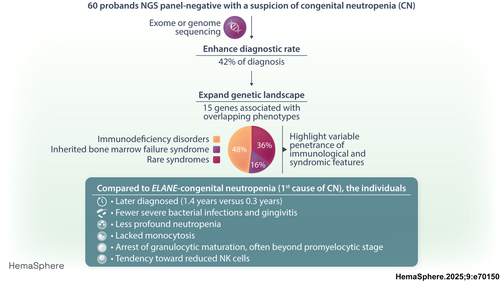
Expanding the phenotypic and genetic landscape of congenital neutropenia through whole-exome and genome sequencing
Graphical Abstract
Abstract
Congenital neutropenia (CN) comprises a heterogeneous group of rare genetic disorders. While some CN cases present only with neutropenia, others present with additional extra-hematological manifestations. The most common cause of CN is variants in ELANE; however, approximately 30 other genes have been implicated. Despite this, the genetic basis remains unknown in roughly 30% of cases. The clinical and genetic heterogeneity of CN makes diagnosis particularly challenging. To address this, we conducted exome or genome sequencing of 60 patients with a suspected diagnosis of CN that remained unresolved following targeted sequencing. A genetic diagnosis was established in 25 patients (42%). Variants were identified in 15 different genes. Half of these cases involved genes traditionally associated with hereditary immunodeficiencies (GINS4, CARD11, ADA2, GINS1, LCP1, SASH3, and WAS). One-third of the cases carried variants in genes linked to syndromic disorders (VPS13B, TAFAZZIN, CLPB, and TONSL), demonstrating variable penetrance of extra-hematological phenotypes. A smaller subset (15%) harbored variants in genes associated with inherited bone marrow failure syndromes (BLM, RPL18, SAMD9, and SRP72), identified incidentally due to atypical presentations. Compared to patients with ELANE-CN, these individuals were diagnosed later, had fewer severe bacterial infections and gingivitis, exhibited less profound neutropenia, lacked monocytosis, and had a granulocytic maturation arrest, often beyond the promyelocytic stage. A shared feature among these cases was a tendency toward reduced lymphocyte subsets, particularly NK cells. This study highlights the significant contribution of exome and genome sequencing in diagnosing CN, given the phenotypic overlap, genetic heterogeneity, and variable penetrance of immunological and extra-hematological features.
INTRODUCTION
Congenital neutropenia (CN) is an extremely rare clinical condition, with an estimated incidence of 10 per 1,000,000 individuals.1 Most CN cases manifest during the neonatal or childhood period. Neutropenia ranges from moderate (<0.5–1.0 × 109/L) to severe (<0.5 × 109/L), and diagnosis may occur incidentally through routine blood tests or in the context of recurrent mouth ulcers, gingivitis, or severe bacterial infections.2 In profound neutropenia, bone marrow examinations typically reveal abnormal neutrophil precursor maturation, often showing a maturation arrest at the promyelocytic stage.3 Since the identification of ELANE variants in cyclic neutropenia and CN,4, 5 around 30 additional gene defects have been identified.6 These findings have highlighted various modes of inheritance of CN, autosomal dominant and recessive, or X-linked, and have underscored the diversity of molecular pathways leading to defective granulopoiesis.6, 7 Notably, identifying these genetic entities of CN has expanded the phenotypic spectrum of CN. In addition to neutropenia, CN may be associated with defects of other hematological lineages, immunodeficiency, and extra-hematological manifestations defining various syndromic neutropenia.6, 7 Moreover, patients with CN face an elevated risk of developing hematological malignancies such as myelodysplastic syndromes or acute myeloid leukemia, with a risk level varying significantly depending on the underlying gene.6 The rarity and broad clinical variability of CN make its etiological diagnosis particularly challenging.
Furthermore, the knowledge of molecular etiologies has highlighted the genetic complexity of diagnosing CN. In addition to the substantial genetic heterogeneity, some CN genes are associated with both autosomal dominant and recessive mode of inheritance resulting in distinct molecular impacts on gene function. Thus, recessive and dominant variants in the same gene may result in distinct clinical outcomes. For instance, bi-allelic loss-of-function (LOF) variants in CLPB cause a severe syndrome characterized by 3-methylglutaconic aciduria, extensive neurological symptoms, cataract, and neutropenia.8, 9 Conversely, monoallelic gain-of-function (GOF) variants in CLPB result in severe neutropenia with additional features such as cataract and epilepsy only in a minority of patients.10 Monoallelic variants in the same gene may also have different functional protein impacts as for LOF variants in the WAS gene, located on chromosome X, leading to Wiskott-Aldrich syndrome or X-linked thrombocytopenia11 whereas GOF variants in WAS cause X-linked severe CN.12 Finally, recessive and dominant variants of the same gene may result in completely different phenotypes as for the TCIRG1 gene causing osteopetrosis and CN, respectively.13 These diverse scenarios make it challenging to assess the pathogenicity of novel variants. Most of them are considered of uncertain significance requiring functional characterization to establish their causality in the disease.
In this study, we conducted exome and genome sequencing on 60 unrelated patients with a suspected clinical diagnosis of CN who remained undiagnosed after targeted sequencing. Our goal was to assess the benefits of genomic sequencing approaches in routine genetic diagnostics. We identified disease-causing defects in 42% of patients. Half of them had variants in genes typically related to immunodeficiency disorders. One-third had variants in genes responsible for rare syndromic disorders revealing variable penetrance and clinical expression of extra-hematological features. A smaller subset had variants in genes associated with inherited bone marrow failure syndrome (IBMFS) and identified incidentally due to atypical presentations. This study underscores the importance of genomic approaches in improving diagnostic accuracy and subsequently guiding clinical management.
METHODS
Participants
We studied 60 probands who were referred by French clinicians from routine clinical care for CN genetic testing at the Department of Medical Genetics (Pitié-Salpêtrière Hospital) between May 2017 and July 2024. All probands had a suspected clinical diagnosis of CN defined as chronic neutropenia with absolute neutrophil count (ANC) below 1.0 × 109/L, after excluding autoimmune neutropenia with positive anti-granulocyte antibodies. In all cases, the referring clinician did not suspect a diagnosis of immunodeficiency, inherited bone marrow failure or syndromic neutropenia, and the clinical features provided at referral did not support genetic testing for a specific disorder.
Disease-causing variants in at least 15 known CN genes (CSF3R, CXCR2, CXCR4, DNAJC21, EFL1, ELANE, G6PC3, GATA2, HAX1, GFI1, JAGN1, SBDS, SRP54, SRP68, and WAS) had been excluded by targeted next-generation sequencing. In some cases, two sequencing gene panels were performed following the identification of novel neutropenia genes.
Clinical and biological characteristics and family history of probands were provided at the time of referral. Proband's data were registered into the French Severe Chronic Neutropenia Registry, which was labeled in 2008 by the French health authorities and approved by the French national data protection agency (CNIL certificate no. 97.075). When available, the parents and affected family members were also included in the study. All participants, or the parents in the case of children, provided written informed consent for inclusion in the registry and for genetic analysis that may explain their symptomatology. This study was performed in accordance with the Declaration of Helsinki and national regulations for genetic testing.
As a comparison cohort, we included 105 patients with ELANE-related CN, who were referred to the Department of Medical Genetics and registered in the French Severe Chronic Neutropenia Registry. The ELANE-related CN cohort has been previously reported.2
Whole-exome and genome sequencing
Genomic sequencing was performed on DNA extracted from blood samples. If possible, it was done on a trio-based approach (proband–parents). Exome capture was carried out with the KAPA HyperCap and HyperExome kits (Roche). The exome libraries were sequenced in paired-end 100 bp reads on a NovaSeq 6000 Sequencing System (Illumina). Whole-genome sequencing was carried out at the national sequencing laboratory SeqOIA (LBM, SeqOIA). Genome library preparation was done with the Illumina® DNA PCR-Free Library Prep, Tagmentation (Illumina), with sequencing performed in paired-end 150 bp reads on a NovaSeq 6000 instrument.
Bioinformatics analyses for both exome and genome sequencing were conducted using in-house pipelines and detailed in Supporting Information S1.
We classified single-nucleotide variants and small indels following the guidelines from the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP).14 In addition to in-house pipelines, we assessed the pathogenicity of structural variants using both ClassifyCNV ACMG score15 and AnnotSV score16 available via CNV-Hub website (https://cnvhub.net/).
Family member testing
When available, biological samples from additional affected family members of individuals with disease-causing variants underwent Sanger sequencing. When a de novo variant was identified, parental relationships were confirmed using exome and genome trio data.
Statistical analysis
GraphPad Prism 7.2 software was used for all the statistical analyses. Differences between groups of individuals were analyzed using the Fisher's exact test if the variable was discrete and unpaired nonparametric Mann–Whitney test for quantitative variables (unpaired nonparametric Kruskal–Wallis test for multiple comparisons). Two-way ANOVA was used for grouped analyses.
RESULTS
Sixty unrelated patients with a suspected clinical diagnosis of CN who remained without definitive diagnosis after targeted sequencing of known genes in CN were selected by a multidisciplinary team. All had a diagnostic risk score of at least 20% of having CN.17 Their clinical characteristics are detailed in Table 1. Their median age at diagnosis of neutropenia was 0.7 years (interquartile range [IQR]: (0.06–6.6 years), with 60% of patients presenting with severe neutropenia (<0.5 ×109/L). To unravel the etiology of their neutropenia, we performed exome sequencing (38/60) and genome sequencing (22/60) for the most recent cases. A monogenic cause was identified in 25 probands, accounting for 42% of the overall cohort. No clinical and hematological features differentiated the 25 patients with disease-causing variants from the 35 with inconclusive exome or genome analysis (Table 1).
| All | Study group | Without molecular etiology | ||
|---|---|---|---|---|
| N = 60 | N = 25 | N = 35 | p | |
| Female sex, % (n) | 47% (28) | 32% (8) | 57% (20) | 0.07 |
| Multiplex families, % (n) | 20% (12) | 28% (7) | 14% (5) | 0.21 |
| Consanguinity, % (n) | 12% (7) | 8% (2) | 14% (5) | 0.69 |
| Clinical profile | ||||
| Age at diagnosis (years), median (IQR) | 0.7 (0.06–6.6) | 1.4 (0.03–3.6) | 0.61 (0.07–7.5) | 0.60 |
| Newborn–infants onset neutropenia, % (n) | 52% (31) | 48% (12) | 54% (19) | 0.77 |
| Children–adult onset neutropenia, % (n) | 48% (29) | 52% (13) | 46% (16) | 0.79 |
| Mouth ulcers/gingivitis, % (n) | 47% (28) | 40% (10) | 51% (18) | 0.44 |
| Severe bacterial infections, % (n) | 62% (37) | 48% (12) | 71% (25) | 0.11 |
| Severe viral infections, % (n) | 10% (6) | 16% (4) | 6% (2) | 0.22 |
| Hematological parameters at diagnosis, median (IQR) | ||||
| Neutrophils (x109/L) | 0.37 (0.20–0.59) | 0.40 (0.27–0.57) | 0.37 (0.18–0.60) | 0.78 |
| Severe neutropenia (ANC < 0.5 × 109/L), % (n) | 60% (36) | 56% (14) | 63% (22) | 0.60 |
| Monocytes (x109/L) | 0.66 (0.27–1.33) | 0.80 (0.45–1.22) | 0.50 (0.20–1.69) | 0.44 |
| Hemoglobin (g/dL) | 11.3 (10.4–13.3) | 11 (10.2–13.3) | 11.6 (10.6–13.4) | 0.53 |
| Platelets (x109/L) | 305 (262–382) | 326 (277–419.5) | 289.5 (254–330) | 0.04 |
| Myeloid maturation arrest, % (n) | 63% (38) | 68% (17) | 61% (20) | 0.59 |
- Note: Results are expressed in median and interquartile ranges (IQR) for quantitative variables, and percentages with the number of patients in brackets for qualitative variables. For each variable, results do not include patients with missing values. p refers to comparisons between patients with disease-causing variants (study group) and those without molecular etiology after exome or genome sequencing.
- Abbreviation: ANC, absolute neutrophil count.
Variant description and classification, and associated mode of inheritance
We identified 30 distinct variants, consisting of single-nucleotide variants (SNV) or small indels, and classified as probably pathogenic (LP) or pathogenic (P) according to ACMG/AMP criteria (Figure 1 and Supporting Information S2: Table 1).14 There was sufficient evidence to support pathogenicity of these variants. This included the absence of the variant in gnomAD v4.1 database, de novo occurrence, segregation analysis within families, in silico bioinformatics predictions of potential effects on mRNA splicing or protein function, functional characterization for previously reported variants, and clinical features (Supporting information S2: Table 1).
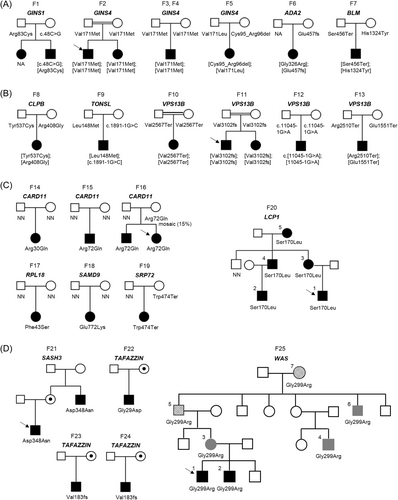
The 30 (P/LP) variants affected 25 probands and involved 15 genes. Biallelic variants in seven different genes (ADA2, BLM, CLPB, GINS1, GINS4, TONSL, and VPS13B; Figure 1A) were identified in half of pedigrees (13 of 25, 52%). Parental segregation analysis revealed that seven probands had compound heterozygous genotypes, while the other six were homozygous and found in pedigrees where consanguinity was either known or suspected based on ethnicity. Heterozygous variants in five different genes (CARD11, LCP1, RPL18, SAMD9, and SRP72; Figure 1B) were found in seven probands (28%). Among them, five carried de novo variants, one (F16) inherited the variant from an asymptomatic mother who was a germline mosaic carrier (Supporting Information S1: Figure 1), and another one (F19) inherited the variant from a clinically asymptomatic parent. The remaining five probands (20%) were hemizygous for X-linked variants linked to three genes (SASH3, TAFAZZIN, and WAS; Figure 1C).
Additionally, neutropenia was diagnosed in 15 affected family members of seven probands. Genetic testing confirmed that all but one, for whom no biological sample was available (F1), had the same genotype as the proband (Figure 1).
Additionally, we identified three de novo heterozygous copy-number variants (CNV): two deletions and one complex CNV comprising a triplication and a duplication of the contiguous region, all located on chromosome 16. These CNVs were classified as pathogenic structural variants based on different classifier tools (Supporting Information S2: Table 2). However, no autosomal dominant gene within these CNVs could be definitively linked to the neutropenia, leaving these three cases inconclusive and grouped with patients for whom no definitive etiology was determined.
Gene defect classification
Our study revealed (P/LP) variants in 15 genes that would not have been previously analyzed as they involved genes predisposing to immunodeficiency disorders, IBMFS, or rare syndromes for which neutropenia is not the primary manifestation (Figure 2). Nearly half of resolved cases (12 of 25, 48%) were due to variants found in genes typically associated with immunodeficiency disorders (Figures 1 and 2). Recessive variants in GINS4 were the most frequent genetic subtype, followed by dominant variants in CARD11. Two of these variants were found in multiple families: one homozygous variant p.(Val171Met) in GINS4 was detected in three pedigrees (F2, F3, and F4) with a shared geographical origin, and a heterozygous variant p.(Arg72Gln) in CARD11 was identified in two unrelated families (F15 and F16). Private variants in five other genes (ADA2, GINS1, LCP1, SASH3, and WAS) linked to immunodeficiency disorders were responsible for the remaining cases.
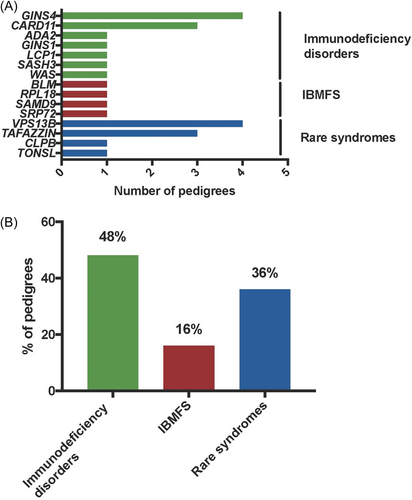
Pathogenic variants in four genes (BLM, RPL18, SAMD9, and SRP72) responsible for IBMFS (4 of 25, 16%) were each identified in a single proband who exhibited clinical and hematological features at the time of CN diagnosis that were not suggestive a priori of Bloom syndrome (BLM), Diamond-Blackfan anemia (RPL18), and myelodysplastic syndrome (SAMD9 and SRP72). Lastly, recessive variants in four genes (CLPB, TAFAZZIN, TONSL, and VPS13B) responsible for rare disorders associated with syndromic features were unexpectedly identified in one-third of probands with atypical clinical presentation (9 of 25, 36%). Biallelic variants in VPS13B linked to Cohen syndrome were the most frequent cause in this subgroup (4 of 9), followed by hemizygous variants in TAFAZZIN responsible for the Barth syndrome (3 of 9).
Clinical and hematological characteristics
Given the initial diagnostic hypothesis of CN, we compared the clinical and hematological characteristics of the 25 probands resolved in this study (study group) to those of CN patients with ELANE variants (ELANE-CN), which is the most common genetic cause of CN (Table 2, Supporting Information S1: Figure 2, and Supporting Information S2: Table 3). We used the clinical and biological data from 105 ELANE-CN probands registered in the French Severe Chronic Neutropenia Registry and previously reported.2
| ELANE-CN | Study group | Immunodeficiency disorders | IBMFS | Rare syndromes | ||
|---|---|---|---|---|---|---|
| N = 105 | N = 25 | p | N = 12 | N = 4 | N = 9 | |
| Female sex, % (n) | 50% (53) | 32% (8) | 0.12 | 33% (4) | 50% (2) | 22% (2) |
| Consanguinity, % (n) | 0% (0) | 8% (2) | 0.036 | 0% (0) | 0% (0) | 22% (2) |
| Clinical profile | ||||||
| Age at diagnosis (years), median (IQR) | 0.34 (0.12–1.4) | 1.4 (0.03–3.5) | 0.64 | 3.2 (0.02–11.5) | 2.35 (0.64–3.1) | 0.2 (0.02–0.85) |
| Newborn–infants onset neutropenia, % (n) | 68% (71) | 48% (12) | 0.10 | 33% (4) | 25% (1) | 78% (7) |
| Children–adult onset neutropenia, % (n) | 32% (34) | 52% (13) | 0.10 | 67% (8) | 75% (3) | 22% (2) |
| Mouth ulcers/gingivitis, % (n) | 84% (88) | 40% (10) | <0.0001 | 58% (7) | 50% (2) | 11% (1) |
| Severe bacterial infections, % (n) | 82% (86) | 48% (12) | 0.0012 | 25% (3) | 75% (3) | 67% (6) |
| Severe viral infections, % (n) | 0% (0) | 16% (4) | 0.0011 | 25% (3) | 25% (1) | 0% (0) |
| G-CSF treatment, % (n) | 89% (93) | 44% (11) | <0.0001 | 67% (8) | 25% (1) | 22% (2) |
| Hematological parameters at diagnosis, median (IQR) | ||||||
| Neutrophils (x109/L) | 0.16 (0.09–0.32) | 0.40 (0.27–0.57) | 0.0001 | 0.46 (0.31–0.58) | 0.53 (0.23–0.74) | 0.30 (0.22–0.47) |
| Severe neutropenia (ANC < 0.5 × 109/L), % (n) | 88% (91) | 56% (14) | 0.0009 | 50% (6) | 25% (1) | 78% (7) |
| Monocytes (x109/L) | 1.86 (1.01–3.10) | 0.80 (0.45–1.22) | <0.0001 | 0.62 (0.31–1.23) | 0.82 (0.52–1.31) | 0.84 (0.66–1.25) |
| Lymphocytes (x109/L) | 5.40 (3.47–6.41) | 3.60 (2.27–6.23) | 0.0747 | 2.43 (0.99–4.06) | 4.91 (3.30–5.37) | 6.99 (3.27–9.08) |
| Hemoglobin (g/dL) | 10.8 (9.8–11.9) | 11 (10.2–13.3) | 0.20 | 12.7 (10.3–14.7) | 10.1 (9.85–10.5) | 11.2 (10.5–12.6) |
| Platelets (x109/L) | 437 (332–529) | 326 (277–419) | 0.016 | 296 (230–368) | 343 (248–506) | 424 (339–522) |
| Myeloid maturation arrest, % (n) | 87% (86) | 68% (17) | 0.025 | 92% (11) | 50% (2) | 44% (4) |
- Note: Results are expressed in median and interquartile ranges (IQR) for quantitative variables, and percentages with the number of patients in brackets for qualitative variables. p refers to comparisons between patients with disease-causing variants (study group) and those with ELANE-CN.
- Abbreviations: ANC, absolute neutrophil count; CN, congenital neutropenia; IBMFS, inherited bone marrow failure syndrome.
The median age at diagnosis for the 25 Study group patients was 1.4 years (IQR 0.03–3.5 years), with half diagnosed before the age of 1. Although ELANE-CN patients were diagnosed earlier (0.34 years), there was no significant difference in the median age at diagnosis between the two groups. In terms of diagnostic circumstances, patients in the study group experienced fewer severe bacterial infections and episodes of gingivitis and mouth ulcers compared to ELANE-CN patients (48% vs. 82%, p = 0.001, and 40% vs. 84%, p < 0.0001, respectively; Table 2).
All patients in the study group exhibited either severe (ANC < 0.5 × 109/L) or moderate neutropenia (ANC 0.5–1.0 × 109/L). However, neutropenia severity was lower in the study group compared to ELANE-CN group (median ANC: 0.4 × 109/L vs. 0.16 × 109/L). Severe neutropenia was present in 56% of study group patients compared to 88% in the ELANE-CN group (p = 0.0009). Notably, the monocytosis that is a typical feature of ELANE-CN was absent in the study group. Hemoglobin and platelet counts were not different between both the study and ELANE-CN groups.
Bone marrow examinations revealed a myeloid maturation arrest in most patients (Table 2). However, in ELANE-CN patients, the maturation arrest occurred at the promyelocyte–myelocyte stage, whereas in the study group, it occurred later. This was indicated by a normal proportion of promyelocytes and myelocytes but a lower proportion of metamyelocytes and mature neutrophils (Figure 3).
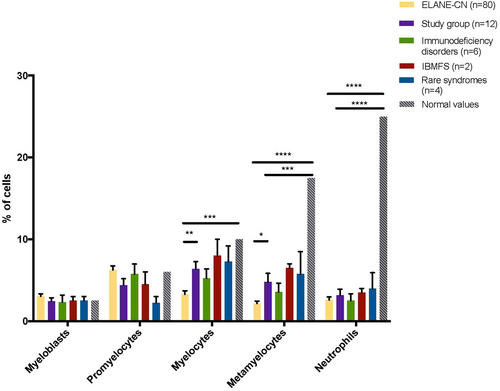
Overall, these findings indicate that patients in the study group were diagnosed later, had less severe neutropenia, and were less symptomatic compared to ELANE-CN patients.
Lymphocytes and immunological profile
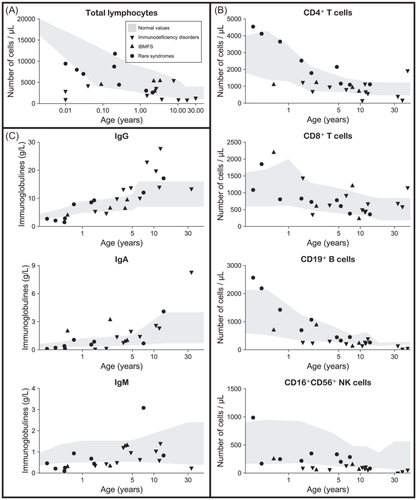
At diagnosis, overall lymphocyte counts did not differ significantly between the study group and ELANE-CN group (Table 2). However, median absolute lymphocyte counts were lower in the 11 patients from the immunodeficiency disorders subgroup (2.42 × 109/L vs. 5.40 × 109/L in ELANE-CN, p < 0.01) and in comparison, to age-matched controls as shown in Figure 4A. We analyzed lymphocyte subsets and immunoglobulin levels (Figure 4 and Supporting Information S2: Tables 3 and 4) in comparison to age-matched controls. CD4+ and CD8+ T-cell counts were normal in nearly all study group patients (Figure 4B). In contrast, CD19+ B-cell and CD16+CD56+ NK-cell counts were low in 82% and 100% of patients with immunodeficiency disorders, respectively (Figure 4B). The analysis of immunoglobulin levels showed that most patients had normal serum IgG levels except four with variants in syndromic genes (VPS13B, TONSL, and TAFAZZIN) displaying low IgG values (Figure 4C and Supporting Information S2: Table 3). Half of patients had low IgM values independently of the involved gene defect. Three patients with low levels of IgG had also low IgA values. Besides, an adult patient with a dominant-negative CARD11 variant p.(Arg72Gln) later developed lymphoproliferative disease (large granular lymphocytic leukemia) and multiple myeloma at age 51.
Family studies highlighted the broad hematological and immunological variability
The analysis of two large pedigrees (F20 and F25), with five and seven affected cases, respectively, revealed the wide clinical variability among affected family members. In family F20 (Figure 1B and Supporting Information S2: Table 3), an autosomal dominant form of neutropenia linked to LCP1 gene, the proband was diagnosed at birth with profound neutropenia and lymphopenia. Family screening revealed another pediatric case with severe neutropenia but also three adults, carriers of the LCP1 missense variant and presenting with mild (1–1.5 × 109/L) to moderate asymptomatic neutropenia associated with deafness. All affected individuals also displayed variable levels of lymphocyte subsets and immunoglobulins.
In family F25 (Figure 1C and Supporting Information S2: Table 3), a novel hemizygous missense variant in the X-linked WAS gene was initially identified through targeted sequencing and classified as LP according to ACMG/AMP guidelines. The variant was inherited by two male siblings from their affected mother (F25-3) who displayed moderate neutropenia and thrombocythemia with platelet anisocytosis. Extended family screening revealed two additional male cases (F25-4 and F25-6) with a phenotype similar to F25-3, and a myelodysplastic syndrome in the proband's maternal grandfather (F25-5) and great-grandmother (F25-7). All affected individuals had a reduced number of B lymphocyte cells, and some had low CD8+ T-cell and/or NK cell counts (Supporting Information S2: Table 3). Further genomic investigations were pursued since the family history was compatible with an autosomal dominant inheritance and the broad spectrum of hematological phenotypes. Genome sequencing conducted in four affected cases did not reveal additional molecular causes beyond the WAS variant.
Genetic etiological diagnosis brought to light syndromic features
Two-thirds of probands (17/25) exhibited non-specific additional features at diagnosis or at referral (Table 3). Growth retardation (intra and/or extra-uterine) was the most common feature (8 of 17, 47%). Unexpectedly, causative variants were identified in genes responsible for syndromic disorders associated with recessive inheritance and for which neutropenia is not the presenting symptom, as VPS13B (Cohen syndrome, OMIM 216550), TAFAZZIN (Barth syndrome, OMIM 302060), CLPB (3-methylglutaconic aciduria type VIIB, OMIM 616271), and TONSL (spondyloepimetaphyseal dysplasia, OMIM 271 510) genes. These patients lacked initial clinical features suggestive of these syndromes, but follow-up confirmed consistent phenotypes (Table 3). In the five patients linked to VPS13B and CLPB, intellectual disability that is part of the clinical spectrum of Cohen and CLPB syndromes was absent at diagnosis of neutropenia (mean age of diagnosis 0.3 years) but were present after a median follow-up of 5 years in all patients. Among the three patients with a fortuitous diagnosis of Barth Syndrome associated with TAFAZZIN variants, two (F22 and F23) had a left ventricular non-compaction associated with normal cardiac function revealed by cardiac ultrasound performed after the genetic testing result. The third patient (F24) initially suspected of having Cohen syndrome due to neurodevelopmental delay and obesity was later identified with Barth syndrome after genomic analysis. In patient F9 presenting moderate neutropenia and hypogammaglobulinemia, the identification of biallelic variants in TONSL was unexpected as this gene was associated skeletal dysplasia phenotypes.18 Further investigation after the genetic testing showed facial dysmorphy, growth retardation, and metaphyseal striations in favor of a moderate form of SPONASTRIME dysplasia.
| Family ID | Gene | Age at CN diagnosis (years) | Age at genetic WES/WGS testing (years) | Extra-hematological features reported | Age at last follow-up | |
|---|---|---|---|---|---|---|
| At referral | After genetic testing, with follow-up information | |||||
| F1 | GINS1 | 30.2 | 32.5 | Intra and extrauterine growth retardation, facial dysmorphy, glaucoma, autoimmune hemolytic anemia | None | 34.9 |
| F2 | GINS4 | 0.04 | 2.8 | Growth retardation | None | 10.0 |
| F3 | GINS4 | 0.003 | 4.8 | None | None | 12.8 |
| F4 | GINS4 | 2.7 | 7.6 | Growth retardation | None | 8.0 |
| F5 | GINS4 | 0.005 | 3.7 | None | None | 11.6 |
| F6 | ADA2 | 17.8 | 41.6 | Erysipelas, condyloma | Enzymatic ADA2 deficiency | 42.8 |
| F7 | BLM | 2.4 | 2.9 | Intra and extrauterine growth retardation, eczema (face), hypospadias | Microcephalya | 3.1 |
| F8 | CLPB | 1.4 | 3.9 | Intrauterine growth retardation | Intellectual disability, methylglutaconic aciduria | 9.7 |
| F9 | TONSL | 0.003 | 4.8 | None | Facial dysmorphy, growth retardation, mild radiological osteopenia unconfirmed by osteodensitometry, metaphyseal striations | 7.4 |
| F10 | VPS13B | 0.03 | 0.2 | None | Global hypotonia, strabismus, intellectual disability | 1.5 |
| F11 | VPS13B | 0.02 | 0.4 | Intrauterine growth retardation | Intellectual disability | 7.3 |
| F12 | VPS13B | 0.2 | 2.1 | None | Ligament hyperlaxity, intellectual disability | 5.5 |
| F13 | VPS13B | 0.008 | 0.5 | Ponderal stagnation, rethrognathism | Intellectual disability, microcephaly, axial hypotonia | 2.4 |
| F14 | CARD11 | 1.5 | 18.4 | None | Crohn-like disease | 19.3 |
| F15 | CARD11 | 7.8 | 8.3 | Abdominal pain, diarrhea | None | 8.3 |
| F16 | CARD11 | 11.8 | 45.4 | Fatigability, syndromic pronosupination of left forearm with synostosis, eczema with prurit | Severe atopic dermatitis, large granular lymphocytic leukemia, heavy chain multiple myeloma | 51.2 |
| F17 | RPL18 | 2.3 | 9.7 | Thumb malformation | None | 15.4 |
| F18 | SAMD9 | 3.4 | 7.8 | Profuse warts | None | 11.1 |
| F19 | SRP72 | 0.09 | 0.5 | None | None | 2.3 |
| F20 | LCP1 | 0.01 | 7.1 | Deafness | None | 13.2 |
| F21 | SASH3 | 10.5 | 10.7 | Warts | None | 15.7 |
| F22 | TAFAZZIN | 0.2 | 0.6 | Fatigability | Left-ventricular non-compaction (cardiac ultrasound), normal cardiac function | 8.9 |
| F23 | TAFAZZIN | 2.1 | 7.4 | Microcephaly, neuro-developmental disorder, fatigability | Left-ventricular non-compaction (cardiac ultrasound), normal cardiac function | 8.4 |
| F24 | TAFAZZIN | 0.3 | 8.0 | Neonatal cardiomyopathy, growth retardation (GH treatment), obesity, severe myopia, neuro-developmental disorder | None | 15.0 |
| F25 | WAS | 3.7 | 9.0 | None | None | 10.9 |
- Note: The referral corresponds to the WES/WGS genetic testing.
- Abbreviation: WES/WGS, whole-exome sequencing/whole-genome sequencing.
- a Known by the clinician but not reported at referral.
Patients (F7, F17, and F18) with pathogenic variants in BLM, RPL18, and SAMD9 leading to an unexpected diagnosis of IBMFS had atypical presentations at referral. Retrospective analysis showed few clinical features supporting their final genetic diagnosis. For instance, the patient (F17) presenting profound neutropenia with granulocytic maturation arrest and diagnosed with a RPL18 variant, a rare cause of Diamond-Blackfan anemia,19 lacked the key diagnostic criteria (in particular, anemia, elevated level of erythrocyte adenosine deaminase, and fetal hemoglobin) but presented with thumb hypoplasia, leading to an initial investigation for Fanconi anemia.
Overall, these findings underscored the complexity of CN diagnoses, highlighting the importance of comprehensive phenotypic and genomic investigations to identify underlying genetic causes.
DISCUSSION
Our study strongly supports routine genetic testing based on exome or genome sequencing in patients with suspected CN. We demonstrated that 42% of patients with at least a 20% risk of having CN, as estimated by the Bejjani et al. diagnostic score,17 and unresolved after targeted CN gene sequencing, carried causative variants in genes associated with immunodeficiency disorders, IBMFS, or rare syndromes. These diagnoses would have been missed or delayed under the current diagnostics strategy that relies on gene panel sequencing specific to CN, immunodeficiency disorders, or IBMFS and is limited to patients presenting with characteristic clinical features.
Genomic approaches now benefit from evidence-based virtual gene panels that are regularly updated by scientific experts and made available through resources such as PanelApp.20, 21 For example, the “Immunological Disorders_SuperPanel,” which comprises 17 overlapping disorder panels (986 genes, version 12.23), is available via PanelApp Australia. Additionally, using Human Phenotype Ontology (HPO) annotations can help identify rare syndromes where neutropenia is not a primary diagnostic feature and involves genes not typically linked to hematological or immunological disorders.22 Finally, using exome or genome sequencing for patients with suspected CN removes the need for clinicians to have exhaustive knowledge of all immunological disorders or rare syndromes that may include hematological features in their clinical spectrum. This broader approach significantly enhances diagnostic efficiency and accuracy.
We identified only SNV or small delins located in exons or canonical splice sites, except for one variant in the 5'UTR region of GINS1, which had been previously reported.23 In cases that remain without identified molecular etiology, we searched for deep non-coding variants as such molecular etiologies have been reported in GATA2, GINS1, and more recently in ELANE.23-25 However, genome sequencing performed in the most recent cases did not reveal any non-coding variants predicted to affect RNA splicing or regulatory regions. The challenges in predicting the impact of non-coding variants may account for their absence in our study.
We also analyzed structural variants and identified three de novo heterozygous CNVs classified as pathogenic. However, none of the autosomal dominant genes within these CNVs could be definitively associated with neutropenia, leaving these cases unresolved. Nevertheless, it is noteworthy that each of these CNV includes a gene (IRF8, CORO1A, and GINS2) associated with recessive forms of hereditary immune deficiencies. Interestingly, neutropenia has been documented in defects of IRF8 and CORO1A.26, 27 Furthermore, GINS2 is part of the GINS complex, which includes GINS1 and GINS4, whose gene defects have been linked to CN.23, 28 We cannot exclude the possibility that monoallelic defects in these genes may result in neutropenia, as demonstrated in other CN genes such as CLPB.10 Alternatively, it is possible that a second pathogenic event contributing to these cases was missed.
The primary etiologies identified were gene defects associated with immunodeficiency disorders with nearly half of the investigated patients receiving a definitive diagnosis.29, 30 Most cases were identified in childhood, showing profound neutropenia with granulocytic maturation arrest in half of them. Clinically, these patients resembled those with ELANE-related CN but had fewer severe bacterial infections and gingivitis. A common feature in this subgroup was a tendency toward low NK cell counts. NK cell deficiency is typically linked to herpes viral infections,31 but this characteristic was absent at referral. Consequently, clinicians did not associate the low NK cell count and moderate lymphocyte subset reduction with neutropenia.
Interestingly, we identified several genetic causes involving replication factor genes, notably GINS1 and GINS4, which have been previously implicated in NK cell deficiencies.23, 28 These genes are responsible for extremely rare conditions, with only one family described for each in the literature. We identified four new families with recessive GINS4 variants. Additionally, two other causes (ADA2 and SASH3), also associated with NK cell deficiency, were identified.32, 33
We also found heterozygous CARD11 variants in three families (four cases). CARD11 encodes an adaptor protein with dominant-negative and GOF variants linked to distinct immunodeficiencies.34 The identified variants, located in the N-terminal CARD domain, were functionally characterized as dominant-negative LOF mutations.35 Initially associated with severe atopy, a broader study revealed that beyond atopy, which may be absent, neutropenia and immunodeficiency can also be present.34 Among our cases, two exhibited severe atopy retrospectively. These CARD11 variants were not initially suspected due to asymptomatic parents; however, mosaicism in the mother of two siblings explained the inheritance.
Additionally, we identified a three-generation pedigree with variable neutropenia and lymphopenia linked to LCP1, encoding L-plastin, an F-actin binding protein involved in immune and hematological functions.36-38
Patients with variants in genes responsible for immunodeficiency disorders are likely underdiagnosed, due to absent or unrecognized primary phenotypic manifestations. These findings highlight the importance of comprehensive immunological workups to guide etiological diagnosis.
In one-third of patients, we identified variants in genes associated with rare syndromes (CLPB, TAFAZZIN, TONSL, and VPS13B). In over half of these cases, syndromic features were absent at referral due to the early onset of neutropenia.8, 9, 39, 40 Two distinct situations emerged: First, follow-up revealed that patients later developed extra-hematological manifestations or had subclinical symptoms highlighting variable penetrance. For example, intellectual disability, absent at referral, was later observed in all subjects with VPS13B variants (Cohen syndrome) and in a patient with CLPB syndrome. Among three patients with TAFAZZIN-related Barth syndrome, two had left ventricular non-compaction identified only after genetic testing.
Second, atypical clinical presentations made syndromic diagnoses challenging. For instance, Barth syndrome typically presents with early-life dilated cardiomyopathy (96% incidence),41, 42 yet we identified an 8-year-old patient with a milder cardiac condition initially suspected of having Cohen syndrome due to overlapping features (neurodevelopmental delay, growth retardation, obesity, and severe myopia). As demonstrated in cross-sectional studies, Barth syndrome survival improves significantly beyond age 3, making late-presenting cases harder to identify.41, 42 This case also illustrated broad phenotypic variability: the hemizygous TAFAZZIN variant found in our patient had previously been reported in an infant with early cardiomyopathy, muscle hypotonia, and growth delay, who died at age 2 from sudden cardiac arrest without neutropenia.43
The most unexpected syndromic diagnosis was linked to biallelic TONSL variants, responsible for SPONASTRIME dysplasia and other skeletal dysplasia phenotypes.18 This rare disorder, with fewer than 15 reported cases, exhibits significant phenotypic variability. Notably, hypogammaglobulinemia and neutropenia were documented in three patients, two of whom lacked the hallmark feature of short stature. TONSL is not included in CN- or immunodeficiencies-specific gene panels, and its identification relied on Human Phenotype Ontology-based variant filtering. Follow-up investigations revealed mild skeletal dysplasia and dysmorphic features, consistent with a subtle syndromic form.
This incidental diagnosis underscores the challenge of identifying rare syndromes when neutropenia is not the primary phenotype. Genetic testing for syndromic genes is often pursued only when classic features are present, emphasizing the need for detailed clinical phenotyping. Identifying extra-hematological traits strengthens diagnostic hypotheses, especially when novel variants require cautious interpretation. Reporting unexpected syndromic cases necessitates collaboration with clinical experts to minimize patient anxiety and ensure tailored management and genetic counseling.
Finally, in a small subset of patients, we identified variants in genes typically associated with IBMFS, such as SAMD9 and SRP72 (linked to inherited myelodysplastic syndrome),44, 45 and RPL18 (linked to Diamond-Blackfan anemia).19 These conditions have been reported in children and adults with chronic cytopenia, often presenting atypically.46 Given the heterogeneity of both IBMFS and CN, making an accurate genetic diagnosis remains particularly challenging.
Patients with CN also have an increased risk of leukemic transformation, explaining why neutropenia and IBMFS may co-exist within families.6 This was evident in a large family in our study with a broad spectrum of immuno-hematological manifestations, associated with a GOF variant in WAS, responsible for X-linked neutropenia.12, 47 The intra-familial observation of multiple chronic neutropenia cases, whether associated with thrombocytopenia, lymphopenia, or MDS, alongside hematological involvement in multiple obligatory female carriers, made the diagnosis particularly complex. This is especially true for WAS-related X-linked neutropenia, an extremely rare entity, as GOF variants are associated with variable phenotypes. Additionally, an attenuated hematological phenotype has been documented in female carriers with a GOF WAS variant,47 and only one MDS case linked to X-linked WAS neutropenia has been reported to date.48
Our study has several limitations. First, not all probands underwent whole genome sequencing, as this approach was only recently implemented in routine diagnostic care. As a result, rare non-coding variants or structural variants may have been missed in patients who remained without a definitive diagnosis after exome sequencing. Second, we considered only variants classified as (P/LP) according to ACMG/AMP guidelines. Some unresolved cases involved variants of unknown significance that require functional studies to confirm their role in the disease. This limitation explains the absence of novel gene identification in our study. Third, our analysis focused on Mendelian forms of CN, and we could not exclude the possibility of oligogenic contributions to disease predisposition.
In conclusion, our study underscores the value of genomic approaches in enhancing diagnostic rate of CN. We expanded the genetic landscape of CN by identifying molecular etiologies that overlap with other disorders, particularly immunodeficiency disorders. The study highlights the genetic heterogeneity of CN, with variants identified in 15 distinct genes. Although each genetic defect is individually rare, together they represent a substantial proportion of CN cases. Accurate molecular diagnosis provides crucial prognostic insights and prompts clinicians to screen for additional features, enabling earlier and tailored medical management according to the specific gene involved.
ACKNOWLEDGMENTS
This research was made possible through access to the data generated by the France Genomic Medicine Plan 2025 for genome sequencing. The authors thank all patients who consented to the sharing and publication of their anonymized clinical data, as well as Pierre Chantelot for his practical support in creating the figures.
AUTHOR CONTRIBUTIONS
S. M. and C. B.-C. designed the study, analyzed the data, and wrote the manuscript. S. M. and C. B.-C. analyzed the exome and genome sequencing. P. P., S. C., L. D., and J. B. performed and analyzed the genetic analyses. J. B. and A. L. performed bioinformatics pipelines and variant annotations. B. B. collected the clinical data and registered the clinical data in the French Registry of Chronic Neutropenia, and J. C.-T. performed data extraction from the Registry. Y. R., N. A., S. A., W. A.-C., G. B., D. B., B. B., C. D., V. G., A. G., A. M.-C., P. D., G. V., B. N., L. N., M. N., M. P., L. T., and J. D. participated in the medical follow-up of the patients and provided clinical data and biological samples. M. D. discussed the results. P. H. performed the analysis of myeloid gene panel sequencing. S. M. performed statistical analysis. S. M., C. B. C., I. P., and J. D. reviewed the manuscript. All authors approved the final version of the manuscript.
CLINICAL TRIAL REGISTRATION
This study has not been registered on ClinicalTrials. All the data collected were gathered for diagnostics purposes and do not require clinical trial registration.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
CONSENT
The patients or the parents for participants under 18 years signed an informed consent for the genetic diagnosis of congenital neutropenia as well as for prospective research related to their disease. All individuals have consented to sharing and publishing their anonymized clinical data.
DNA samples are stored in the Department of Genetics under the responsibility of C. Bellanné-Chantelot (biological collection no. DC2009-957).
ETHICS STATEMENT
The French Severe Chronic Neutropenia Registry, coordinated by Dr Jean Donadieu, is located in the Department of Pediatric Hematology and Oncology, in Trousseau Hospital, Assistance Publique-Hôpitaux de Paris (Paris) and was certified by the French health authorities in 2008. Clinical and biological information is collected at diagnosis and during the course of the disease. Data monitoring, based on medical charts, is done by a clinical research associate. To be included, patients or their parents for children, give their written informed consent in accordance with the Declaration of Helsinki. Patients are anonymously registered in the database in compliance with the current national legislation (CNIL certificate no. 97.075).
FUNDING
This study was supported by the Assistance Publique-Hôpitaux de Paris (AP-HP), Sorbonne University, Institut National de la Santé et de la Recherche Médicale (INSERM), the Association pour la Recherche contre le Cancer (SM), and the Agence nationale de la recherche (ANR-20-CE14-02; L. D., I. P., and C. B. C.). The French Chronic Neutropenia Registry is supported by grants from X4 Pharma, Prolong Pharma, and Chugai SA.
PERMISSION TO REPRODUCE MATERIAL FROM OTHER SOURCES
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the Supporting Information.