (100%) | 86 (100%) | 56 (100%) |
| HBcrAg | 142 (100%) | 86 (100%) | 56 (100%) |
| HBV RNA (log10 U/mL) | 4.92 (3.35-6.84) | 6.62 (4.92-7.19) | 3.27 (2.20-4.21) |
| HBV DNA (log10 IU/mL) | 7.15 (5.41-8.04) | 7.90 (6.95-8.04) | 5.28 (4.15-6.01) |
| qHBsAg (log10 mIU/mL) | 3.39 (2.82-4.16) | 3.94 (3.31-4.42) | 3.08 (2.59-3.35) |
| HBcrAg (log10 U/mL) | 4.66 (2.93-5.49) | 5.23 (4.36-5.63) | 2.03 (1.36-2.7) |
| qHBeAg (log10 PEI U/mL) | — | 2.50 (1.05-3.09) | — |
Note:
- Median value (IQR).
| All Patients (n = 142) | TAF (n = 37) | TDF (n = 31) | Tenofovir (TAF + TDF; n = 68) | ETV (n = 74) | P Value (Between Tenofovir vs. ETV) | |
|---|---|---|---|---|---|---|
| Male | 74 (52.1%) | 18 (48.6%) | 14 (45.2%) | 32 (47.1%) | 42 (56.8%) | 0.313 |
| Age | 45.5 (33.8-56) | 42 (32.5-55.5) | 46 (33-60) | 45.5 (33-56) | 45.5 (34-56) | 0.943 |
| ALT | 76 (47-137) | 73 (56-123) | 80 (51-116) | 74 (55-118) | 79 (37-186) | 0.945 |
| HBeAg-positive | 86 (60.6%) | 25 (67.6%) | 18 (58.1%) | 43 (63.2%) | 43 (58.1%) | 0.607 |
| HBV RNA (log10 U/mL) | 4.92 (3.35-6.84) | 5.6 (3.66-7.05) | 5.31 (3.75-7) | 5.41 (3.76-7) | 4.5 (2.9-6.64) | 0.09 |
| HBV DNA (log10 IU/mL) | 7.15 (5.41-8.04) | 7.46 (5.67-8.04) | 7.82 (5.92-8.04) | 7.64 (5.79-8.04) | 6.56 (5.16-7.90) | 0.034 |
Note:
- Median value (IQR).
Dynamic Changes of HBV Biomarkers
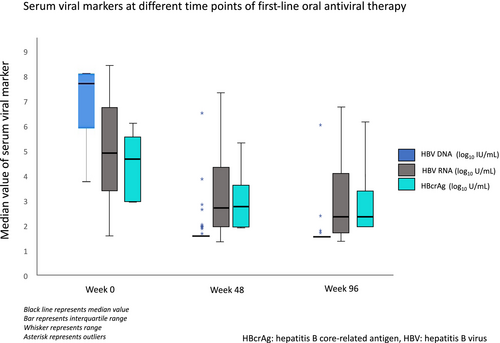
At week 48 of antiviral therapy, median serum HBV DNA was undetectable, but serum HBV RNA and HBcrAg were still detectable at median level of 2.85 (IQR: 2.07-4.46) log10 U/mL and 2.97 (IQR: 2.0-3.78) log10 U/mL, respectively. At week 96 of antiviral therapy, median serum HBV DNA remained undetectable, whereas serum HBV RNA was still detectable at a median level of 2.47 (1.77-4.15) log10 U/mL. Serum HBcrAg was detected at a median level of 2.23 log10 U/mL (Fig. 3). The median reduction of HBV RNA and HBcrAg levels from baseline were 1.46 (IQR: 0.32-2.63) log10 U/mL and 1.37 (IQR: 0.53-1.95) log10 U/mL at week 48 and 1.77 (IQR: 0.50-3.10) log10 U/mL and 1.81 (IQR: 0.85-2.76) log10 U/mL at week 96, respectively. Among all patients who were NA-treated with undetectable HBV DNA, 87.3% and 48.3% had quantifiable HBV RNA and HBcrAg, respectively, at week 48 and 77.5% and 30%, respectively, at week 96. Serum qHBsAg slowly decreased from week 0 to week 48 and week 96 at 3.39, 3.24, and 3.20 log10 mIU/mL, respectively (P < 0.001).
Correlation Between Viral Biomarkers
The correlation coefficients between the viral biomarkers are shown in Table 5. HBV RNA showed good linear correlation with HBV DNA, HBcrAg, and qHBsAg before NA treatment (r = 0.749, 0.892, and 0.692, respectively; all P < 0.001). The correlation between HBV RNA and HBcrAg was still preserved on 48 and 96 weeks of NA therapy (r = 0.806 and 0.795, respectively; both P < 0.001) but weakened between HBV RNA and HBV DNA at week 48 (r = 0.414; P < 0.001) and week 96 (r = 0.152; P = 0.084) and also between HBV RNA and qHBsAg (r = 0.374 at week 48 and r = 0.293 at week 96; both P < 0.001). The linear correlation between serum HBV RNA and HBcrAg and qHBsAg was stronger in patients who were HBeAg-positive than patients who were HBeAg-negative (Supporting Tables S1 and S2). In patients who were HBeAg-positive, qHBeAg correlated strongly with HBcrAg (at baseline: r = 0.805, week 48: r = 0.915, and week 96: r = 0.808; all P < 0.001) and with HBV RNA (at baseline: r = 0.725, week 48: r = 0.625, and week 96: r = 0.57; all P < 0.001) (Supporting Table S1).
| HBV DNA | HBcrAg | qHBsAg | |
|---|---|---|---|
| All Patients Who Were NA-Treated at Baseline (n = 142) | |||
| HBV RNA | 0.749 (P < 0.001) | 0.892 (P < 0.001) | 0.692 (P < 0.001) |
| HBV DNA | — | 0.879 (P < 0.001) | 0.537 (P < 0.001) |
| Week 48 of NA Therapy | |||
| HBV RNA | 0.414 (P < 0.001) | 0.806 (P < 0.001) | 0.374 (<0.001) |
| HBV DNA | — | 0.374 (P = 0.004) | 0.289 (P = 0.001) |
| Week 96 of NA Therapy | |||
| HBV RNA | 0.152 (P = 0.084) | 0.795 (P < 0.001) | 0.293 (<0.001) |
| HBV DNA | — | 0.174 (P = 0.199) | 0.062 (0.486) |
Subgroup Analysis of Serum HBV RNA Dynamics Between Tenofovir and ETV Treatment
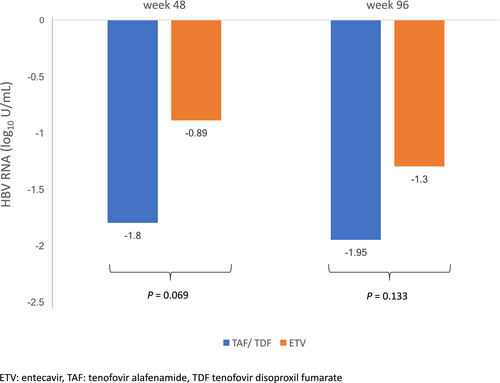
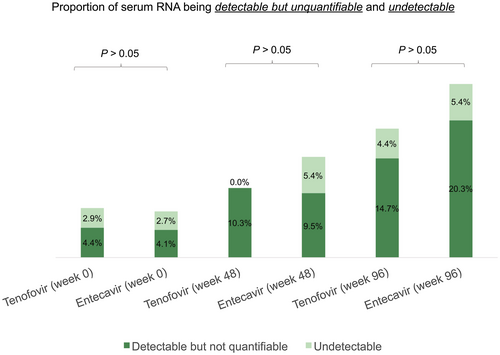
The baseline median serum HBV RNA and DNA level was higher for the patients who were tenofovir-treated compared with patients who were ETV-treated (5.41 vs. 4.50 log10 U/mL; P = 0.09, and 7.64 vs. 6.56 log10 IU/mL; P = 0.034, respectively). The differential reductions in serum HBV RNA at weeks 48 and 96 of NA therapy are shown in Fig. 4. There was no significant difference in the magnitude of decline in serum HBV RNA for patients who were treated with tenofovir (TAF or TDF) compared with ETV at week 48 (−1.8 vs. -0.89 log10 U/mL, respectively; P = 0.069) and at week 96 (−1.95 vs. −1.3 log10 U/mL; P = 0.133). The percentage reduction in serum HBV RNA from baseline was calculated to adjust for the higher baseline serum HBV RNA in the tenofovir-treated group compared with the ETV-treated group. At week 48, serum HBV RNA reduced by 36.6% and 21.9% for tenofovir and ETV group, respectively (P = 0.11). At week 96, serum HBV RNA reduced by 38.8% and 33.4% for tenofovir and ETV group, respectively (P = 0.29). The subgroup analysis for on-treatment kinetics (HBeAg-positive vs. HBeAg-negative) of HBV RNA showed similar results and is shown in Supporting Figs. S6 and S7. At week 96, only 19.1% patients who were tenofovir-treated and 25.7% patients who were ETV-treated had unquantifiable serum HBV RNA (Fig. 5). The higher proportion of serum HBV RNA unquantifiability for ETV treatment might be explained by a lower baseline serum HBV RNA to start with for patients who were ETV-treated.
Discussion
The current study describes the serum HBV RNA profiles in a large cohort of Chinese patients who were treatment-naïve and those who were NA-treated with predominant genotype B and C CHB infection. The pattern of serum HBV RNA profile in different phases of the disease mirrors that of serum HBV DNA, qHBsAg, and HBcrAg. This correlation indirectly gives confidence that the HBV RNA assay is a reliable method for assessing disease phases. The reverse transcriptional efficiency of pgRNA, as reflected by serum HBV RNA to DNA ratio, was higher in patients who were HBeAg-positive than those who were HBeAg-negative. However, this ratio was similar between HBeAg-negative chronic hepatitis and HBeAg-negative chronic infection, a finding that is opposite to the findings of a recently published study that showed a significantly higher ratio in the HBeAg-negative chronic hepatitis group compared with the HBeAg-negative chronic infection group (1.04 vs. 0.85; P = 0.009).(7) In that study, the LLOD of serum HBV DNA and RNA were 500 copies/mL and 100 copies/mL, respectively, in contrast to the assay used in our current study, which has LLOD of 1.65 log10U/mL for HBV RNA and 1 log10 IU/mL for HBV DNA. The differences in LLODs of viral nucleic acid between these two studies will affect the calculated HBV RNA to DNA ratio, and thus the two studies could not be directly compared. This emphasizes the importance of developing a standardized assay in the future. Consequently, to differentiate chronic hepatitis from chronic infection in patients who are HBeAg-negative, serum HBV RNA is not recommended, and serum HBV DNA should be used instead, as also demonstrated in this study by the better AUROC of using HBV DNA in patients who were HBeAg-negative. In contrast, for HBeAg-positive phases, HBV RNA outperforms other viral markers to differentiate chronic active hepatitis compared with chronic infection.
In patients with HBsAg seroclearance, 15.8% and 15.8% still had detectable serum HBV RNA and HBcrAg, respectively, confirming that HBsAg seroclearance does not necessarily equate to cessation of ongoing viral transcription and translation, respectively. This study reports the profile of HBV RNA in patients with HBsAg seroclearance, i.e., functional cure. Such a finding highlights the fact that replication-competent cccDNA and downstream viral replicative steps still persist despite seronegativity for HBsAg. The major difference between complete cure and functional cure is demonstrated well: for complete cure, elimination of cccDNA is required, an endpoint which is still almost impossible to be achieved.(22) With the availability of more sensitive quantitative assays for HBV biomarkers, it is anticipated that viral products will be detected more easily in patients with HBsAg seroclearance, thus potentially aiding identification of those with persistent risks of liver-related complications. In addition, testing for HBV RNA and/or HBcrAg may add an additional tool for diagnosing occult hepatitis B infection.
This large study reports the profiles and correlation of two important HBV biomarkers—serum HBV RNA and serum HBcrAg—in patients with CHB on first-line oral NA therapy. At baseline, serum HBV RNA correlates extremely well with serum HBcrAg in both HBeAg-positive and HBeAg-negative phases. Such correlation is still maintained when patients were treated with NA for up to 96 weeks (r = 0.795-0.892).
Early after NA therapy, HBV RNA would become higher than HBV DNA, as reported in a study involving 16 subjects with CHB who were treated with lamivudine for 12 weeks.(8) This is due to the NA-induced inhibition of reverse transcription of pgRNA. While DNA synthesis is impaired, pgRNA accumulates and is released into the circulation as pgRNA-containing virions. As NA is continued for a longer duration, nuclear recycling of HBV DNA via nucleocapsids into the host nucleus is impaired, leading to gradual decline in cccDNA pool and reduction in number of infected hepatocytes,(23) and thus serial reduction in pgRNA formation. Therefore, although serum HBV RNA would still be higher than serum HBV DNA during NA treatment, the absolute concentration of HBV RNA will gradually decline with longer duration of NA therapy. In the era of very effective suppression of HBV DNA, alternative biomarkers such as HBV RNA and HBcrAg are necessary to gauge the duration of therapy and determine risk of post-NA cessation relapse (6, 14, 16, 17). The findings from this study consolidate that treatment for up to 96 weeks is not sufficient to completely suppress pgRNA production, a surrogate marker for cccDNA transcriptional activity, despite the use of potent first-line oral NAs. Longer treatment duration will be needed, even when the serum HBV DNA is already undetectable, before consideration of stopping NA therapy.
Subgroup analysis was performed in patients who received tenofovir and ETV treatment. There were no significant differences in the magnitude of absolute reduction and percentage reduction in serum HBV RNA in the tenofovir group compared with the ETV group at weeks 48 and 96. However, because this study was limited by a small number of patients in each treatment subgroup, more studies involving larger numbers of patients should be conducted to assess the relative efficacies of tenofovir and ETV in RNA suppression.
The proportion of unquantifiable serum HBV RNA (either detectable but quantifiable or undetectable) was numerically higher for ETV than tenofovir at all time points. This may be attributed to a lower baseline serum HBV RNA in the patients who were ETV-treated compared with patients who were tenofovir-treated (4.5 vs. 5.41 log10 U/mL, respectively; P = 0.09), likely because of a higher proportion of patients with cirrhosis in the ETV group compared with the tenofovir group. Nevertheless, both tenofovir and ETV treatment only led to unquantifiable serum HBV RNA in about one-quarter of the patients at 96 weeks. This suggests that the risks of liver-related complications, theoretically proportional to residual intrahepatic cccDNA pool, could only be reduced after long-term NA therapy. Monitoring of HBV RNA levels, as a surrogate marker for residual intrahepatic cccDNA transcriptional activity, may be of great value to assess the individual risk under NA treatment. Because the current assay detects full-length pgRNA, it would be of interest to develop an assay that can detect various other HBV RNA species, such as spliced HBV variant, X gene RNA, and smaller fragments.(20)
Although serum HBV RNA and HBcrAg show similar profiles(5) and correlate well with each other during NA therapy, their individual roles in patients receiving antiviral agents are evolving.(24) Core protein allosteric modulators (CpAMs) and RNA interfering (RNAi) gene silencers (small interfering RNA [siRNA] and ASO) are forthcoming antiviral agents.(22) These two groups of agents are usually used in combination with NA therapy. The findings of this study provide basic understanding of the dynamics of serum HBV RNA under NA therapy, which is the backbone of combination treatment regimens with CpAMs or RNAi gene silencers, to enhance better interpretation of RNA level reductions in these treatment settings.
Short-term treatment of CpAMs can only reduce HBV RNA but not the viral antigens, including HBsAg and HBcrAg,(25-27) whereas siRNAs have suppressive effects on HBV RNA and viral antigens (mainly HBsAg reduction with less pronounced effects on HBcrAg).(28) It seemed that HBV RNA may be a better tool to assess and compare different antiviral agents, at least for early-phase studies with short-term treatment. Further studies are needed to define the role of HBcrAg in patients receiving more long-term treatment. It would also be interesting to determine whether HBcrAg adds any predictive value in patients who were HBeAg-negative in whom we expect that a greater proportion of HBsAg levels may be contributed by integrated HBV genome.
There are two limitations of the current study. Firstly, no liver biopsies were performed to measure intrahepatic cccDNA and to differentiate chronic hepatitis from chronic infection. However, liver biopsy is associated with the risk of complications and sampling error. Moreover, there is not yet a standardized assay for cccDNA quantification. ALT was used to define chronic hepatitis and chronic infection, which is recommended by international guidelines.(3) Secondly, there was only cross-sectional data for the patients who were treatment-naïve, and the follow-up period for patients who were NA-treated was only up to 96 weeks. Long-term follow-up would explain the significance of serum biomarkers in the clinical outcomes of these patients.
To conclude, serum HBV RNA and HBcrAg levels change dynamically during the natural course of CHB infection in a large cohort of Chinese patients. These two biomarkers correlate well with each other in patients who were treatment-naïve and those who were NA-treated. Persistent detectability of serum HBV RNA and HBcrAg in patients with HBsAg seroclearance suggests that these patients still need monitoring of viral biomarkers and surveillance for liver-related complications.
Acknowledgment
HBV RNA and HBcrAg measurements are provided by Abbott Laboratories and Fujirebio, respectively.
Author Contributions
L.Y.M. was responsible for data interpretation, statistical analysis, and drafting of the manuscript. D.K.H.W. was responsible for data acquisition and study design. G.C. and J.G. were responsible for data acquisition and data analysis. W.K.S. and J.F. were responsible for critical revision of the manuscript. M.F.Y. was responsible for study concept, study design, overall supervision of study, and critical revision of the manuscript.




