PRDM8 exhibits antitumor activities toward hepatocellular carcinoma by targeting NAP1L1
Potential conflict of interest: Nothing to report.
Supported by grants from National Natural Science Foundation of China (81270553, 81300363, 81521004 and 81572262), Fund of State Key Laboratory of Reproductive Medicine, Nanjing Medical University (SKLRM-K201706), Jiangsu Youth Medical Talents (QNRC2016580), Excellent Youth Teacher Support Program of Nanjing Medical University (2015RC19), Natural Science Foundation of Jiangsu Province (BK20131023, BK20151583), Six Talents Peak Project of Jiangsu Province (2014-wsw-004, 2013-wsn-032), Jiangsu Province's Key Provincial Talents Program (ZDRCA2016028), 333 High Class “Talented Man Project” (BRA2016516).
Abstract
Hepatocellular carcinoma (HCC) is a major leading cause of cancer mortality worldwide. PRDI-BF1 and RIZ homology domain containing 8 (PRDM8) is a key regulator in neural development and testis steroidogenesis; however, its role in liver carcinogenesis remains to be investigated. In this study, PRDM8 was found to be down-regulated in HCC, which was linked with shorter recurrence-free survival. Lentiviral-based overexpression and knockdown approaches showed that PRDM8 inhibited HCC cell proliferation, migration, and invasion. PRDM8 caused G1/S cell cycle arrest and induced apoptosis. An in vivo tumor model confirmed the antitumor role of PRDM8 in HCC growth and metastasis. Mechanistic study showed that PRDM8 suppressed the PI3K/AKT/mTOR signaling cascade through the regulation of nucleosome assembly protein 1-like 1 (NAP1L1). Conclusion: PRDM8 as a functional tumor suppressor is frequently down-regulated in HCC. Through regulating NAP1L1, PRDM8 inhibits PI3K/AKT/mTOR signaling in HCC. PRDM8 is a potential target for therapies of HCC. (Hepatology 2018).
Abbreviations
-
- CCK-8
-
- Cell Counting Kit-8
-
- HCC
-
- hepatocellular carcinoma
-
- HMTase
-
- histone lysine methyltransferase
-
- IHC
-
- immunohistochemistry
-
- IP
-
- immunoprecipitation
-
- NAP1L1
-
- nucleosome assembly protein 1-like 1
-
- PRDM8
-
- PRDI-BF1 and RIZ homology domain containing 8
-
- shRNA
-
- small hairpin RNA
Hepatocellular carcinoma (HCC) is the fifth most common cancer in males and the second leading cause of cancer-related mortality worldwide. Approximately 782,500 new cases and 745,500 deaths occur annually, with China alone accounting for about half of the total number of cases and deaths.1 Although treatments have greatly improved over the last decades, HCC's outcome is still unfavorable. It will be of paramount importance to elucidate the molecular changes in HCC in order to identify new therapeutic strategies for this deadly malignant disease.
The PRDI-BF1 and RIZ homology domain (PRDM) family of proteins have been reported to be involved in pathological conditions, particularly in cancer.2, 3 PRDM1 is silenced in human diffuse large B cell lymphoma.4 Loss of functional PRDM2 has also been found in many types of solid tumors, and forced PRDM2 expression causes apoptosis and/or cell cycle arrest in cancer cell lines and inhibits growth in vivo.5, 6 PRDM4 is located within a tumor suppressor locus that is frequently deleted in cancer.7 PRDM5 is suggested to act as a tumor suppressor, and epigenetic silencing of the PRDM5 promoter has been found in several different cancer cell lines and clinical samples from different tissue origins including breast, liver, ovary, cervix, and the gastrointestinal tract.8-10 Overexpression of PRDM5 in cancer cells induces cell cycle arrest and apoptosis. PRDM11 overexpression induces apoptosis and delayed lymphoma onset.11 PRDM12 exerts an antiproliferative effect on chronic myeloid leukemia.12
The role of PRDM family involvement in malignancies gives rise to a hypothesis that PRDI-BF1 and RIZ homology domain containing 8 (PRDM8) may play a role in human carcinogenesis. According to previously published studies, PRDM8 is mainly demonstrated as a crucial regulatory molecule during neural development and testis steroidogenesis.13-16 Recent whole-exome sequencing results indicated decreased mRNA level of PRDM8 in invasive pituitary adenoma specimens compared with noninvasive ones,17 suggesting the connection between PRDM8 and human malignancies. However, the function and mechanism of PRDM8 in carcinogenesis remain elusive.
In this study, we revealed that PRDM8 was expressed at low levels in HCC tissues compared with its expression in the associated noncancerous tissues, and low PRDM8 expression was closely correlated with HCC recurrence. PRDM8 attenuated HCC cell proliferation, migration, invasion, and metastasis. Furthermore, PRDM8 induced cell cycle arrest and enhanced cell apoptosis. Based on mechanistic analysis, PRDM8 inhibited PI3K/AKT/mTOR signaling pathway via suppressing oncogenic nucleosome assembly protein 1-like 1 (NAP1L1). Our findings indicated that PRDM8 may serve as a potential diagnostic biomarker and therapeutic target for HCC.
Patients and Methods
PATIENTS AND SPECIMENS
Two independent cohorts of HCC patients were enrolled in this study. For the first cohort (cohort 1), 60 pairs of HCC tissues and corresponding adjacent nontumorous tissue samples and related clinical information were obtained from patients who received hepatectomy at Department of Hepatobiliary Surgery, the First Affiliated Hospital of Nanjing Medical University between January 2015 and December 2016 with the patients' consent, and 12 paired HCC samples in cohort 1 were randomly selected for western blotting analysis. For the second cohort (cohort 2), tissue microarrays (TMAs) containing 398 pairs of HCC tissues with complete clinicopathological and follow-up data were purchased from Shanghai Zhuoli Biotech (Shanghai, China).
- Definitive HCC diagnosis by pathology.
- Surgical resection, defined as complete resection of all tumor nodules with the cut surface being free of cancer by histological examination.
- No radiotherapy or chemotherapy received before surgery.
The study protocol which conformed to the ethical guidelines of the 1975 Declaration of Helsinki was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University.
STATISTICAL ANALYSIS
All data are shown as means and standard errors of the mean. Prism 6 (GraphPad Software, La Jolla, CA, USA) and SPSS 21.0 (IBM SPSS software, NY, USA) were used for the Wilcoxon matched-pairs test, Student t test, chi-square test, Fisher exact test, or Pearson correlation test when appropriate. Kaplan-Meier analysis and Cox proportional hazards regression model were used for survival analysis. Values of P < 0.05 were considered statistically significant.
Detailed methodology can be found in supplementary materials and methods.
Results
PRDM8 EXPRESSION WAS DOWN-REGULATED IN HCC AND CORRELATED WITH A POOR OUTCOME
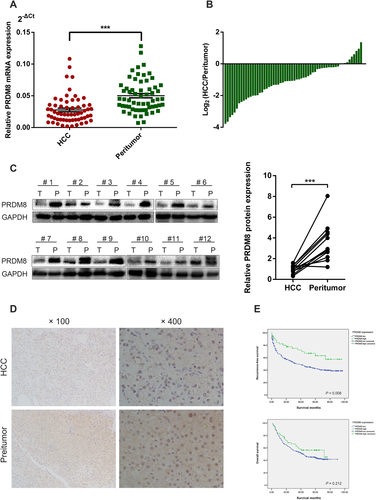
We examined the mRNA expression level of PRDM8 in 60 human HCC tissue samples and their matched nontumorous liver specimens by quantitative reverse transcription polymerase chain reaction (RT-qPCR). The results revealed that PRDM8 mRNA level was markedly down-regulated in HCC tissues (Fig. 1A,B). Decreased protein expression level of PRDM8 in HCC tissue was further confirmed in 12 paired samples randomly obtained from cohort 1 using western blotting (Fig. 1C). Immunohistochemistry (IHC) also confirmed that the protein expression level of PRDM8 was down-regulated in HCC tissues in cohort 1 (Fig. 1D). The clinicopathological analysis revealed that PRDM8 expression was closely correlated with tumor multiplicity, Edmonson stage, tumor size, tumor encapsulation, vascular invasion, and tumor-node-metastasis (TNM) stage (Supporting Table S1). In addition, TMA results confirmed that the expression level of PRDM8 protein was significantly lower in HCC than that in peritumoral tissues (Supporting Fig. S1A,B). Further survival analysis on cohort 2 showed that low PRDM8 group had obvious higher recurrence probability than those in high PRDM8 group, whereas no significant effect on overall survival was observed (Fig. 1E). Multivariate Cox regression analysis demonstrated that low PRDM8 expression was an independent predictive indicator for recurrence-free survival (hazard ratio [HR] = 1.742, 95% confidence interval [CI], 1.056-2.874; P = 0.030) for HCC patients (Table S2).
PRDM8 INHIBITED CELL PROLIFERATION, MIGRATION AND INVASION IN VITRO
We performed RT-qPCR and western blotting to investigate the expression of PRDM8 in HCC cell lines and noncancerous liver cell lines. We found the mRNA and protein expression levels of PRDM8 were reduced in HCC cell lines compared with noncancerous cell lines (Supporting Fig. S2A,B). According to the expression level of PRDM8 in HCC cell lines, we stably overexpressed PRDM8 in low PRDM8-expressing YY8103 and SMMC7721 cells by lentivirus, named YY8103-PRDM8 and SMMC7721-PRDM8, respectively (Supporting Fig. S2C). High PRDM8-expressing Focus and HepG2 cells were selected to knock down the expression level of PRDM8 using lentivirus carrying small hairpin RNA (shRNA). The knockdown efficiency of three shRNAs for PRDM8 was confirmed through comparison with negative control at both mRNA and protein levels (Supporting Fig. S2D,E). Among the three shRNAs tested, shRNA#1 generated the most consistent knockdown results across Focus and HepG2 cell lines, and was thus chosen for subsequent studies.
To determine whether PRDM8 is functionally involved in HCC cells, we performed a variety of in vitro assays to evaluate the effect of PRDM8 overexpression on cell functions including proliferation, migration, and invasion. The results of Cell Counting Kit-8 (CCK-8) assays showed that up-regulation of PRDM8 significantly suppressed cell proliferation of YY8103 and SMMC7721 (Fig. 2A). Colony formation assays confirmed the inhibitory effect of PRDM8 overexpression on cell proliferation (Fig. 2B). In wound healing migration assay, PRDM8-overexpressed cells exhibited delayed wound healing (Fig. 2C). Transwell assays demonstrated that cell migration and invasion were strongly impaired by overexpression of PRDM8 in YY8103 and SMMC7721 cells (Fig. 2D).
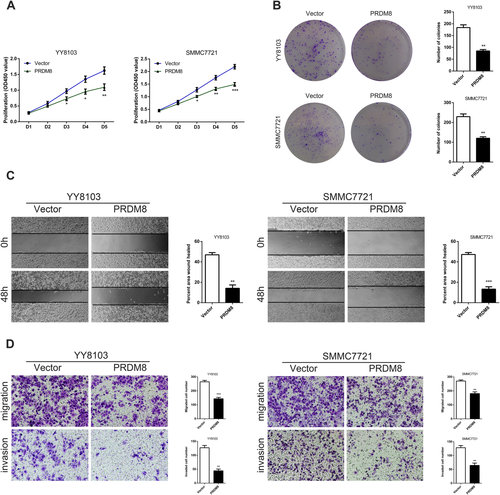
Then we assessed the impact of PRDM8 knockdown on HCC cell functions. The CCK-8 and colony formation assays revealed that down-regulation of PRDM8 significantly promoted cell proliferation of Focus and HepG2 (Supporting Fig. S3A,B). As shown in Supporting Fig. S3C, the migration ability of Focus and HepG2 cells with PRDM8 knockdown was increased compared with the control groups. Results from Transwell assays further showed that PRDM8 deficiency enhanced the abilities of HCC cells to migrate and to invade (Supporting Fig. S3D). These results were consistent with the PRDM8 overexpression data, and they indicated that PRDM8 could be a key player in the regulation of cell functions of HCC.
PRDM8 INDUCED G1/S CELL CYCLE ARREST AND ENHANCED APOPTOSIS IN VITRO
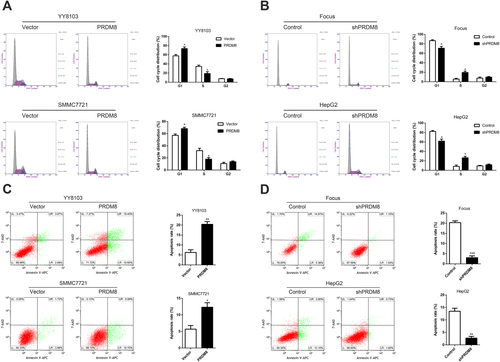
We quantified cell cycle distribution using flow cytometry and found that the number of cells was increased at G1 phase and decreased at S phase in PRDM8-overexpressed YY8103 and SMMC7721 cells (Fig. 3A). In contrast, the percentage of G1 phase cells in HepG2-shPRDM8 and Focus-shPRDM8 cells was significantly lower than that in control groups (Fig. 3B). Flow cytometry analysis using Annexin V-APC/7-AAD staining demonstrated that cell apoptosis was induced by up-regulating PRDM8 in HCC cells (Fig. 3C), whereas knockdown of PRDM8 in HepG2 and Focus inhibited cell apoptosis (Fig. 3D).
PRDM8 SUPPRESSED TUMOR GROWTH AND METASTASIS IN VIVO
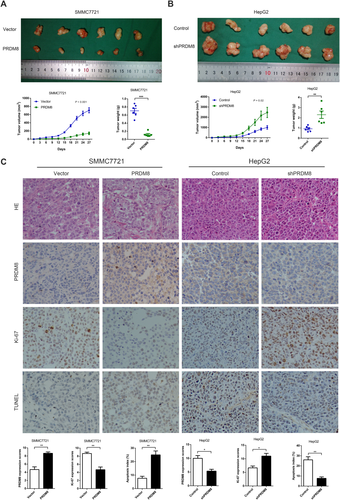
The HCC cells with stable overexpression or knockdown of PRDM8 were implanted into nude mice to examine the expansion and migration of tumor cells in vivo. Each mouse was monitored once every three days, and the mice were euthanized after four weeks. Tumors derived from SMMC7721-PRDM8 cells grew slower, and the average tumor weight was significantly lower in mice inoculated with PRDM8-overexpressed SMMC7721 cells at the fourth week (Fig. 4A). In mice inoculated with HepG2-shPRDM8 cells, average tumor volume was markedly higher at the fourth week than in control mice, and tumors were also heavier in mice inoculated with HepG2-shPRDM8 (Fig. 4B). Hematoxylin and eosin, IHC, Ki-67, and terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end-labeling (TUNEL) staining of the xenograft tumors are shown in Fig. 4C, and the data further confirmed the antitumor activities of PRDM8 toward HCC.
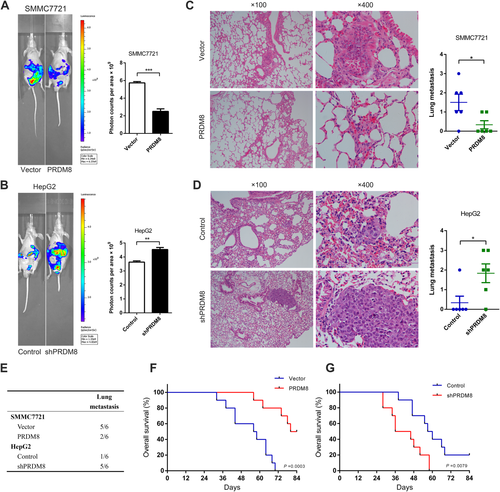
Detection of metastasis in a nude mouse model transplanted with HCC cells indicated that overexpression of PRDM8 was significantly related to reduced metastasis of SMMC7721 according to photon flux detection (Fig. 5A). In mice injected with HepG2-shPRDM8 cells, we observed increased metastasis of tumor cells when compared to controls (Fig. 5B). Histological analysis of lung tissue showed that incidence of lung metastases in SMMC7721-PRDM8 group mice was decreased, while the incidence was higher in HepG2-shPRDM8 group mice compared with the controls (Fig. 5C–E). In addition, the SMMC7721-PRDM8 group had a longer overall survival time than the SMMC7721 control group (Fig. 5F). Consistently, HepG2-shPRDM8 group had worse survival outcome compared to control group (Fig. 5G).
PRDM8 INHIBITED HCC PROGRESSION VIA PI3K/AKT/MTOR SIGNALING
Considering the pivotal role of PI3K/AKT/mTOR signaling in tumor progression, we investigated the effect of PRDM8 on the specific signaling cascade. We found that PRDM8 overexpression reduced phosphorylation of AKT and mTOR in HCC cells (Fig. 6A), indicative of inhibition of PI3K/AKT/mTOR signaling. Conversely, PRDM8 knockdown enhanced the activity of PI3K/AKT/mTOR signaling (Fig. 6B).
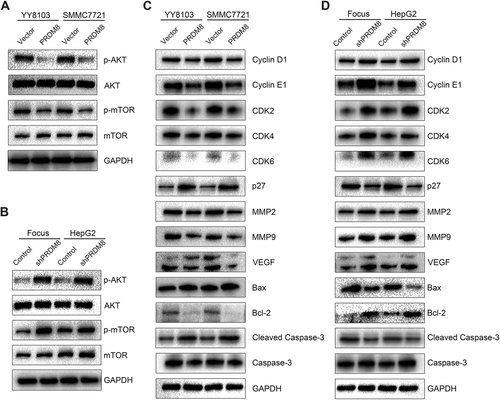
In addition, PRDM8 overexpression altered the expression pattern of the key regulators that are indispensable in cell cycle regulation, tumor metastasis, and cell apoptosis. Western blotting results indicated that the expression levels of Cyclin D1, Cyclin E1, CDK2, CDK4, CDK6, MMP2, MMP9, VEGF, and Bcl-2 were reduced, while p27, Bax, and cleaved caspase-3 levels were up-regulated in PRDM8-overexpressed cells (Fig. 6C). In Focus-shPRDM8 and HepG2-shPRDM8 cells, the levels of Cyclin D1, Cyclin E1, CDK2, CDK4, CDK6, MMP2, MMP9, VEGF, and Bcl-2 expression were increased, and down-regulated expression levels of p27, Bax, and cleaved caspase-3 were also observed (Fig. 6D). According to the results of previous study, the expression of Prdm8 is complementary to that of Hes5 in mouse neurogenesis.18 Thus, we attempted to examine the effect of PRDM8 overexpression or knockdown on HES5 mRNA expression in HCC. However, no significant results were found (data not shown). In addition, previous works reported that reduced expression of PRDM8 was caused by hypermethylation in patients with dyskeratosis congenital, aplastic anemia, and Down syndrome.19, 20 Based on our results, down-regulated PRDM8 expression in HCC cell lines was not significantly restored by treatment with 5-aza-2′-deoxycytidine (data not shown). Prediction of upstream microRNAs was performed using TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org). Several microRNAs including miR-302, miR-372, miR-373, miR-425-5p, and miR-520 were suggested as potential upstream regulators of PRDM8.
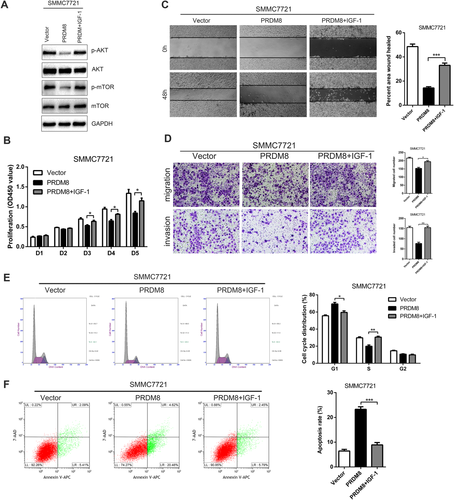
We promoted the activity of AKT signaling by IGF-1 treatment, and found that AKT induction restored the decreased proliferation, migration, and invasion in PRDM8 overexpressed SMMC7721 cells (Fig. 7). Consistently, pharmacological inhibition of AKT (MK-2206) abrogated the increased proliferation, migration, and invasion results from PRDM8 knockdown in HepG2-shPRDM8 cells (Supporting Fig. S4). Taken together, these data indicated that PRDM8 suppressed HCC progression through blocking PI3K/AKT/mTOR signaling.
PRDM8 SUPPRESSED PI3K/AKT/MTOR SIGNALING AND HCC PROGRESSION VIA NAP1L1
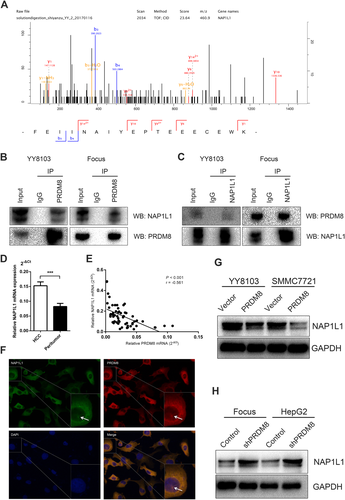
The immunoprecipitation (IP) assay was performed to explore the potential binding protein of PRDM8. As presented in Fig. 8A, further mass spectrometry identification indicated that NAP1L1 was the captured protein by PRDM8. Co-IP verified the specificity of this interaction, suggesting that PRDM8 could bind with NAP1L1 (Fig. 8B,C). Next, we analyzed the mRNA expression of NAP1L1 in the tissue samples of patients in cohort 1. An increased level of NAP1L1 was found in HCC tissues compared with the corresponding peritumor tissues (Fig. 8D). Person correlation analysis also presented a negative correlation between the expression of PRDM8 and NAP1L1 (Fig. 8E). Immunofluorescence results showed that PRDM8 co-localized with NAP1L1 in cytoplasm and nucleus (Fig. 8F). PRDM8 ectopic expression led to reduced NAP1L1 protein level in YY8103 and SMMC7721 cells (Fig. 8G). Conversely, PRDM8 knockdown caused increased NAP1L1 protein (Fig. 8H). To determine whether altered NAP1L1 expression played a role in PRDM8 suppression of HCC progression, we up-regulated the expression of NAP1L1 in SMMC7721-PRDM8 cells using lentivirus (Supporting Fig. S5A). The results revealed that NAP1L1 overexpression rescued the attenuated PI3K/AKT/mTOR signaling activity (Supporting Fig. S5B), and increased proliferation, migration, and invasion of PRDM8-overexpressed SMMC7721 cells (Supporting Fig. S5C–H). In addition, we knocked down NAP1L1 in HepG2-shPRDM8 cells (Supporting Fig. S6A). NAP1L1 knockdown blunted the ability of PRDM8 shRNA to promote phosphorylation of AKT and mTOR (Supporting Fig. S6B), and restored the increased HCC cell proliferation, migration, and invasion (Supporting Fig. S6C–H). Moreover, based on the results of CCK-8 and transwell assays, NAP1L1 alone could exert an antiHCC effect, and overexpressing PRDM8 when NAP1L1 was silenced had an obvious synergistic effect (Supporting Fig. S7A,B).
According to a recent study, depletion of NAP1L1 in hepatocytes leads to the down-regulation of 358 genes, and Ingenuity Pathway Analysis of the downregulated genes indicates that PI3K/AKT signaling is one of the top canonical pathways. GAB1, an important upstream regulator of PI3K/AKT/mTOR, is among those downregulated genes.21 We therefore speculated that NAP1L1 might activate PI3K/AKT/mTOR signaling through GAB1. As shown in Supporting Fig. S7C, western blotting results validated that HCC cells overexpressing NAP1L1 exhibited a higher GAB1 expression level, while HCC cells silencing NAP1L1 presented a decreased GAB1 expression. GAB1 might be the key mediator that activated PI3K/AKT/mTOR in the downstream of NAP1L1. Collectively, these observations indicated that PRDM8 negatively regulated NAP1L1 expression, which was important for PRDM8 to suppress the growth, migration, and invasiveness of HCC cells.
Discussion
HCC is one of the most lethal malignancies in the world, and the high mortality and recurrence of HCC is due to the uncontrolled growth and metastasis of cancer cells. It is of great importance to identify factors that contribute to the uncontrolled expansion and metastasis of HCC. Here, we indicated that PRDM8 was frequently down-regulated in HCC, and PRDM8 expression was an independent predictive factor for recurrence-free survival. PRDM8 exhibited antitumor activities toward cancer cell progression by its targeting of NAP1L1 and regulating the PI3K/AKT/mTOR signaling pathway. Furthermore, PRDM8 induced cell arrest in G1 phase and enhanced cell apoptosis in HCC. Therefore, our results indicated PRDM8 as a tumor suppressor in HCC and a potential biomarker for clinical therapy.
PRDM8 contains an N-terminal PR domain, followed by three classical C2H2 zinc fingers towards the C-terminus. Previous studies demonstrated that PRDM8 is mainly involved in neural development and male fertility.13-16 Mutations in human PRDM8 may cause congenital stationary night blindness22 and early-onset Lafora body disease.23, 24 Our data indicated that PRDM8 exhibited antiHCC activities via regulating NAP1L1. PRDM8 could bind to NAP1L1, thus preventing NAP1L1 from binding its downstream molecules. Besides, PRDM8 has been suggested to possess intrinsic histone lysine methyltransferase (HMTase) activity, acting as repressive HMTase by catalyzing di-methylated lysine 9 of histone H3.13 However, whether PRDM8 functions directly or indirectly to methylate histones is uncertain.16, 23 Future studies are warranted to evaluate the HMTase activity of PRDM8 in human malignancies. It has been shown that PRDM8 could also function as a transcription repressor. Loss of Prdm8 function leads to an increased expression of PRKCA, and ChIP assay showed that PRDM8 is present at Prkca promoter.22 In neural circuit assembly, Prdm8 and Bhlhb5 bind together to form a repressor complex, and Bhlhb5 is one of the target genes of Prdm8. In Prdm8 mutant mice, upregulated Bhlhb5 mRNA is observed.25 Phylogenetic analysis reveals that Prdm8 is most closely related to Prdm13.26 Previous research suggested that Prdm13 binds to the promoter of Nap1l2, and loss of Prdm13 function results in an increased Nap1l2.27 Therefore, it is possible that PRDM8 could transcriptionally suppress NAP1L1 expression.
As demonstrated in previous study, Prdm8 expression in early telencephalic development is complementary to that of the Hes5 gene.18 We investigated the influence of PRDM8 on HES5 mRNA expression in HCC; however, no significantly altered HES5 expression was detected. It is also possible that HES5 might be an upstream regulator of PRDM8. Aberrant hypermethylation of PRDM8 is observed in dyskeratosis congenital, aplastic anemia, Down syndrome, and Werner syndrome.19, 20, 28 PRDM8 has hypomethylated CpG islands in endometrial cancer.29 Association between in utero phthalate exposure and altered DNA methylation of PRDM8 has also been identified recently.30 Of note, expression of PRDM8 is down-regulated in a subset of colorectal and gastric cancer cell lines, but the expression is not significantly restored by treating cells with 5-aza-2′-deoxycytidine.9 Moreover, our data suggested that down-regulated PRDM8 expression in HCC cell lines could not be restored by treatment with 5-aza-2′-deoxycytidine, indicating that DNA methylation is likely not the primary mechanism by which PRDM8 was silenced. Predicted upstream microRNAs of PRDM8 included miR-302, miR-372, miR-373, miR-425, and miR-520. Future studies are needed to determine the potential upstream molecules of PRDM8.
PI3K/AKT/mTOR signaling pathway has a broad range of action and plays a pivotal role in HCC.31 An upregulation of PI3K/AKT/mTOR activity is observed in 40-50% of HCCs.32 Independent of the underlying etiology of HCC, activation of the PI3K/AKT/mTOR pathway is associated with less differentiated tumors and earlier recurrence.33 PI3K/AKT/mTOR pathway occupies a central position in the network of dysregulated signaling pathways in HCC. Inhibiting PI3K/AKT/mTOR can prevent abnormal cell proliferation, cellular metabolism, and tumor angiogenesis, thus providing potential molecular targeted therapeutics.34 Considering the important role of PI3K/AKT/mTOR signaling pathway in HCC, we examined the phosphorylation of AKT and mTOR, and found that PRDM8 exhibited its antitumor activities through PI3K/AKT/mTOR signaling. Enhanced PI3K/AKT/mTOR activity rescued the suppressed proliferation, migration, and invasion in PRDM8-overexpressed cells, whereas treatment of MK-2206, an AKT inhibitor, down-regulated the increased HCC progression caused by PRDM8 knockdown.
According to the mechanistic study results, we showed that upregulation of NAP1L1 enhanced PI3K/AKT/mTOR activity and HCC progression, while NAP1L1 knockdown inhibited the PI3K/AKT/mTOR signaling and the oncogenic capacity in HCC. Moreover, GAB1 might be the key mediator that activated PI3K/AKT/mTOR in the downstream of NAP1L1. NAP1L1 is the human counterpart of the yeast NAP-I protein, a histone-binding factor involved in the maintenance of cumulative nucleosome formation.35 In humans, NAP1L1 belongs to a family that is considered to be involved in nucleosome assembly and transcriptional regulation.36 Levels of both NAP1L1 mRNA and protein increase rapidly in conjunction with cell proliferation in a T-lymphoid cell model.37 NAP1L1 has been identified to be overexpressed in fetal liver compared to adult liver, and in hepatoblastoma compared with normal adult livers.38, 39 NAP1L1 is upregulated in small-intestinal neuroendocrine cell-derived neoplastic tissue,40 colon cancer,41 and malignant adenocarcinoid, compared with normal mucosa.42 NAP1L1 is indicated to be involved in ovarian cancer cell responses to cytotoxic gold compounds.43 In pancreatic neuroendocrine neoplasms, NAP1L1 promotes tumor cell proliferation and regulates cell entry into the S phase via inhibition of the mTOR pathway and epigenetic regulation of p57 promoter methylation.44 The potential use of NAP1L1 for differentiation between glioblastoma and lower grade gliomas has also been demonstrated.45 Reportedly, Nap1l1 promotes the proliferation of murine induced pluripotent stem cell attributable to G2/M transition caused by downregulation of p27 and p21, and upregulation of cyclin B1, and the activation of AKT signaling is involved in the process.46 In addition, Tanaka, et al. showed that NAP1L1 increases the Ser32 phosphorylation of IκB, thereby activating NF-κB pathway and subsequently promoting antiapoptotic Mcl-1.47 AKT could also lead to NF-κB activation through phosphorylating IKKα at Thr23.48 Hence, NAP1L1 may have an intricate influence on its downstream signaling transduction networks. On one hand, NAP1L1 may directly upregulate NF-κB signaling via IκB Ser32 phosphorylation, thus exerting an oncogenic effect on HCC. On the other hand, NAP1L1 may affect AKT signaling, thus activating NF-κB signaling in an AKT-dependent way.
In summary, our study showed the downregulation of PRDM8 in HCC, and further demonstrated that PRDM8 played a crucial role in HCC through regulating NAP1L1, thus fine tuning the PI3K/AKT/mTOR signaling pathway. According to the survival analysis, low expression of PRDM8 was associated with recurrence-free survival in HCC patients. PRDM8 may be a promising biomarker for the prognosis of HCC, and is a potential therapeutic target for HCC. More studies are warranted for further investigation of the function of PRDM8 in the future.
Acknowledgment
We thank the Analysis Center of Nanjing Medical University for assistance in proteomics.
REFERENCES
Author names in bold designate shared co-first authorship.




