Hepatocyte nuclear factor 4α-nuclear factor-κB feedback circuit modulates liver cancer progression
Potential conflict of interest: Nothing to report.
See Editorial on Page 1466
Supported by grant Key Program (81230011), Creative Research Groups (81221061), Distinguished Young Scholars (30825020), Excellent Young Scholars (81222034) and Young Scholar (81201938) from the National Natural Science Foundation of China, and grant from Shanghai Science and Technology Committee (12ZR1439300).
Abstract
Hepatocyte nuclear factor 4α (HNF4α) is a liver enriched transcription factor and is indispensable for liver development. However, the role of HNF4α in hepatocellular carcinoma (HCC) progression remains to be elucidated. We report that reduced HNF4α expression correlated well with the aggressive clinicopathological characteristics of HCC and predicted poor prognosis of patients. HNF4α levels were even lower in metastatic HCCs, and ectopic HNF4α expression suppressed the metastasis of hepatoma cells both in vitro and in vivo. Forced HNF4α expression attenuated the expression and nuclear translocation of RelA (p65) and impaired NF-κB activation through an IKK-independent mechanism. Blockage of RelA robustly attenuated the suppressive effect of HNF4α on hepatoma cell metastasis. MicroRNA (miR)-7 and miR-124 were transcriptionally up-regulated by HNF4α, which repressed RelA expression by way of interaction with RelA-3′ untranslated region (UTR). In addition, nuclear factor kappa B (NF-κB) up-regulated the expression of miR-21 in hepatoma cells, resulting in decreased HNF4α levels through down-regulating HNF4α-3′UTR activity. Conclusions: Collectively, an HNF4α-NF-κB feedback circuit including miR-124, miR-7, and miR-21 was identified in HCC, and the combination of HNF4α and NF-κB exhibited more powerful predictive efficiency of patient prognosis. These findings broaden the knowledge of hepatic inflammation and cancer initiation/progression, and also provide novel prognostic biomarkers and therapeutic targets for HCC. (Hepatology 2014;60:1607-1619)
Abbreviations
-
- ChIP
-
- chromatin immunoprecipitation
-
- DFS
-
- disease-free survival
-
- EMT
-
- epithelial-mescenchymal transition
-
- HCC
-
- hepatocellular carcinoma
-
- HNF4α
-
- hepatocyte nuclear factor 4α
-
- NF-κB
-
- nuclear factor-κB
-
- OS
-
- overall survival
-
- PVTT
-
- portal vein tumor thrombus
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors and the third leading cause of cancer death globally.1 Despite the advances in diagnosis and treatment, HCC remains one of the most deadly cancers at present, with a very dismal outcome. Surgical resection and chemotherapy exhibit limited effect for patients at advanced stages due to frequent relapse and metastasis.2 It is therefore critical to identify novel therapeutic targets so that new strategies for HCC therapy can be developed. Substantial functional studies of pro-oncogenes and tumor suppressor genes have greatly contributed to the advance of knowledge on the molecular mechanism of HCC development.3 However, the detailed mechanism of HCC development remains far from fully understood, and few biomarkers for the prognosis of patients following surgical treatment are available in the clinic so far.
Hepatocyte nuclear factor 4 (HNF4), first identified in nuclear extracts of rat liver, is a member of the nuclear receptor superfamily (NR2A1) of ligand-dependent transcription factors.4 As a key member of HNF4 family, HNF4α is enriched in hepatocytes and plays a pivot role in hepatic epithelium formation during embryonic development and epithelial phenotype maintenance of hepatocytes in mature liver.5 Our previous studies also demonstrated that HNF4α delivery could suppress liver fibrosis in distinct experiment models and induce the differentiation of hepatoma cells.6, 7 Moreover, we reported that the level of HNF4α was gradually decreased during DEN (diethylnitrosamine)-induced rat hepatocarcinogenesis and dramatically down-regulated in human HCC tissues, suggesting its significance in the initiation of HCC.8, 9 Nevertheless, the role of HNF4α in HCC metastasis and its significance in patient prognosis are obscure, and the molecular mechanism of HNF4α reduction in HCC development remains elusive.
Substantial evidence has elucidated the close association between inflammation and cancer.10 Chronic inflammation elicited by viral infection, toxic substance intake, or autoimmune disorders render sustained liver damage, liver cirrhosis, and eventually give rise to HCC.11 In mammals, transcription factor nuclear factor kappa B (NF-κB) is activated by many proinflammatory cytokines and is a central component of the cellular response to inflammation.12 NF-κB heterodimer, predominantly RelA:p50, or homodimer such as RelA:RelA is sequestrated by I-κB protein and maintained in an inactive state in cytoplasm. Canonical NF-κB activation is mediated by its upstream I-κB kinase (IKK) upon various extracellular cytokines.13 The hepatic inflammatory microenvironment including cytokines and chemokines produced by hepatocytes and liver-infiltrating macrophage has a significant impact on HCC development.14 Activation of NF-κB in hepatocytes or hepatoma cells is a critical molecular event response to chronic hepatic inflammation.15 A number of cytokines, chemokines, and enzymes are transcriptionally regulated by NF-κB, which are closely correlated with HCC initiation and progression.16
In the present study we report an HNF4α-NF-κB feedback circuit consisting microRNA (miR)−124, miR-7, and miR-21, which not only modulate HCC initiation and progression, but also predict the prognosis of HCC patients.
Materials and Methods
Patient Tissues and Microarray Analysis
In all, 429 HCC tissues were obtained from patients who underwent surgical resections in Eastern Hepatobiliary Surgery Hospital, Shanghai, China from 2001 to 2007 (Supporting Tables 1, 2). Forty-two pairs of portal vein tumor thrombus (PVTT) samples resected from HCC patients were diagnosed by a pathologist. A tissue microarray block containing the 181 HCCs and paired noncancerous surrounding tissues was constructed using a tissue microarrayer (Outdo Biotech, Shanghai, China). Immunostaining was performed on tissue microarray slides. The antibodies used are listed in Supporting Table 3. Assessment of staining was based on the percentage of positively stained cells and the nuclear staining intensity using Image-scope software (Aperio Technologies,). Overall survival (OS) was defined as the interval between the dates of surgery and death. Disease-free survival (DFS) was defined as the interval between the dates of surgery and recurrence; if recurrence was not diagnosed, patients were censored on the date of death or the last follow-up. The procedure of patient sample collection was approved by the Ethical Committee of Eastern Hepatobiliary Surgery Hospital.
In Vitro Cell Behavior Assay
For the cell adhesion assay, 2 × 104 SMMC-7721 cells infected with AdGFP or AdHNF4α were seeded into 96-well plates coated with 10 μg/mL fibronectin (Calbiochem, La Jolla, CA) or 200 μg/mL Matrigel (BD Biosciences, San Jose, CA).8, 17 After rinsing at least three times, adhered cells were measured using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). For the wound-healing assay, monolayers of SMMC-7721 cells infected with AdGFP or AdHNF4α were wounded by scraping with a plastic pipette tip and rinsed several times with medium to remove dislodged cells. Cells that had migrated into the wound area were photographed. For the migration assay, SMMC-7721 cells (2 × 104) infected with AdGFP or AdHNF4α were plated into the upper chamber of a polycarbonate transwell filter chamber and incubated for 24 hours. Cell counts are expressed as the mean number of migrated cells per field of view.
In Vivo Metastasis Assay
For in vivo metastasis assay, 2×106 SMMC-7721 cells stably expressing luciferase were injected into the tail vein of nude mice. AdGFP or AdHNF4α (2 × 109 pfu) was then injected by the tail vein once per week for 6 weeks.17, 18 Ten weeks postinoculation, the mice were subjected to in vivo luciferase analysis every week to monitor lung metastasis. Following survival analysis, the remaining mice were sacrificed and the lungs were excised for hematoxylin and eosin (H&E) staining. The study was approved by the Ethical Committee of the Second Military Medical University.
DEN-Induced Rat Carcinogenesis
Wistar rats were intraperitoneally injected with DEN (Sigma-Aldrich, St. Louis, MO) at 70 mg/kg body weight once a week. After 10-week DEN administration, the rats were infused with phosphate-buffered saline (PBS), 5 × 109 pfu AdGFP, or AdHNF4α by way of a tail vein every other week. Hepatoma was detected in all rats of the model group 22 weeks after the first DEN administration, and the remaining mice were then sacrificed. Five age-matched normal rats were used as control.
Luciferase Reporter Assay
Hepatoma cells cultured in 24-well plates were cotransfected with RelA-3′ untranslated region (UTR) reporter and 400 ng/well psiCHECK2 plasmid and 20 pmol/well microRNA mimic or inhibitor using 2 μL/well Lipofectamine 2000. Renilla and firefly luciferase activities were measured by Dual-Luciferase Reporter Assay (Promega) using a luminometer (Synergy 4 Hybrid Microplate Reader, BioTek, VT). NF-κB luciferase reporter assay was performed as described previously.8
Chromatin Immunoprecipitation (ChIP) Assay
Hepatoma cells were cross-linked and processed according to the Millipore Chromatin Immunoprecipitation (ChIP) Assay Kit protocol. A mouse antihuman HNF4α monoclonal antibody (R&D Systems) or control IgG (Santa Cruz) was used for immunoprecipitation. The putative binding sites of HNF4α were analyzed by JASPAR. Real-time polymerase chain reaction (PCR) analysis was carried out using the primers listed in Supporting Table 4. At least three independent experiments were performed.
Statistics
Differences among variables were assessed by 2-tailed Student t tests. Kaplan-Meier and log-rank analysis was used to assess patient survival between subgroups. Pearson correlation analysis was performed to determine the correlation statistics between two variables. Data are presented as the mean ± SEM unless otherwise indicated. All data in this study were calculated with SPSS16.0 (SPSS). P < 0.05 was considered statistically significant.
A detailed description of the materials and methods can be found in the Supporting Information.
Results
Reduced HNF4α Expression in HCCs Predicts Poor Prognosis of Patients
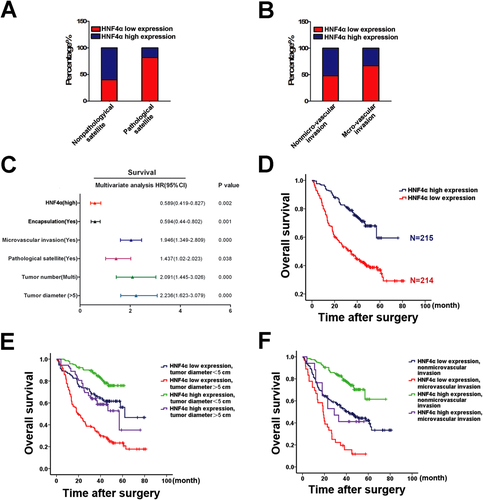
To explore the clinical significance of HNF4α expression, immunohistochemistry was performed to determine HNF4α expression in HCC tissues from 429 patients. As shown in Supporting Table 1, reduced HNF4α levels correlated well with the malignant phenotype of HCCs, in particular tumor metastasis. HCC with pathological satellites or microvascular invasion exhibited relatively low HNF4α levels (Fig. 1A,B; Supporting Fig. 1A). Multifactor Cox regression analysis revealed that HNF4α reduction is one of the independent risk factors for prediction of patient prognosis (Fig. 1C; Supporting Table 2). As expected, patients with high HNF4α expression displayed superior OS (median OS 38 and 31 months, respectively, difference >7 months, P < 0.05) and longer DFS (median DFS 32 and 25 months, respectively, difference >7 months, P < 0.05) (Fig. 1D; Supporting Fig. 1B). In addition, the combination of HNF4α expression and tumor microvascular invasion, encapsulation, tumor diameter, or tumor number significantly increased the prediction accuracy of patient prognosis (Fig. 1E,F; Supporting Fig. 1C,D). Moreover, expression of HNF4α was even less in 27 of 42 metastatic HCCs (PVTT) compared with their paired primary tumors, which further indicates the potential role of HNF4α in HCC metastasis (Supporting Fig. 1E,F).
HNF4α Suppresses Metastatic Potential of Hepatoma Cells
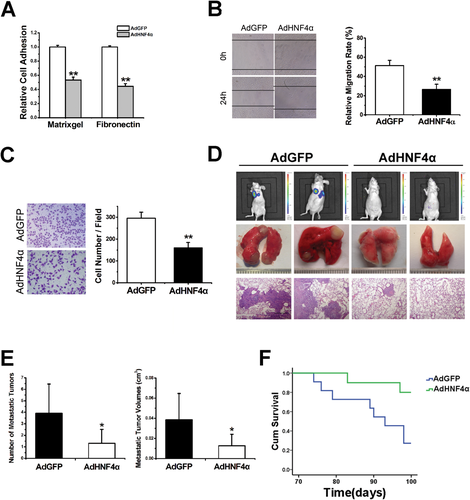
Adhesion of tumor cells to extracellular matrix (ECM) is one of the essential steps in HCC metastasis, manipulating a necessary niche for subsequent invasion. As shown in Fig. 2A and Supporting Fig. 2A, SMMC-7721 cells infected by adenovirus expressing HNF4α exhibited impaired adherence capacity to both Matrigel and fibronectin as compared with their counterparts. Moreover, the migration capability of SMMC-7721 cells with forced HNF4α expression was notably diminished (Fig. 2B,C). Hepatoma cell lines including Hep3B, LM-3, YY, and MHCC-H were also used and consistent results were achieved (data not shown). To imitate the hepatoma cells with constitutive high HNF4α expression, stable transfectant of SMMC-7721 cells expressing HNF4α (SMMC-7721 HNF4α) was constructed (Supporting Fig. 2B), and a suppressive role of HNF4α on the metastatic potential of hepatoma cells was also observed (Supporting Fig. 2C,D). SMMC-7721 cells stably expressing luciferase were established and injected into nude mice followed by systematic AdHNF4α delivery (Supporting Fig. 2E). As shown in Fig. 2D,E, HNF4α delivery significantly suppressed the lung metastasis of hepatoma cells and prolonged the survival time of animals (Fig. 2F). Moreover, the health condition of AdHNF4α-treated mice was much better than that of control mice, implying the therapeutic significance of HNF4α in HCC metastasis (Supporting Fig. 2F). In addition, a stable transfectant of SMMC-7721 cells expressing HNF4α and control cells were injected into nude mice and consistent results were achieved (Supporting Fig. 2G).
HNF4α Inhibits NF-κB Activity by Way of Suppression of RelA Expression
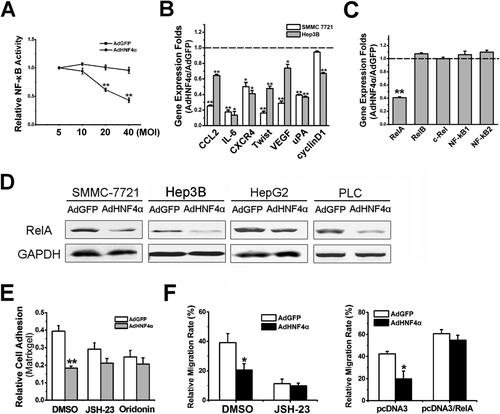
Accumulating evidence suggests that the NF-κB pathway plays a critical role in HCC development and metastasis.19, 20 To investigate the effect of HNF4α on NF-κB activation, a stable transfectant of hepatocytes expressing NF-κB luciferase reporter was used.17 Interestingly, HNF4α introduction not only suppressed NF-κB basal activity in a dose-dependent manner, but also diminished tumor necrosis factor alpha (TNF-α)-stimulated NF-κB activation, which indicates the inhibitory role of HNF4α on NF-κB signaling (Fig. 3A; Supporting Fig. 3A). More important, metastasis-associated NF-κB target genes were significantly down-regulated by HNF4α in hepatoma cells, suggesting NF-κB could be involved in the suppressive effect of HNF4α on HCC metastasis (Fig. 3B). Intriguingly, protein levels of I-κB were not increased in hepatoma cells with HNF4α overexpression compared with control cells (Supporting Fig. 3B). Moreover, Bay 11-7082, a selective inhibitor for I-κBα phosphorylation and subsequent degradation, significantly repressed NF-κB signaling and hepatoma cell migration, but failed to attenuate the inhibitory effect of HNF4α on NF-κB activation or hepatoma cell metastasis (Supporting Fig. 3C-E), suggesting that HNF4α could suppress NF-κB signaling through an IKK-independent mechanism. To further elucidate the regulation of NF-κB signaling by HNF4α, levels of NF-κB family members were determined in hepatoma cells with ectopic HNF4α expression. Among the five subunits of NF-κB, only RelA expression was dramatically repressed by HNF4α in a time-dependent manner (Fig. 3C; Supporting Fig. 3F), which was further confirmed by western blot (Fig. 3D; Supporting Fig. 3G). Consistently, inverse correlation of HNF4α and RelA was observed in various HCC cell lines (Supporting Fig. 3H). Translocation of RelA from cytoplasm to nucleus is considered a critical event during NF-κB activation. Nuclear extract analysis revealed reduced RelA protein in the nucleus of hepatoma cells infected by AdHNF4α compared with control cells (Supporting Fig. 3I). As expected, forced HNF4α expression could not impair the adhesion capacity of hepatoma cells pretreated with JSH-23 or oridonin, which are RelA-specific inhibitors that block RelA nuclear translocation or transactivation without affecting I-κB degradation (Fig. 3E; Supporting Fig. 3J).21 Consistently, HNF4α delivery exhibited no suppressive effect on the migration of hepatoma cells with RelA overexpression or blockage, indicating that HNF4α represses the hepatoma cell metastasis by way of down-regulation of RelA expression (Fig. 3F).
HNF4α Attenuates Hepatic Inflammation and Hepatoma Cell EMT
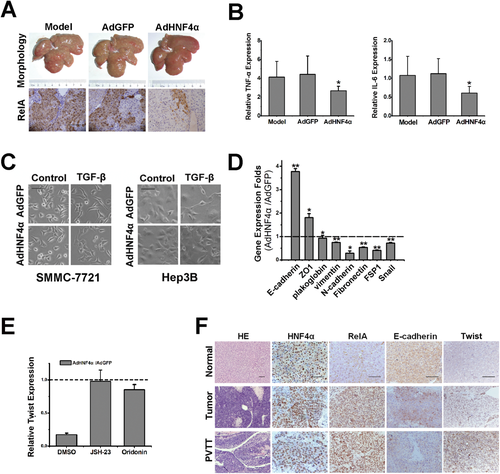
It has been accepted that inflammation plays a pivotal role in cancer initiation and progression, and NF-κB is a key mediator of hepatic inflammation. We therefore explored the influence of NF-κB suppression by HNF4α on hepatic inflammation. The pathological process of DEN-elicited rodent HCCs includes chronic inflammation, which is similar to those of human HCCs.8, 22 As shown in Supporting Fig. 4A, expression of RelA was up-regulated in rat liver concomitant with the down-regulation of HNF4α during DEN-induced hepatocarcinogenesis. Furthermore, adenovirus-mediated HNF4α delivery robustly suppressed RelA expression in the liver of rat exposed to DEN treatment (Fig. 4A). Microarray analysis suggested that expression of TNF-α and interleukin (IL)-6, predominant inflammation-associated NF-κB target genes, were transcriptionally down-regulated in the liver of rats with HNF4α delivery during DEN-induced hepatocarcinogenesis, which was further validated by real-time PCR assay (Fig. 4B; Supporting Fig. 4B). As expected, long-term DEN exposure caused hepatic damage characterized by the induction of alanine aminotransferase (ALT) and aspartate aminotransferase (AST), and subsequent inflammation was significantly ameliorated by HNF4α delivery (Supporting Fig. 4C,D). Consistently, as shown in Supporting Fig. 4E, in parallel with the attenuated inflammation and hepatic damage, compensated hepatocyte proliferation was also diminished, which is currently considered the key step of inflammation-promoted HCC development.23, 24 Morphological analysis of SMMC-7721 and Hep3B cells exposed to TGF-β, a classic EMT inducer, showed that forced HNF4α expression dramatically inhibited the EMT of hepatoma cells (Fig. 4C). As shown in Fig. 4D, hepatoma cells with ectopic HNF4α expression exhibited induction of epithelial markers, and reduction of mesenchymal markers, which was further confirmed by in vivo HNF4α delivery to xenograft tumor of mice (Supporting Fig. 4F). Similar results were achieved in SMMC-7721 and HepG2 cells (data not shown). Moreover, forced HNF4α expression not only inhibited RelA expression, but also suppressed the expression of twist (Supporting Fig. 4G,H). Consistently, overexpression of RelA remarkably enhanced twist expression in hepatoma cells (Supporting Fig. 4I). As shown in Fig. 4E, HNF4α delivery could not down-regulate twist expression in hepatoma cells with pretreatment of RelA inhibitors. HNF4α reduction in parallel with the increase of RelA/twist was observed in metastatic HCCs as compared with primary tumors (Fig. 4F; Supporting Fig. 4J,K). These data suggest that the inhibitory effect of HNF4α on HCC metastasis could be, at least partially, attributed to its suppression of hepatoma cell EMT through repression of RelA/twist cascade.
HNF4α Suppresses RelA Expression in an miR-7 and miR-124-Dependent Manner
To explore the detailed mechanism underlying the suppression of RelA by HNF4α, hepatoma cells were exposed to DNA methylation-specific inhibitor 5-AZA-CdR or histone deacylase-selective inhibitor TSA. As shown in Supporting Fig. 5A,B, neither 5-AZA-CdR nor TSA could block the down-regulation of RelA by HNF4α. Intriguingly, HNF4α delivery significantly suppressed the activity of RelA−3′UTR in hepatoma cells (Fig. 5A). It has been well documented that microRNA interacts with 3′UTR of the target genes and down-regulates their expression through induction of mRNA degradation and suppression of protein translation. JASPAR and TargetScan website prediction indicated that miR-7 and miR-124 might be target genes of HNF4α and could bind the 3′UTR of RelA. As shown in Fig. 5B and Supporting Fig. 5C, HNF4α could interact with the promoter of miR-7-2 and miR-124-1, and enhanced the expression of pri-miR-7-2 and pri-miR-124-1 in hepatoma cells. Consistently, expression of miR-7 and miR-124 was increased in hepatoma cells with ecotopic HNF4α expression (Fig. 5C). Delivery of miR-7 or miR-124 mimics dramatically inhibited the activity of RelA−3′UTR and reduced RelA expression (Supporting Fig. 5D,E). Moreover, delivery of miR-7 or miR-124 inhibitor increased RelA expression and attenuated the down-regulation of RelA by HNF4α (Fig. 5D; Supporting Fig. 5F). Point mutation of miR-7 or miR-124 binding sequence in RelA−3′UTR diminished HNF4α-mediated down-regulation of RelA−3′UTR activity (Fig. 5E). In addition, suppression of hepatoma cell migration by HNF4α was robustly affected by miR-7 or miR-124 inhibitor and completely abolished upon the combination of two inhibitors (Fig. 5F).
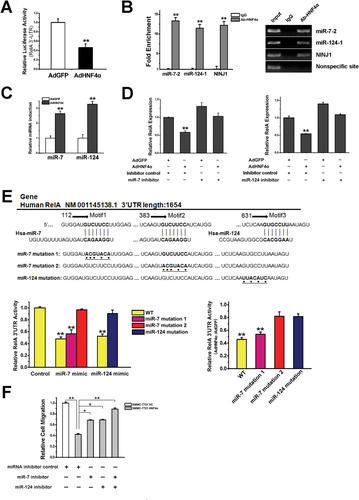
MiR-21 Is Required for the Feedback Regulation of HNF4α by NF-κB
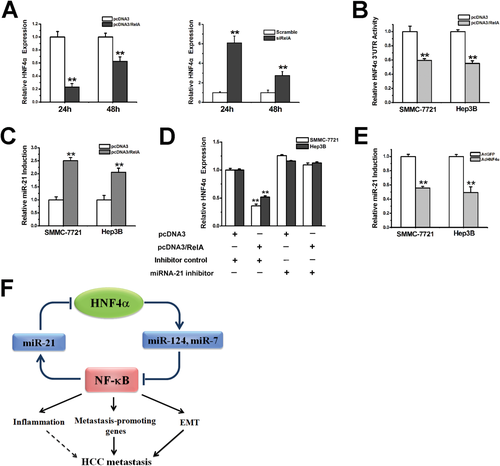
Overexpression or interference of RelA remarkably decreased or increased HNF4α expression in hepatoma cells, respectively, suggesting a feedback regulation of HNF4α by NF-κB (Fig. 6A; Supporting Fig. 6A,B). Interestingly, ecotopic RelA expression dramatically reduced the activation of HNF4α−3′UTR, indicating that microRNA could be involved in the feedback regulation (Fig. 6B). Among the nine microRNAs that were reported to regulate HNF4α expression,25 only miR-21 might be transcriptionally regulated by NF-κB according to TargetScan analysis. We then transfected RelA in hepatoma cells and found that overexpression of RelA evidently enhanced the expression of miR-21 (Fig. 6C). As shown in Supporting Fig. 6C-E, delivery of the miR-21 mimic notably suppressed HNF4α−3′UTR activity and HNF4α expression. Treatment of miR-21 inhibitor increased HNF4α levels and attenuated the down-regulation of HNF4α by RelA (Fig. 6D; Supporting Fig. 6F). Negative correlation of miR-21 and HNF4α was also observed in human HCC samples (Supporting Fig. 6G). As expected, HNF4α overexpression reduced miR-21 expression in hepatoma cells (Fig. 6E), which disclosed an HNF4α-NF-κB feedback circuit consisting miR-124, miR-7, and miR-21 involved in HCC progression (Fig. 6F).
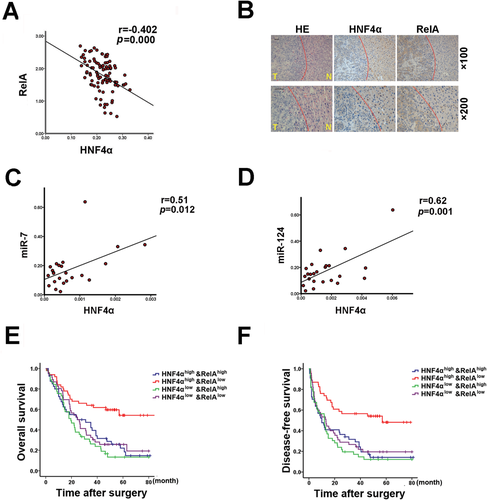
Combination of HNF4α and RelA Exhibits Improved Prognostic Accuracy for Patients
Tissue microarray analysis of a cohort of HCC specimens clearly demonstrated the inverse correlation between HNF4α and RelA in patients (Fig. 7A). Moreover, immunohistochemical assay using consecutive sections identified the reciprocal expression pattern of HNF4α and RelA in human HCCs and paracancerous tissues (Fig. 7B). As expected, HNF4α expression was positively correlated with the levels of both miR-7 and miR-124 in HCCs of patients (Fig. 7C,D). Previous laboratory studies have provided various potential biological markers to predict the clinic outcome of patients. Accumulating evidence indicates that, with appropriate combination, multiple markers might be more accurate than any one alone for predicting the prognosis of patients. In this study we found that HCC patients with elevated HNF4α expression and reduced RelA levels exhibited distinctly longer OS and DFS than those patients with reduced HNF4α expression or elevated expression of both HNF4α and RelA (Fig. 7E,F). These data further support that HNF4α suppresses HCC progression by way of the inhibition of RelA expression, and indicates that a combination of HNF4α and RelA is a more powerful predictor of good prognosis of HCC patients than HNF4α alone.
Discussion
The dismal prognosis of HCC has been attributed to postsurgical recurrence and metastasis-related de novo tumor development.26 Hepatoma cells detached from primary lesions usually invade the major branches of the portal vein, and PVTT is considered as an indication for poor prognosis of patients.27, 28 Distinct HCC-associated proteins or microRNAs have also been proposed as biomarkers for patient prognosis.29, 30 However, there are few predictive biomarkers for HCC prognosis available in the clinic to date. Herein, we for the first time report the down-regulation of HNF4α in metastatic HCC and clarify the correlation between HNF4α expression and the prognosis of patients. More important, we found that HNF4α delivery dramatically inhibited the metastasis of hepatoma cells by way of suppression of NF-κB signaling, implying its clinical significance in HCC targeted therapy. In further study, an HNF4α-NF-κB feedback circuit was identified, which indicates the essential role of HNF4α in inflammation-associated HCC initiation and progression.
HCC predominantly arises from cirrhotic liver, where there usually has been a repeated wound-healing response upon chronic inflammation.31 Chronic nonresolving inflammation is one of the distinct features of hepatocarcinogenesis. In parallel with oxidative damage-induced genetic and/or epigenetic changes, inflammation-driven compensated hepatocytes proliferation contributes to HCC development.32, 33 Substantial studies have established the unique role of NF-κB in inflammation-elicited liver cancer.34 Moreover, through up-regulation of metastasis-promoting chemokines, cytokines, and enzymes, NF-κB also functions as a critical promoter of HCC metastasis.20, 35 In the current study, AdHNF4α delivery attenuated DEN-triggered hepatic inflammation, while we did not observe expression of HNF4α in rat Kupffer cells. We speculated that this might be partially due to the suppression of IL-6 and TNFα expression in hepatocytes by AdHNF4α. Cytokines such as IL-6 produced by hepatocytes or HCC progenitor cells has also been reported to be involved in hepatocarcinogenesis.24, 36 Furthermore, long-term DEN exposure-caused hepatic damage was significantly ameliorated by HNF4 delivery, which also led to the reduced hepatocyte death and the subsequent reduction of inflammation.24, 33, 37 Herein, our data reveal that HNF4α could attenuate the production of inflammatory cytokines and chemokines, and suppress the inflammatory response of hepatoctyes or hepatoma cells through repressing NF-κB activity, which implies that the inhibitory effect of HNF4α on HCC initiation/progression could be, at least partially, due to its suppression of hepatic inflammation. More important, our results also illustrate that HNF4α remarkably inhibited the expression of NF-κB target genes including CCL2, CXCR4, μPA, and VEGF, which play pivotal roles in cancer metastasis, indicating that HNF4α could suppress HCC metastasis by way of down-regulation of NF-κB activity. Epithelial-mesenchymal transition of tumor cells is considered a central mechanism responsible for cancer metastasis.38 Weinberg and colleagues39 reported that Twist is a master regulator of morphogenesis and plays an essential role in EMT and tumor metastasis. Consistent with our previous study which demonstrated that HNF4α could attenuate hepatocyte EMT during chemical-induced hepatocarcinogenesis, herein we report that HNF4α was able to inhibit the EMT of hepanoma cells through down-regulation of Twist by way of an NF-κB-dependent manner, which at least in part accounts for the suppressive role of HNF4α on HCC metastasis.8
Activation of NF-κB signaling is a hallmark of inflammatory response of hepatocyte, and sustained NF-κB activation was considered a key event in the pathogenesis of inflammation-associated cancers, in particular HCC.23 With this report, we elucidate that HNF4α attenuates the expression and nuclear translocation of RelA, the most essential subunit of the NF-κB family, and dramatically impaired the transactivation of NF-κB. To unveil the detailed mechanism underlying the down-regulation of RelA by HNF4α, we first tested the epigenetic regulation. Treatment of hepatoma cells with TSA or 5-AZA-CdR exhibited no influence on HNF4α-mediated RelA reduction, which excludes the involvement of a histone acetylation and DNA methylation-associated mechanism. Accumulating studies have demonstrated that dysregulation of miRNA expression plays an important regulatory role in the pathogenesis of HCC development.40 Through bioinformatic analysis, we found that miR-7 and miR-124 could be regulated by HNF4α and might bind to the 3′UTR of RelA. In further study, our data demonstrated that HNF4α was able to stimulate the transcription of miR-7 and miR-124 in hepatoma cells, and both miR-7 and miR-124 could significantly repress RelA expression and NF-κB activation. Taken together, we clarified a critical link between HNF4α and NF-κB, which are two of the most important mediators in chronic liver disease.
Recent studies posit that microRNAs and transcription factors may cooperate to tune gene expression in distinct biological and pathological processes.41 The microRNA-transcription factor feedforward or feedback circuit plays an important regulatory role in carcinogenesis.42 Herein we found that miR-21, an important microRNA in solid tumor, was transcriptionally regulated by NF-κB and could down-regulate HNF4α expression in hepatoma cells. Collectively, an HNF4α-NF-κB feedback circuit consisting of specific miRNAs was disclosed in HCC. NF-κB activation during chronic hepatic inflammation repressed HNF4α expression by way of miR-21 induction, which contributes to the hepatocyte dedifferentiation and hepatocarcinogenesis. Down-regulation of HNF4α in hepatoma cells enhances NF-κB activation through miR-124 and miR-7-dependent machinery, which facilitates the metastasis of hepatoma cells. Considering the importance of HNF4α and NF-κB in chronic liver disease, this feedback circuit is not only essential in connecting hepatic inflammation and tumorigenesis, but also indicates a novel strategy in HCC prevention and therapy.




