- 1
There were different rates of viral genotype 3 in the two studies (15.2%1 versus 9.2%2). According to McCarthy et al., patients infected with genotype 3 have the highest rate of the CC genotype.
- 2
There were differences in the sample sizes of the non-G1–infected groups (66/2761 versus 144/6492). These differences affected the 95% confidence intervals for the rates (54.0%-77.8%1 versus 41.6%-58.4%2).
- 3
There were differences in the genetic backgrounds of the populations (Spanish versus American Caucasian). In fact, the frequency of CC was also slightly higher among our G1-infected patients (39.1%1 versus 33.5%2).
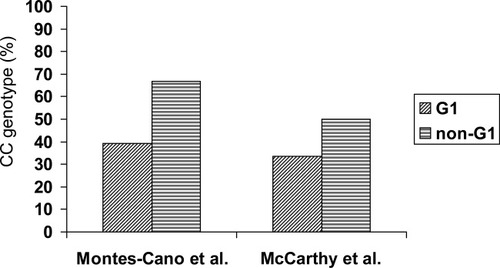
Rates of the IL-28B rs12979860CC genotype among patients infected with G1 HCV or non-G1 HCV in two different studies.
In any case, it is important to emphasize that this interesting finding has been reported by independent teams working with two different HCV patient cohorts. Analyzing our non-G1 data with their model, Lindh et al. remarked that “a 67% CC genotype rate … should not develop in an uninfected population with a 45% CC genotype rate … unless the clearance rate in patients carrying a non-CC genotype is higher than that in the CC genotype group.” In our opinion, their model is not entirely appropriate because it does not take into account the different 95% confidence intervals for the rates: the interval for our non-G1 group (54.0%-77.8%) is larger than the interval for our uninfected group (39.6%-49.9%). Lindh et al.'s model suggests a counterintuitive hypothesis similar to that proposed by McCarthy et al., which is based on an interaction model between IL-28B and viral genotypes. This explanation would be unexpected a priori because the IL-28B rs12979860CC genotype and viral genotypes 2 and 3 are good predictors of treatment-induced viral clearance. Moreover, IL-28B rs12979860CC is associated with spontaneous viral clearance, whereas the viral genotype has not been consistently described as a predictor of spontaneous resolution.3 Other explanations for this unexpected overrepresentation of rs12979860CC among non-G1–infected patients, such as epidemiological, virological, and/or immunological factors promoting a positive selection of individuals bearing the rs12979860CC genotype among non-G1–infected patients, are possible. Therefore, further studies in patient cohorts and in vitro assays are needed to test these hypotheses.
References
Marco Antonio Montes-Cano M.D.*, José Raúl García-Lozano M.D.*, Cristina Abad-Molina M.D.*, Manuel Romero-Gómez M.D. , Natalia Barroso M.D. , José Aguilar-Reina M.D. , Antonio Núñez-Roldán M.D.*, María Francisca González-Escribano M.D.*, * Servicio de Inmunología ,Hospital Universitario Virgen del ocío/Instituto de Biomedicina de Sevilla Seville, Spain, Sección Hepatología Hospital Universitario de Valme Seville, Spain, Servicio de Digestivo Hospital Universitario Virgen del Rocío Seville, Spain.




