Influence of a single nucleotide polymorphism in the P1 promoter of the furin gene on transcription activity and hepatitis B virus infection†
Potential conflict of interest: Nothing to report.
Abstract
Hepatitis B e antigen (HBeAg) is a viral strategy of immune response evasion associated with hepatitis B virus (HBV) persistence. Spontaneous HBeAg seroconversion is usually accompanied by liver disease remission. Unfortunately, this goal is difficult to achieve and requires expensive and time-consuming treatment. Furin, a proprotein convertase, is involved in HBeAg maturation and is therefore a potential therapeutic target or indicator for predicting disease progression and antiviral response. Here we demonstrate that healthy Han Chinese from southern China (an endemic area of HBV infection) harbor a common single nucleotide polymorphism (SNP; −229 C/T) in a 1268-bp region of the P1 promoter of the furin gene [FES upstream region (Fur)]. A luciferase reporter gene assay showed that transcription activity is about 3 times higher in allele T carriers than in allele C carriers of this SNP. Allele T includes a suboptimal transcription factor NF-E2 [i.e., nuclear factor (erythroid-derived 2)]–binding motif according to bioinformatics and studies using site-directed mutagenesis. We also observed that individuals carrying allele T were more likely to become persistently infected. When persistently infected patients were divided into subgroups according to recent guidelines and HBeAg-defective virus infection was taken into account, patients with allele T or genotype TT had a decreased likelihood of HBeAg seroconversion or an increased likelihood of progressing to HBeAg-negative chronic hepatitis B or liver cirrhosis if accompanied by HBeAg-defective virus infection. Conclusion: The common SNP in the P1 promoter of the Fur gene affects furin transcription activity and HBV infection outcome, possibly by increasing furin messenger RNA expression, and this suggests that furin is a potential therapeutic target and that this SNP is a potential predictor of disease progression or therapeutic response. (HEPATOLOGY 2009.)
The clinical outcome of hepatitis B virus (HBV) infection varies from acute, self-limited hepatitis to persistent infection that may progress to cirrhosis or hepatocellular carcinoma.1 Natural history studies have linked poor prognosis to persistent hepatitis B e antigen (HBeAg) or increased serum alanine aminotransferase levels after HBeAg seroconversion.2-4 Most patients with HBeAg-positive chronic hepatitis B (CHB) become inactive hepatitis B surface antigen (HBsAg) carriers (IHCs) with low levels of serum HBV DNA and normal liver function profiles after HBeAg seroconversion.1, 5, 6 HBeAg seroconversion is also an important therapeutic milestone for patients with HBeAg-positive CHB, and to reach it requires an expensive and prolonged therapeutic course.7, 8
It has been postulated that HBeAg subverts the host immune response to establish chronic HBV infection.9, 10 The precore region of HBV encodes a secretion sequence that signals translocation of the HBeAg precursor into the endoplasmic reticulum. There, 19 amino acids in the precore region are cleaved by signal peptidase. The products then bind to the endoplasmic reticulum membrane and are transported to the Golgi apparatus. Some are further cleaved by furin, a subtilisin-like proprotein convertase, and secreted as an immunosuppression modulator9-12; the rest are transported to the plasma membrane, where they become targets of antiviral immunity.13, 14 Because HBeAg production in vitro is significantly influenced by furin activity when either down-regulated by the furin inhibitor or up-regulated by the supertransfecting furin encoding gene11, 15 and the cleaved and uncleaved products have opposite effects on the immune system, interindividual differences in furin activity may significantly influence HBV infection outcome.
The gene of furin [FES upstream region (Fur)] is located in chromosome 15q26.1. At least three distinct promoters (P1, P1A, and P1B) mediate transcription of the Fur gene. The messenger RNAs (mRNAs) differ in their 5′ end but are all translated from the same AUG to generate identical furin proteins.16, 17 The P1 promoter is the most active promoter in hepatocytes and contributes to the high level of furin mRNA expression in liver.16, 18 The P1 promoter has TATA and putative CCAAT elements in the proximal region16 Six single nucleotide polymorphisms (SNPs) in and around the P1 promoter have been registered in the Single Nucleotide Polymorphism Database (dbSNP) of the National Center for Biotechnology Information (accessed July 14, 2008 at http://www.ncbi.nlm.nih.gov). However, their distribution in large populations, effects on transcription activity, and relationship to given diseases have not been well studied. To date, there is one other report of the furin promoter SNP, and it found no correlation to cardiac allograft vasculopathy formation.19
In recent years, growing data have demonstrated that HBV infection outcome is significantly influenced by interindividual differences in human genetics.20, 21 It has been further linked to differences in SNPs of many genes.22-26 Thus, this study examined the effects of a common SNP in the P1 promoter of the Fur gene on transcription activity and HBV infection outcome. The results suggest that furin plays an important role in HBV infection and may even be a promising therapeutic target in the future.
Abbreviations
BCP, basal core promoter; CD-PCR, competitively differentiated polymerase chain reaction; cDNA, complementary DNA; CHB, chronic hepatitis B; CHC, chronic hepatitis B virus carrier; dbSNP, Single Nucleotide Polymorphism Database; DIG, digoxigenin; FITC, fluorescein isothiocyanate; Fur, FES upstream region; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; HRP, horseradish peroxidase; IHC, inactive hepatitis B surface antigen carrier; LC, liver cirrhosis; mRNA, messenger RNA; ND, not determined; NF-E2, nuclear factor (erythroid-derived 2); PCR, polymerase chain reaction; SNP, single nucleotide polymorphism.
Patients and Methods
Populations.
- 1
Chronic HBV carriers: individuals without clinical symptoms but with HBeAg and a high level of HBV DNA in serum, a normal serum aminotransferase level, and other liver function tests within normal limits for at least 1 year.
- 2
Patients with CHB: individuals with clinical symptoms, elevated serum aminotransferase levels, and/or abnormalities of other liver functions.
- 3
IHCs: individuals without HBeAg (with antibody to hepatitis B e antigen mostly) in serum, without clinical symptoms, and with normal serum aminotransferase levels and other liver function tests within normal limits for at least 1 year.
- 4
Patients with HBsAg-related liver cirrhosis (LC): individuals with serum HBsAg, abnormal liver function profiles, and portal hypertension detected by ultrasound scanning and barium swallow study.
Furthermore, CHB and LC were divided into HBeAg-positive and HBeAg-negative forms. Persistent HBV infection was divided into four phases: immune tolerant phase, immune response phase, inactive carrier state phase, and reactivation phase. These are also designated phases I, II, III, and IV in this study.
Genomic DNA Isolation.
Whole blood was collected from all participants and centrifuged to separate the plasma for HBV marker detection. The cell pellet was resuspended in a volume of normal saline equal to the plasma volume. Genomic DNA was extracted from 200 μL of the cell suspension with Omega kits (Omega-Tek, Mansfield, OH). All DNA preparations were stored at −70°C until further use.
Polymerase Chain Reaction (PCR) Direct Sequencing.
To obtain a 1268-bp fragment consisting of the P1 promoter and its upstream sequence of the Fur gene (GenBank accession no. NC 000015), genomic DNAs from 84 healthy individuals were amplified with primers Furin-PCRS1 and Furin-PCRA1 (see Table 1). The PCR bands of interest were excised from agarose gel, and the DNA fragments were purified with gel extraction kits (Omega-Tek). The purified fragments were sequenced with sense (Furin-DSS1) and antisense (Furin-DSA2) sequencing primers from Shanghai Invitrogen Biotechnology Co., Ltd. (Shanghai, China).
| Denomination | DNA Sequence |
|---|---|
| Furin-PCRS1 | 5′-ACCACCATCCTGTGACGTTCC |
| Furin-PCRA1 | 5′-CTGCTGCTTCCCCGCCACTC |
| Furin-DSS1 | 5′-AAGGGACATTTGGCAATACC |
| Furin-DSA2 | 5′-TCCAGTTCTGGCTAAACCAC |
| Furin-P2CC | 5′-GCAAGCTTAGGCAGAAGGTAAGTGCAGACCC |
| Furin-P2TC | 5′-GCAAGCTTAGGCAGAAGGTAAGTGCAGACTC |
| Furin-PS | 5′-CAGAGAATTATCCGACCCAAGATG |
| Furin-PA | 5′-CTGCTGCTTCCCCGCCACTC |
| Furin-P2C | 5′-DIG-CTGACGCATCAGCGTGGCAGAAGGTAAGTGCAGAgC |
| Furin-P2T | 5′-FITC-GTCCGTCGTGCTACTGGCAGAAGGTAAGTGCAGAgT |
| Furin-P2A | 5′-TGGGTGCTGCTGCAACAGTCAGGCTCCTATCC |
| Furin-P2P | 5′-Amino linker-C6-GTGATGTGGTGGTGACATATCTGGTCGCAC |
| Furin-PCS | 5′-CGACGCGTAAGGGACATTTGGCAATACC |
| Furin-PCA | 5′-CCCAAGCTTTCCAGTTCTGGCTAAACCAC |
- The sequences were derived from the furin gene (GenBank accession no. NC 000015:6375561-6394064). The italic characters are the differentiating sequences of no relationship to the sequence of the furin gene. The nucleotides marked by a single line are loci of restriction endonucleases. The nucleotides marked by double lines are polymorphism nucleotides. The lowercase letters indicate mismatched nucleotides designed to increase the selective efficiency of the primers.
- Abbreviations: DIG, digoxigenin; FITC, fluorescein isothiocyanate.
Transcription Activity Assays.
The fragments carrying the C or T allele based on SNP −229 C/T [the location was determined from the genomic sequence (GenBank accession no. NC 000015) and the mRNA sequence (GenBank accession no. NM 002569)] were cloned into pGL3-Basic (Promega, Madison, WI) to generate pFur-229C and pFur-229T. After being confirmed by DNA sequencing, these constructs were then transfected into HepG2, HuH7, and HeLa cells. Cells were transfected with the FuGENE HD transfection reagent (Roche Applied Science, Indianapolis, IN); according to the manufacturer's protocol, 600 ng/well of the pGL3 construct and 100 ng/well of pRL-tk (Renilla luciferase expression construct; Promega) were used for normalization of the transfection efficiencies. Each construct was transfected four times in triplicate. The firefly luciferase reporter and Renilla luciferase internal control activities were measured with a Turner Designs luminometer (model TD-20/20) with the Dual-Luciferase reporter assay system (Promega).
To further test the effect of the C and T type promoter on the transcription of the Fur gene itself, we generated two constructs, pFur-229C-Fur and pFur-229T-Fur, by inserting a complete complementary DNA (cDNA) copy of furin into pFur-229C and pFur-229T at their Hind III sites (between the P1 promoter and the reporter gene). After being confirmed by DNA sequencing, the constructs and pGL-3-Basic were then transfected into HepG2 cells with the FuGENE HD transfection reagent (Roche Applied Science). After 24 hours of culturing, total RNA was isolated with the TRIZOL reagent (Invitrogen), and then the isolate was treated with DNase to remove contaminating DNA. First-strand cDNA was synthesized with M-MuLV reverse transcriptase (Fermentas). Real-time PCR was performed in triplicate with furin primers (forward primer, CCTACAGCAGTGGCAACCAGAA; reverse primer, CCTCCAGGGTGAGAGCAATGA) in an ABI-Prism 7000 sequence detection system with the Power Syber Green kit (Applied Biosystems). The ΔΔCt method was used to semiquantify Fur mRNA levels according to the manufacturer's protocol. Glyceraldehyde 3-phosphate dehydrogenase was used as the internal standard for normalization.
Site-Directed Mutagenesis.
pFur-229C and pFur-229T, generated from pGL3-Basic, were used to construct site-directed mutants. Oligonucleotides were custom-synthesized by Shanghai Invitrogen Biotechnology Co., Ltd. Site-directed mutants were generated with the QuikChange Lightning site-directed mutagenesis kit (Stratagene, La Jolla, CA), and the resulting constructs were confirmed by DNA sequencing by Shanghai Invitrogen Biotechnology Co., Ltd.
Genotyping of the Common SNP in the P1 Promoter of the Fur Gene.
Individuals with self-limited or persistent HBV infections were genotyped for SNP −229 C/T with our previously published method of competitively differentiated polymerase chain reaction (CD-PCR).28 This technique is a specific and inexpensive method for SNP genotyping.25, 26 Briefly, primers Furin-P2CC (for allele C) and Furin-P2TC (for allele T; see Table 1) were used to construct positive control recombinant plasmids. Conventional PCR was performed with primers Furin-PS and Furin-PA. The reaction mixture and the cycling conditions were similar to those in our previous reports. CD-PCR was performed with primers Furin-P2C, Furin-P2T, and Furin-P2A. The reaction mixture and the cycling conditions were also similar to those of our previous reports. CD-PCR products were hybridized with solidified probe Furin-P2P in two wells of a microtiter plate. Horseradish peroxidase–labeled anti-digoxigenin (Roche Diagnostic, Ltd.) and then horseradish peroxidase–labeled anti–fluorescein isothiocyanate (Roche Diagnostic) were added to these wells. The assay of the color reaction was as previously reported. PCR direct sequencing was performed in about 1% of randomly selected samples to evaluate the accuracy of the CD-PCR results.
Detection of HBeAg-Defective Variants.
The HBeAg-defective variants are mainly characterized by one G-to-A substitution at nucleotide 1896 (G1896A) and by a dual mutation (A1762T, G1764A) in the basal core promoter (BCP) region. These variants were detected in serum samples from patients with HBeAg-negative CHB or LC by CD-PCR performed according to a previously published protocol.28
Statistical Analyses.
The difference in the allele distribution and SNP −229 C/T genotype distribution was statistically analyzed with the χ2 test or Fisher's exact probability analysis. The age, liver function profiles, and relative luciferase activity were analyzed with the Student t test. P < 0.05 was considered statistically significant. All statistical analyses were conducted with SPSS software (version 11).
Results
SNPs in the P1 Promoter of the Fur Gene in Non–HBV-Infected Individuals.
The P1 promoter fragments of the Fur gene in all 84 non–HBV-infected individuals were successfully amplified and had the expected molecular weight. Although six SNPs have been registered for the DNA sequence of this fragment, only two, −229 C/T (rs4932178) and −660 –/A (rs34397663), were actually found, and only −229 C/T was common. The typical performance parameters used in PCR direct sequencing are shown in Fig. 1A. The sequence shown in Fig. 1A is antisense to the Fur gene (genotypes CC, CT, and TT are expressed as GG, GA, and AA) since optimal sequencing results are obtained with an antisense sequencing primer because SNP −229 C/T is located in the 3′ terminal of the fragment. The frequencies of alleles C and T were 80.9% (136/168) and 19.1% (32/168), and the distribution frequencies of genotypes CC, CT, and TT were 67.9% (57/84), 26.2% (22/84), and 5.9% (5/84) in these individuals, respectively. Their genotype distribution was in Hardy-Weinberg equilibrium.
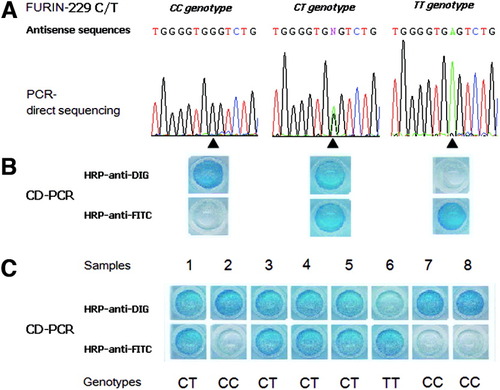
Single nucleotide polymorphism −229 C/T in the P1 promoter of the Fur gene was genotyped with (A) PCR direct sequencing and (B,C) CD-PCR. The sequence here is antisense to the Fur gene. That is, genotypes CC, CT, and TT are expressed as GG, GA, and AA in sequencing. Numbers 1 to 8 indicate the different samples. Abbreviations: CD-PCR, competitively differentiated polymerase chain reaction; HRP, horseradish peroxidase; DIG, digoxigenin; FITC, fluorescein isothiocyanate; Fur, FES upstream region; PCR, polymerase chain reaction.
Transcription Activity of the P1 Promoter Carrying Allele C or T.
The pGL3 constructs (pFur-229C and pFur-229T) containing either allele C or T were successfully cloned (Fig. 2A). To assess the impact of these two alleles of SNP −229 C/T on transcription, two hepatocyte-derived cell lines (HepG2 and HuH7) and one non–hepatocyte-derived cell line (HeLa) were used. Compared with pFur-229C, the pFur-229T construct stimulated transcription activity 3.02-fold (t = 12.09, P < 0.0001), 2.97-fold (t = 4.46, P < 0.001), and 3.23-fold (t = 14.99, P < 0.0001) in HepG2, HuH7, and HeLa cells, respectively (Fig. 2B). When furin cDNA was cloned directly after the C or T type of P1 promoter, the furin mRNA level of pFur-229T-Fur was 2.29 times that of pFur-229C-Fur (Fig. 2C; the 2−ΔΔCt values of each construct against an empty control were compared). On the basis of ΔCt, they had significant differences (−0.61 ± 0.08 versus 0.57 ± 0.16, t = −15.09, P = 0.0001).
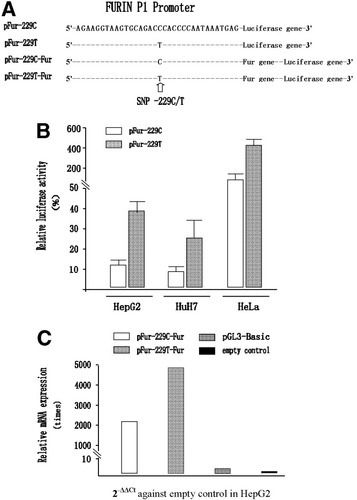
Characterization of the promoter activity of −229C and −229T alleles based on the P1 promoter of the Fur gene from different cell lines. (A) A description of alleles C and T (pFur-229C and pFur-229T) of the P1 promoter placed into the luciferase reporter vector pGL3-Basic. (B) An assessment of the potential of allele T within the P1 promoter within different human cell lines with the luciferase reporter assay system. There was an approximately 3-fold increase in the transcriptional activity of the allele T promoter (pFur-229T) in comparison with pFur-229C in these cell lines. (C) An assessment of the transcription effect of allele T within the P1 promoter on the furin gene itself. The 2−ΔΔCt values of each construct against an empty control were compared. The mRNA level of the furin gene of pFur-229T-Fur was 2.29 times that of pFur-229C-Fur. Abbreviations: Fur, FES upstream region; mRNA, messenger RNA; SNP, single nucleotide polymorphism.
Transcription Factor Binding of the P1 Promoter Carrying Allele C or T.
Although there is a putative CCAAT box near the SNP, no transcription factor binds to the motif according to a Web site (www.cbrc.jp/research/db/tfsearch.html). However, it indicates that the T type P1 promoter of the Fur gene is more efficient for binding to the transcription factor NF-E2 [i.e., nuclear factor (erythroid-derived 2); Fig. 3A]. To test this possibility, we generated eight constructs with an optimized NF-E2 binding element (pFur-M1, pFur-M2, pFur-M5, and pFur-M6) or destroyed one (pFur-M3 and pFur-M7) and destroyed the CCAAT box (pFur-M4 and pFur-M8) with site-directed mutagenesis based on pFur-229C and pFur-229T, respectively (Fig. 3A). The luciferase reporter assay showed that the transcription activities of the P1 promoter carrying alleles C and T were significantly decreased in 75.0% (Fig. 3B; pFur-M3 versus −229C, P = 0.002) and in 40.6% (Fig. 3B; pFur-M7 versus −229T, P = 0.015), respectively, by NF-E2 element destruction, but they were not significant reduced by CCAAT box destruction (Fig. 3B; pFur-M4 versus −229C, P = 0.291; pFur-M8 versus −229T, P = 0.231). On the other hand, although those carrying optimized allele C, pFur-M1 and pFur-M2, had comparable transcription activities (Fig. 3B; 98.4% and 190.6% versus pFur-229C, for both P > 0.05), those carrying optimized allele T, pFur-M5 and pFur-M6, had significantly increased transcription activities (Fig. 3B; increased 3.5 times and 15.6 times versus pFur-229T, for both P < 0.01). These results suggest that the higher transcription activity of the T type of P1 promoter is possibly based on its higher NF-E2 binding activity and adjacency to the CCAAT box. The T nucleotide at position −229 seems to play an important role in NF-E2 binding.
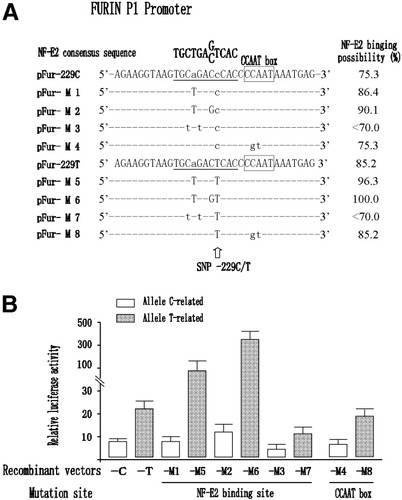
Characterization of the transcription factor binding manner of the P1 promoter carrying allele C or T in position −229 of the Fur gene in the HepG2 cell line with site-directed mutagenesis. (A) A description of the constructs based on pGL3-Basic with a more efficient NF-E2 binding element (pFur-M1, pFur-M2, pFur-M5, and pFur-M6) or damage to one of them (pFur-M3 and pFur-M7) and to the CCAAT box (pFur-M4 and pFur-M8) was generated with site-directed mutagenesis based on pFur-229C and pFur-229T, respectively. The binding possibility was calculated with the engine on a Web site (www.cbrc.jp/research/db/tfsearch.html). (B) Assessment of the potential of the constructs from pGL3-Basic. The difference in transcription activity between pFur-229C and pFur-229T was completely abolished by the damage to the NF-E2 element (pFur-M3 versus pFur-M7) but was not influenced by the damage to the CCAAT box (pFur-M4 versus pFur-M8). Transcription activities were all increased by optimization of the efficiency of binding NF-E2 in the P1 promoter carrying both allele C and allele T (pFur-M1 and pFur-M2 versus pFur-229T; pFur-M5 and pFur-M6 versus pFur-229C). Abbreviations: Fur, FES upstream region; NF-E2, nuclear factor (erythroid-derived 2); SNP, single nucleotide polymorphism.
SNP −229 C/T in Individuals with Self-Limited or Persistent HBV Infections.
SNP −229 C/T from individuals with self-limited and persistent HBV infection was successfully genotyped with CD-PCR. In agarose gel electrophoresis, the molecular weights of the products of basic PCR and CD-PCR were as expected. The CC, CT, and TT genotypes identified by PCR direct sequencing conformed to those genotyped by CD-PCR (Fig. 1B). The results for the rest of the samples were very recognizable (Fig. 1C). The genotyping results for selected samples were confirmed by PCR direct sequencing. The genotyping results for individuals with self-limited and persistent HBV infections are shown in Table 2. The overall frequencies of alleles C and T were 76.1% (932/1224) and 23.9% (292/1224). The frequencies of genotypes CC, CT, and TT were 61.1% (374/612), 30.1% (184/612), and 8.8% (54/612). The genotype distributions of SNP −229 C/T in these populations were not in complete Hardy-Weinberg equilibrium.
| Fur Gene (−229 C/T) | Non–HBV-Infected Individuals | Self-Limited Infection, n = 112 (%) | Persistent Infection | ||||
|---|---|---|---|---|---|---|---|
| Total, n = 500 (%) | CHC, n = 100 (%) | IHC, n = 106 (%) | CHB, n = 114 (%) | LC, n = 180 (%) | |||
| Allele | |||||||
| C | 136 (80.9) | 184 (82.1) | 748 (74.8) | 151 (75.5) | 170 (80.2) | 156 (68.4) | 271 (75.3) |
| T | 32 (19.1) | 40 (17.9) | 252 (25.2)* | 49 (24.5) | 42 (19.8) | 72 (31.6)† | 89 (24.7) |
| Genotype | |||||||
| CC | 57 (67.9) | 77 (68.8) | 297 (59.4) | 59 (59.0) | 67 (63.0) | 59 (51.8)‡ | 112 (62.2) |
| CT | 22 (26.2) | 30 (28.8) | 154 (30.8) | 33 (33.0) | 36 (34.0) | 38 (33.3) | 47 (26.1) |
| TT | 5 (5.9) | 5 (4.5) | 49 (9.8) | 8 (8.0) | 3 (2.8) | 17 (14.9)§ | 21 (11.7)∥ |
- Non–HBV-infected individuals were individuals without serum markers of HBV infection. Self-limited infection includes patients positive for antibody to hepatitis B surface antigen and antibody to hepatitis B core antigen in serum. Persistent infection indicates patients with hepatitis B surface antigen in the serum for more than 6 months.
- Abbreviations: CHB, chronic hepatitis B; CHC, chronic hepatitis B virus carrier; HBV, hepatitis B virus; IHC, inactive hepatitis B surface antigen carrier; LC, liver cirrhosis; SNP, single nucleotide polymorphism.
- * Compared with the self-limited infection group, χ2 = 5.43 and P = 0.019.
- † Compared with the self-limited infection group, χ2 = 11.41 and P = 0.001, and compared with the inactive CHB patients, χ2 = 8.24 and P = 0.004.
- ‡ Compared with the self-limited infection group, χ2 = 6.81 and P = 0.009.
- § Compared with the self-limited infection group, χ2 = 7.02 and P = 0.008, and compared with the inactive CHB patients, χ2 = 9.70 and P = 0.002.
- ∥ Compared with the self-limited infection group, χ2 = 4.42 and P = 0.036, and compared with the inactive CHB patients, χ2 = 6.78 and P = 0.009.
The genotype distribution of SNP −229 C/T in individuals with different clinical outcomes is shown in Table 2. Compared with individuals with self-limited HBV infection, patients with persistent infection were significantly more likely to carry allele T (252/1000 versus 40/224, χ2 = 5.43, P = 0.019) and less likely to carry allele C (748/1000 versus 184/224, χ2 = −5.43, P = 0.019). When persistently infected patients were divided into subgroups according to recent guidelines, a greater frequency of allele T in CHB patients (72/228 versus 40/224, χ2 = 11.41, P = 0.001) or genotype TT in patients with CHB (17/114 versus 5/112, χ2 = 7.02, P = 0.008) and LC (21/180 versus 5/112, χ2 = 4.42, P = 0.036) was found in comparison with individuals with self-limited HBV infection. The distribution frequencies of genotype TT were also significantly higher in patients with CHB and LC than in IHCs. However, the distributions of both allele and genotype frequencies in non–HBV-infected individuals and patients with self-limited HBV infection were similar (see Table 2). A possible explanation is that their case numbers were limited.
HBeAg-Defective Virus in Patients with HBeAg-Negative CHB or LC.
The association of the major HBeAg-defective viruses with G1896A and the BCP double variant is shown in Table 3. G1986A and the BCP double variant were detected in 58.2% (96/165) and 38.8% (64/165), respectively, and HBeAg-defective virus was detected in 89.7% (148/165) of patients with HBeAg-negative CHB or LC.
| Phases of HBV Infection | Phase I, n = 100 (%) | Phase II, n = 129 (%) | Phase III, n = 106 (%) | Phase IV, n = 165 (%) |
|---|---|---|---|---|
| Age (years) | 32.5 ± 10.2 | 39.6 ± 12.3 | 37.8 ± 11.9 | 42.9 ± 13.3* |
| Gender (male/female) | 90/10 | 110/17 | 95/11 | 153/13 |
| Clinical diagnosis | CHC | CHB/LC | IHC | CHB/LC |
| HBeAg state | + | + | − | − |
| HBeAg-defective virus | ||||
| G1896A (+) | ND | ND | ND | 96 (58.2) |
| BCP double variant (+) | ND | ND | ND | 64 (38.8) |
| One or both (+) | 148 (89.7) | |||
| SNP −229 C/T | ||||
| Allele C | 151 (75.5) | 184 (71.3) | 170 (80.2) | 243 (73.6) |
| Allele T | 49 (24.5) | 74 (28.7) | 42 (19.8)* | 87 (26.4) |
| Genotype CC | 59 (59.0) | 74 (57.4) | 67 (63.0) | 97 (58,8) |
| Genotype CT | 33 (33.0) | 36 (27.9) | 36 (34.0) | 49 (29.7) |
| Genotype TT | 8 (8.0) | 19 (14.7) | 3 (2.8)† | 19 (11.5) |
- Abbreviations: BCP, basal core promoter; CHB, chronic hepatitis B; CHC, chronic hepatitis B virus carrier; Fur, FES upstream region; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; IHC, inactive hepatitis B surface antigen carrier; LC, liver cirrhosis; ND, not determined; SNP, single nucleotide polymorphism.
- * Compared with the phase II group, χ2 = 4.93 and P = 0.027.
- † Compared with the phase II group, χ2 = 9.71 and P = 0.002, and compared with the phase IV group, χ2 = 6.53 and P = 0.011.
Relationship Between the Genotype Distribution of SNP −229 C/T and the Status of HBeAg.
Allele T and genotype TT were less frequent in individuals with self-limited infection and in IHCs, providing only indirect evidence that SNP −229 C/T affects HBeAg maturation or seroconversion. To obtain direct evidence, the relationship between SNP −229 C/T genotype distribution and HBeAg status was investigated (see Table 3). Among 500 patients with persistent HBV infection, 129 carried HBeAg in the serum. The genotype distribution of SNP −229 C/T was not significantly different between patients with and without HBeAg. However, when the four phases of HBV persistent infection and the presence of HBeAg-defective virus were considered, IHC patients (in phase III without HBeAg in serum) had a significantly lower frequency of allele T and genotype TT (see Table 3). Compared with patients in phase III, patients in phase II (with HBeAg in serum) and patients in phase IV (without HBeAg in serum but with HBeAg-defective virus infection) had a significantly higher frequency of genotype TT of SNP −229 C/T. However, the allele or genotype distribution between them was similar (see Table 3).
Relationship Between SNP −229 C/T Distribution and Liver Function Profiles.
Patients with LC were inpatients and had well-recorded clinical data and below-normal levels of albumin and prothrombin (important secretion products of hepatocytes). Many papers suggest that the maturation of both albumin and prothrombin is associated with furin. Therefore, determining the effect of SNP −229 C/T on the serum levels of albumin and prothrombin may help determine its function. All variables of interest, including liver function tests listed in Table 4, had no influence on the allele or genotype distribution of SNP −229 C/T.
| Demographic Data and Main Liver Function Profiles | SNP −229 C/T Genotype | ||
|---|---|---|---|
| CC, n = 112 | CT, n = 47 | TT, n = 21 | |
| Age (years) | 46.09 ± 10.78 | 46.96 ± 11.35 | 47.62 ± 10.83 |
| Gender (male/female) | 105/7 | 39/8 | 21/0 |
| HBeAg-positive cases (%) | 27 (24.1) | 9 (19.1) | 7 (33.3) |
| HBV DNA level (log10 copies/mL) | 5.21 ± 1.57 | 4.85 ± 1.30 | 5.00 ± 1.33 |
| Alanine aminotransferase (IU/L) | 267.82 ± 517.88 | 89.15 ± 149.50 | 274 ± 429.50 |
| Aspartate aminotransferase (IU/L) | 271.37 ± 452.23 | 112.96 ± 133.12 | 271.62 ± 271.47 |
| Total bilirubin (μmol/L) | 217.72 ± 205.90 | 146.92 ± 203.52 | 162.24 ± 157.76 |
| Serum albumin (g/L) | 32.45 ± 5.57 | 32.65 ± 5.08 | 30.77 ± 7.39 |
| Prothrombin time (seconds) | 23.81 ± 7.18 | 20.36 ± 6.37 | 22.42 ± 7.24 |
- Abbreviations: HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; SNP, single nucleotide polymorphism.
Discussion
The natural history of HBV infection is generally determined by the interplay between virus replication and the host immune response.27 However, much evidence suggests that genetic factors in humans regulate both virus replication and the host's immune response.20, 21 SNPs are genetic factors that determine interindividual differences in disease progression by influencing viral infection or the vigor of the immune response.26, 29, 30 In this study, a common SNP (−229 C/T) in the P1 promoter of the Fur gene had a significant influence on gene transcription activity and HBV infection outcome. Individuals with allele T or genotype TT had a higher risk of developing persistent HBV infection with HBeAg in the serum or of developing HBeAg-negative CHB or LC with HBeAg-defective virus infection.
Six SNPs are registered in the dbSNP for the 1268-bp region of the P1 promoter of the Fur gene. In 84 healthy Han Chinese individuals, −229 C/T was the only common SNP (with an allele C frequency of 80.9% and an allele T frequency of 19.1%; this was similar to the distribution reported in the dbSNP). Up to now, the function of this SNP has been unclear. The luciferase reporter gene assay in this study showed that transcription activity is about 3 times higher in samples with allele T than in samples with allele C. When furin cDNA was cloned directly after C or T P1 promoter, the mRNA level of the furin gene of pFur-229T-Fur was 2.29 times that of pFur-229C-Fur. A transcription factor binding site search on a Web site (www.cbrc.jp/research/db/tfsearch.html) found that allele T may constitute a suboptimal transcription factor NF-E2–binding motif. In human beings, NF-E2 was first found as a heterodimer of two proteins, p45 and p18, involved in gene transcription in the erythroid and megakaryocytic lineages. p45 is now found to have at least three homologous forms. So-called NF-E2–related factor 2 is expressed in hepatocytes and has anti-inflammatory and anti-oxidative activity.31, 32 In this study, we demonstrated that promoter activities increased as the putative efficiency binding to NF-E2 was optimized and decreased as the element was destroyed. Although this SNP is only 4 bp apart from a putative CCAAT box that may actively control the transcription activity of the P1 promoter,16 the transcription activities of the P1 promoter carrying allele C or T were not significantly influenced when the CCAAT box was destroyed. These results suggest that the difference in the NF-E2 binding efficiency is the mechanism by which this SNP influences the P1 promoter activity. Because NF-E2 is overexpressed in the liver, the SNP −229 C/T of the Fur gene may play an important role in controlling furin mRNA expression and possibly the HBeAg level in the serum of patients with HBV infection. However, more work has to be done in the future to obtain direct DNA-protein interaction evidence and furin mRNA expression data in vivo.
Investigating the relationship of SNP with disease occurrence or progression is also a way to assess SNP function. Allele T or genotype TT is less frequent in individuals with self-limited infection and in IHCs. This finding is indirect evidence that SNP −229 C/T affects HBeAg maturation or seroconversion. However, a finding that SNP −229 C/T influences HBeAg status would constitute direct evidence. Unfortunately, HBeAg status in patients with chronic HBV infection depends largely on the phase of the chronic infection and whether the HBV is HBeAg-defective. Generally, there are four stages of chronic HBV infection8, 27 and two variants of HBeAg-defective viruses (G1896A and the dual BCP variant).33, 34 Taking phases of infection into account, we found that the frequencies of both allele T and genotype TT were less in HBeAg-negative IHCs (phase III) but comparable in HBeAg-positive CHB and LC patients (phase II) and HBeAg-negative CHB and LC patients (phase IV). Most of the latter had HBeAg-defective virus in the serum. Thus, these results do not reject the effect of SNP −229 C/T on HBeAg seroconversion. Rather, they suggest that HBV-infected patients with allele T or genotype TT have a higher risk of developing HBeAg-negative CHB and LC when the virus is HBeAg-defective.
The results of the transcription activity assay and the evidence showing that allele T or genotype TT affects HBV infection suggest that the P1 promoter carrying allele T has higher furin activity. Inhibiting furin activity can inhibit HBeAg maturation and may favorably affect HBeAg seroconversion. Thus, furin is a promising therapeutic target for chronic HBV infection, just as it is for tumors and human immunodeficiency virus infection.35, 36 Recently, furin was found to play an important role in maintaining peripheral immunotolerance by increasing the number of regulatory T cells and the sensitivity of effector T cells to regulatory T cell activities.37 The regulatory T cells maintain peripheral tolerance against self-antigens and foreign antigens and modulate the function and expansion of HBV-specific CD8+ cells in patients with chronic HBV infection.38 Thus, promoting immunotolerance to HBV infection may be an alternative mechanism for increasing the risk of developing persistent HBV infection in patients with the furin high-producer genotype.
Furin expression plays important roles in maintaining normal physiology. For example, albumin and prothrombin maturation in the liver depends on furin.39, 40 In patients with HBV-related LC, we found no significant differences in the prothrombin time and albumin level between genotypes CC and TT, and this supports the conclusion of another report that furin has no role in albumin maturation41 or at least is not the only proprotein convertase involved.42 These facts suggest that the advantages and disadvantages can be reconciled to permit the use of furin as a therapeutic target in the future.
In conclusion, −229 C/T is a common SNP in the P1 promoter of the Fur gene in Han Chinese from southern China. Allele T of this SNP is associated with significantly higher transcription activity, and patients carrying allele T or genotype TT have a decreased risk of HBeAg seroconversion and an increased risk of developing HBeAg-negative CHB and LC when the virus is HBeAg-defective. These observations suggest the future use of furin as a promising therapeutic target and of SNP −229 C/T as a genetic predictor of disease progression or therapeutic response.
Acknowledgements
The authors thank Dr. Gang Li and Dr. Xiao Yan Han for helpful advice and Lin Gu for technical help.