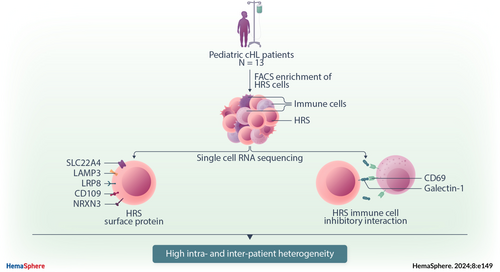
Single-cell RNA sequencing of pediatric Hodgkin lymphoma to study the inhibition of T cell subtypes
Graphical Abstract
Abstract
Pediatric classic Hodgkin lymphoma (cHL) patients have a high survival rate but suffer from severe long-term side effects induced by chemo- and radiotherapy. cHL tumors are characterized by the low fraction (0.1%–10%) of malignant Hodgkin and Reed–Sternberg (HRS) cells in the tumor. The HRS cells depend on the surrounding immune cells for survival and growth. This dependence is leveraged by current treatments that target the PD-1/PD-L1 axis in cHL tumors. The development of more targeted therapies that are specific for the tumor and are therefore less toxic for healthy tissue compared with conventional chemotherapy could improve the quality of life of pediatric cHL survivors. Here, we applied single-cell RNA sequencing (scRNA-seq) on isolated HRS cells and the immune cells from the same cHL tumors. Besides TNFRSF8 (CD30), we identified other genes of cell surface proteins that are consistently overexpressed in HRS cells, such as NRXN3 and LRP8, which can potentially be used as alternative targets for antibody–drug conjugates or CAR T cells. Finally, we identified potential interactions by which HRS cells inhibit T cells, among which are the galectin-1/CD69 and HLA-II/LAG3 interactions. RNAscope was used to validate the enrichment of CD69 and LAG3 expression on T cells near HRS cells and indicated large variability of the interaction strength with the corresponding ligands between patients and between tumor tissue regions. In conclusion, this study identifies new potential therapeutic targets for cHL and highlights the importance of studying heterogeneity when identifying therapy targets, specifically those that target tumor-immune cell interactions.
INTRODUCTION
Classical Hodgkin lymphoma (cHL) accounts for 10%–15% of all lymphoma cases and represents the most commonly diagnosed lymphoma subtype in adolescents and young adults.1 Combined-modality treatment regimens composed of multiagent chemotherapy and involved-site radiation therapy have greatly improved cHL survival, with current cure rates exceeding 90%.1 However, 10%–30% of adult cHL patients and 10% of pediatric cHL patients have refractory or recurrent disease.2-4 Despite the administration of high-dose chemotherapy supported by autologous hematopoietic stem cell transplantation (ASCT), adult refractory/relapsed (R/R) patients have a poor prognosis.5 For pediatric R/R patients, the prognosis is better,6 but long-term toxicity of the treatment is a significant problem. A recent study has shown that pediatric HL survivors had 100 excess deaths per 10,000 person-years 25 years post-diagnosis and nearly 400 excess deaths ≥40 years from diagnosis, making HL the cancer with the second-highest long-term excess mortality after medulloblastoma.7 These excess deaths are primarily attributed to treatment-related secondary malignant neoplasms and symptomatic cardiac/pulmonary toxicities.7 Hence, there is a high demand for novel and innovative treatment approaches that target the tumor more specifically and have reduced side effects while preserving or improving clinical efficacy.
The cellular ecosystem of cHL is unique as it consists of rare malignant Hodgkin and Reed–Sternberg (HRS) cells, which typically represent 0.1%–10% of all cells in the tumor tissue and are surrounded by a dense immune microenvironment consisting of mostly lymphocytes, myeloid cells, and fibroblasts.8 The CD30-positive (CD30+) HRS cells are believed to be derived from proapoptotic germinal center (GC) B cells as they have rearranged (nonfunctional) immunoglobulin genes, gone through somatic hypermutation, and lost the expression of B cell lineage markers such as CD19, CD79a, and immunoglobulin gene transcripts.9-11 There are several lines of evidence suggesting that the rich immune infiltrate in cHL creates an immunosuppressive tumor microenvironment (TME) that is essential for supporting HRS cell survival and growth. Specifically, HRS cells are tightly adhered to surrounding T cells, a phenomenon termed rosetting, which likely is essential for HRS cell survival.12 Additionally, HRS cells do not survive in immunodeficient mice nor grow as solitary cells in vitro, and establishing HRS-derived cell lines has been proven difficult.13, 14 Given that HRS cells depend on complex interactions with different immune cells, breaking or interfering with these interactions presents a promising treatment strategy.
Currently, two immune checkpoint inhibitors have been FDA-approved for R/R cHL patients, namely, nivolumab and pembrolizumab, both of which target the PD-1/PD-L1 signaling axis.15, 16 Of note, pembrolizumab is the only immunotherapy currently approved for pediatric cHL patients. Although the use of these PD-1 inhibitors, in combination with standard chemotherapy regimens, has led to a significant improvement in clinical outcomes, a large portion of patients still relapse, highlighting the need for the development of alternative therapies, for example, those that interfere with interactions between HRS and immune cells that are essential for HRS cell survival.17
Single-cell RNA sequencing (scRNA-seq) provides a unique opportunity to describe the TME in detail through precise molecular profiling of individual cells while simultaneously predicting tumor-immune cell interactions.18 Previous studies applying scRNA-seq to cHL samples have already yielded novel insights into the cHL TME. For example, a LAG3+ regulatory T cell-like subpopulation that contributes to the immunosuppressive phenotype of cHL was identified.19 The same group later characterized a unique CD4+PD-1+CXCL13+ T follicular helper (TFH) cell-like subset in lymphocyte-rich cHL that surrounds HRS cells. The presence of this T cell population was found to be associated with poor clinical outcomes.20 Furthermore, the transcription factors TOX and TOX2 were identified as key regulators of exhaustion in previously reported rosette-forming CD4+CD26− T cell populations.21 Besides T cells, myeloid cells, including dendritic cells (DCs), monocytes, and macrophages were found to be enriched in the close vicinity of HRS cells, all expressing immunoregulatory checkpoints including PD-L1, TIM-3, and the tryptophan-catabolizing protein IDO.22 Recently, Aoki et al. found that CXCR5+ HRS cells and CXCL13+ macrophages form a prominent crosstalk axis in relapsed cHL.23 These discoveries have significantly improved our understanding of the pathogenic mechanisms that are active in the cHL microenvironment. However, there are several important limitations in the aforementioned scRNA-seq studies, including the lack of HRS cells detected in the data sets, and the underrepresentation of pediatric patients included in the cohorts.
Here, we performed flow cytometry-based cell enrichment combined with plate- and chip-based scRNA-seq to specifically capture HRS cells and simultaneously the TME cells of pediatric cHL samples. We used these data to characterize the cellular ecosystem of cHL and investigate the interactions between HRS and immune cells. Specifically, the interaction between HRS cells with LAG3+ CD8 T cells and CD69+ T cells was predicted based on scRNA-seq data and validated by RNAscope. We show that the presence and strength of HRS-immune cell interactions are highly variable not only between patients but also between regions, highlighting the importance of inter- and intrapatient heterogeneity in cHL when identifying novel therapy targets in cHL. In addition, we identified HRS cell genes of surface proteins, other than CD30, that were expressed by the majority of HRS cells but few healthy tissues and thus pose potential new therapy targets.
RESULTS
Cell sorting and data processing
Two pediatric cHL lymph nodes were dissociated into single cells, sorted into 384-well plates, and processed using the single-cell optimized RNA-sequencing technique (SORT-seq) protocol.24 Events with high levels of side scatter (SSC+) were sorted into two columns of each plate to enrich HRS cells. HRS cells could only be identified in the scRNA-seq data of one of the two samples, which had a higher-than-average (~5%) HRS content in the tumor based on diagnostic CD30 immunohistochemistry (IHC) staining. Therefore, the sorting strategy was adjusted to enrich HRS cells using five antibodies based on previous literature25, 26 (SSC+CD20−CD95+CD15+CD30+CD40+; Supporting Information S1: Figure S1). In addition, SSC+CD20− cells were sorted to capture any other potential subset of the HRS cells. Eleven additional pediatric cHL lymph nodes were processed with this panel. Finally, three nonmalignant reactive lymph nodes (RLNs) were included in this study as controls (Table 1). The majority of cells in RLN were CD3+, and therefore SSC+ (nonlymphocytes) and CD20+ (B cells) cells were enriched in these samples. Two scRNA-seq approaches were utilized in this study, namely, the plate-based SORT-seq platform, which links fluorescence-activated cell sorting (FACS) data to scRNA-seq data, and the chip-based Singleron platform, which can achieve higher throughput. After data integration and quality control steps, 8822 cells were included in the final data set (Supporting Information S1: Tables S1 and S2 and Figure S2).
| Patient | Diagnosis | Subtype | Stage | EBV | Location | Age | Sex | scRNA-seq (platform) | RNAscope |
|---|---|---|---|---|---|---|---|---|---|
| PB24752 | cHL | NS | N/A | Neg | N/A | 15 | Male | Yes (SORT-seq) | No |
| PB19568 | cHL | NS | II A | Neg | Cervical | 16 | Female | Yes (SORT-seq) | No |
| PB09287 | cHL | MC | II A | Pos | Cervical | 10 | Male | Yes (SORT-seq) | No |
| PB31727 | cHL | NOS | III B | Neg | Cervical | 12 | Male | Yes (SORT-seq) | No |
| PB26217 | cHL | NS | III A | Neg | Cervical | 17 | Male | Yes (SORT-seq) | Yes |
| PB16107 | cHL | NS | II AE | Neg | Supraclavicular | 16 | Female | Yes (SORT-seq) | No |
| PB11473 | cHL | NS | II A | Neg | Supraclavicular | 9 | Female | Yes (SORT-seq) | Yes |
| PB05088 | cHL | NS | II BE | Neg | Cervical | 14 | Male | Yes (SORT-seq) | No |
| PB09908 | cHL | NS | III AE | Neg | Cervical | 15 | Male | Yes (SORT-seq) | No |
| PB16761 | cHL | NS | III A | Neg | Cervical | 14 | Female | Yes (singleron) | No |
| PB10130 | cHL | NS | IV | Neg | Cervical | 11 | Male | Yes (singleron) | No |
| PB05135 | cHL | NS | II BE | Neg | Cervical | 16 | Female | Yes (singleron) | No |
| PB20799 | cHL | NS | II B | Neg | Cervical | 15 | Male | Yes (singleron) | No |
| PB06422 | cHL | NOS | III B | N/A | N/A | 15 | Male | No | Yes |
| PB27302 | cHL | NS | VI BE | N/A | N/A | 16 | Male | No | Yes |
| PB25394 | RLN | - | - | - | Armpit | 8 | Male | Yes (SORT-seq) | No |
| PB32331 | RLN | - | - | - | Cervical | 13 | Male | Yes (SORT-seq) | No |
| PB32684 | RLN | - | - | - | N/A | 15 | Male | Yes (SORT-seq) | No |
- Abbreviations: cHL, classic Hodgkin lymphoma; EBV, Epstein–Barr virus; MC, mixed cellularity; MS, mixed cellularity; N/A, not available; Neg, negative; NOS, not otherwise specified; NS, nodular sclerosis; RLN, reactive lymph node; scRNA-seq, single-cell RNA sequencing; SORT-seq, single-cell optimized RNA-sequencing technique.
scRNA-seq captures HRS cells
Classification of the cell types was guided by automated cell type identifiers CHETAH27 and SingleR28 (Supporting Information S1: Figure S3A,B). B and T lymphocytes and myeloid cells were identified (Figure 1A). Each of these cell types was processed and clustered separately, and subtypes were identified based on canonical subtype markers (Supporting Information S1: Figure S3C–E). Exhausted and naive CD8 T cells were identified, as well as exhausted, cycling, and naive CD4 T cells and TFH cells. Naive, plasma, and GC B cells were detected, as were monocytes and macrophages. Different subsets of DCs were also identified, including mature, plasmacytoid, CLEC9A+, CD1C+, and CCR6+ DCs (Figure 1A).
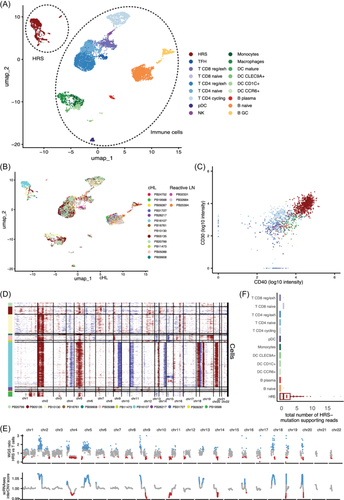
In addition, a few clusters were detected in the scRNA-seq data that separated from the other cell types. These clusters only contained data from cHL samples, and the cells of most individual patients clustered separately (Figure 1A,B). As opposed to the immune cells in the data set, 67% of these cells were classified by CHETAH as an intermediate cell type, indicating they were not of any cell type in the CHETAH cancer TME reference data set (Supporting Information S1: Figure S4A). These cells followed expression patterns found on HRS cells by IHC (Supporting Information S1: Figure S4A,B). Indeed, based on the indexing data from the samples processed with the SORT-seq platform, 93% of those cells that fell within the HRS cluster were sorted by the HRS cell gate, indicating that the HRS markers were expressed both on RNA and protein levels in these cells (Figure 1C). Furthermore, when inferring chromosomal copy numbers using inferCNV,29 large chromosomal gains and losses were detected, which are characteristic of HRS cells (Figure 1D). In particular, we observed recurrent amplifications of chromosomes 2p (n = 8), 5 (n = 7), and 9p (n = 4), and recurrent deletions of chromosomes 13 (n = 5), which is in line with previous studies.10, 30 To validate that these cells were indeed HRS cells, whole-genome sequencing (WGS) was applied to DNA extracted from 3500 sorted HRS cells of patient PB16107 using low-input whole-genome amplification. The copy number variation (CNV) pattern that was inferred from the scRNA-seq HRS cluster of this patient was highly similar to WGS-based CNVs (Figure 1E). To further validate that these cells were HRS cells, 22 WGS-based single-base substitutions (SBS) were identified that had sufficient coverage in the scRNA-seq data; 99.5% of the 980 unique reads that supported HRS cell SBS were from cells in the HRS cluster (Figure 1F). Together, these results validate the HRS cell identity.
As HRS cells have rosetting T cells that can remain attached throughout the cell sorting procedure, we investigated the expression of T cell genes in the HRS clusters. Although some events in the HRS clusters did express a few T cell markers, only CD4 was expressed to similar levels as CD4 T cells, indicating that if present, the T cells contributed relatively few transcripts compared with the HRS cells (Supporting Information S1: Figure S4C,D).
Defining a core set of HRS marker genes
As described earlier, genes that are expressed by most HRS cells in most patients could pose novel targets for cHL treatment. In addition, highly specific HRS markers can potentially simplify the identification of HRS cells, for example, by decreasing the number of antibodies needed for flow cytometry purification. To overcome the potential biases of the single data set from our center and to identify targets that are relevant for pediatric and adult cHL, we performed differential expression analysis (DEA) between HRS cells and healthy B cells in both the scRNA-seq and two microarray data sets of microdissected HRS cells.13, 31 Nine hundred and sixty-five genes were consistently overexpressed in HRS cells in the three data sets and were together termed the “HRS-core” set (Figure 2A,B). As expected, the HRS-core set was depleted for genes located on the recurrently deleted chromosome 13, albeit not significantly (p = 0.08). We validated the HRS-core set using bulk RNA-seq of cHL lymph nodes and RLN obtained from the diagnostics department. The expression of most HRS-core genes correlated well with the expression of HRS marker TNFRSF8 (CD30) in cHL bulk RNA-seq, underlining their specificity for HRS cells (Supporting Information S1: Figure S5A,B). Of the HRS-core set, 145 genes were also differentially expressed between cHL and RLN samples. Of these, 86 were also uniquely overexpressed in bulk cHL compared with other B cell lymphomas.
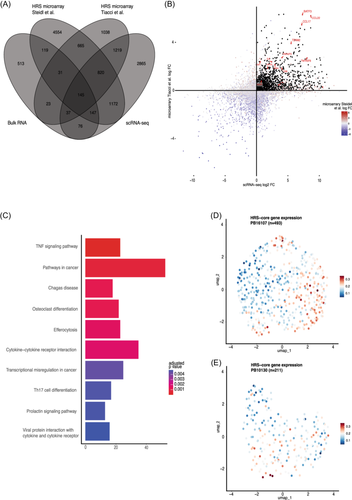
The HRS-core genes were enriched for gene ontology terms involved in pathways that were previously reported to be active in HRS cells, such as extrinsic apoptotic signaling and mononuclear cell migration (Supporting Information S1: Figure S5C). In addition, the tumor necrosis factor (TNF) signaling pathway was found to be the most significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway in HRS cells, which has previously been identified to be particularly active in these tumor HRS cells (Figure 2C).13, 31, 32 The cytokine–cytokine receptor interaction pathway was also upregulated, highlighting the importance of cytokine signaling for tumor cell survival. Indeed, we identified genes encoding well-described signaling molecules that play central roles in cHL pathology, such as IL6, IL13, IL15, CCL17 (TARC), CCL22 (MDC), and LTA (TNF−).33-36 Corresponding receptor genes IL13RA1 and IL15RA were also part of the HRS-core set, suggesting that IL-13 and IL-15 might be the interleukins that are most commonly involved in autocrine HRS cell signaling. These interleukins might play a central role in HRS cell survival and therefore pose potential targets for therapy. Genes encoding other well-described interleukins such as IL4, IL5, IL8, and IL10, and their receptors32, 37, 38 were not consistently overexpressed in HRS cells in all three data sets. Furthermore, genes that are normally only expressed in other tissues were identified, for example, the TENM2/3, ADCY1, BRINP2 (nervous system), and DHRS2 (bladder). These genes are likely expressed due to the chromosomal rearrangements within the HRS cells.39
To identify potential therapeutic targets, HRS-core genes were selected with a high predicted likelihood of being present on the plasma membrane, being expressed in only a few healthy tissues, and being present on the HRS cells of at least 10/12 (≥80%) cHL patients. This resulted in 11 genes including the canonical HRS marker TNFRSF8 (CD30), which is already targeted by the clinically approved drug brentuximab vedotin (anti-CD30 antibody–drug conjugates).16, 40, 41 In addition, neuron-specific NRXN3, lipoprotein receptor LRP8, TNF, lysosome-associated LAMP3, amino acid transporters SLC22A4 and SLC1A3, serine hydrolase ABHD12, ion channel-encoding PIEZO1, immunoglobulin IGSF3, and CD109 were identified. Genes that are proven to be essential in the cHL TME, like TNF, are likely to be useful targets for unconjugated antibodies that block their binding to target cells. Genes that encode proteins that are unlikely to play key roles in HRS signaling pathways, such as NRXN3, could be used as therapeutic targets for antibody–drug conjugates or CAR T cells, or in flow cytometry to identify HRS cells.
Intrapatient HRS cell heterogeneity
Next, we checked whether the expression of the HRS-core genes varied within patients. Investigating intrapatient heterogeneity of HRS cells was not possible for most samples, as the number of HRS cells was too low (9–85 cells). For PB16107 and PB10130, however, 493 and 211 HRS cells were captured, respectively, and were therefore processed separately. Interestingly, the HRS-core genes were variably expressed in PB16107 and PB10130, and formed a continuum in expression (Figure 2D,E). This finding suggests that the general HRS-core expression “program” can have varying levels of activity within the HRS cells of a single patient.
To further analyze diversity, we investigated the inferred CNV profiles. Only one minor subclone was found in PB16107, which had a loss of chromosome 15 (Figure 1C). However, no subcluster of cells could be found by principal component analysis (PCA) or t-distributed stochastic neighbor embedding that had a lower expression of genes positioned on chromosome 15. For PB10130, no subclones were found in the inferred CNV profile. This suggests that chromosomal instability does not drive the gene expression heterogeneity seen in the HRS cells.
Comparing the cellular composition of cHL versus RLNs
To study cell–cell interactions in cHL, it is important to first identify which cell types are enriched and depleted in the cHL TME compared to RLNs (Figure 3A). For this analysis, only unbiased live cells were used that were not enriched for any marker in flow cytometry. Overall, naive B, regulatory/exhausted CD4, and CD8 T cells made up most of the TME in cHL samples (Figure 3B). When compared with RLN, exhausted CD4 T cells were overrepresented (p = 0.05). In contrast, GC B cells, the B cell type that is abundant in normal GCs, were present at lower frequencies in cHL lymph nodes (p = 0.02), as were NK cells (p = 0.02). Plasma cells and TFH cells were also depleted, although not significantly, due to their low numbers in both cHL and controls.
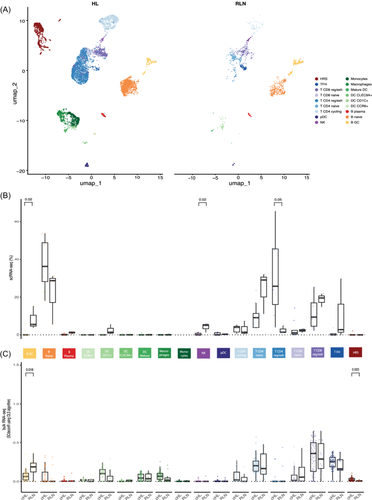
To validate these results in a larger number of samples, the deconvolution method ordinary least squares (OLS) was performed on bulk RNA-seq of 59 cHL and 14 RLN samples obtained from the diagnostics department, using the scRNA-seq data as a reference (Figure 3C). Exhausted CD8+ T cells were found to make up the majority of cells in the bulk RNA-seq of both cHL and RLN samples. GC B cells were again found to be significantly depleted in cHL compared with RLN (p = 0.016), while HRS cells were significantly enriched, as expected (p = 0.023). Taken together, these results showed that NK and GC B cells are depleted from cHL tissue and that exhausted T cells are enriched and make up most of the TME. It seems therefore likely that HRS cells interact most with exhausted T cells.
HRS cells inhibit T cells by a variety of interactions
Our data provide a unique opportunity to investigate in vivo interactions between HRS cells and the TME on a per-patient basis. Interaction scores were calculated by grouping cell types into broader categories and multiplying the expression of a ligand/receptor in a TME cell type of one patient with the expression of the corresponding receptor/ligand in the HRS cells of the same patient (Figure 4A). Most interactions that we identified were only observed in one or a few patients (Supporting Information S1: Figure S6) and 72% of the interactions were found to be more active in cHL compared with RLN.
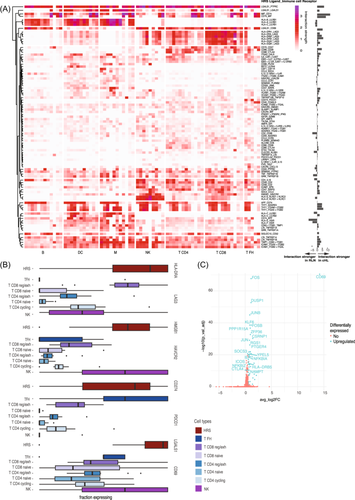
The high number of exhausted T cells in cHL, and the high number of predicted interactions between HRS and T cells suggest an important role for this cell type in HRS survival. Indeed, some of the strongest interactions in cHL had a role in the inhibition/exhaustion of T and NK cells. LAG3, a known exhaustion marker, was overexpressed in the reg/exh CD8 T cells across most patients in our data set (Figure 4B). In addition, NK cells expressed more HAVCR2 (TIM3). Ligands for these receptors, namely, HLA-II and HMGB1, were highly expressed in HRS cells (Figure 4B). PD-L1 (CD274) is highly expressed in HRS cells due to 9p24.1 alterations, which is confirmed in our data.44 However, the PD-L1/PD1 interaction was found to be very weak in our data set. The only subtype of T cells that expressed PD1 in our data set was the TFH cell (Figure 4B). TFH cells were depleted in cHL lymph nodes in our data, which might explain the previous observation that cHL tumors are depleted of PD1-expressing T cells.45
Finally, galectin-1 (LGALS1) is highly and specifically expressed in HRS cells of all samples and is predicted to interact with the CD69 receptor expressed on a subset of cHL T cells (Figure 4B).46 Although CD69 is known as an early activation marker on T cells, there are also indications that CD69 might play an immune inhibitory role since CD69-deficient mice and anti-CD69 antibody administration have shown to induce enhanced antitumor immunity.47 To understand the function of the CD69 receptor in our data set, T cells were divided into two groups based on the CD69 expression. As expected, FOS, FOSB, JUN, and JUNB were upregulated in CD69+ T cells (Figure 4C). These are subunits of the activator protein-1, the main transcription factor that drives CD69 expression.48 Notably, we also found that, compared with CD69− T cells, CD69+ T cells significantly overexpressed CTLA-4 and SOCS3 (Figure 4C). CTLA-4 functions as an immune checkpoint that transmits an inhibitory signal in T cells when stimulated.49 In addition, mouse studies have demonstrated that the deletion of SOCS3 in T cells enhances the antitumor response by promoting the differentiation of exhausted T cells into effector cells, suggesting that SOCS3 expression promotes exhaustion.50 Taken together, these results point toward an inhibitory function of the CD9 receptor in cHL T cells. Therefore, targeting the galectin-1/CD69 interaction might be of interest for developing strategies to treat cHL using targeted approaches.
Intra- and interpatient spatial heterogeneity of commonly detected interactions
To validate the presence of the inhibitory interactions between HRS and T cells, the RNA in situ hybridization assay RNAscope was performed. For four patients, the interaction between HLA-DRA+ HRS cells and LAG3+ CD8 T cells was studied (probes for HLA-DRA, LAG3, CD8, and CD30/TNFRSF8 were used). In a separate panel, the interaction between LGALS1+ HRS cells with CD69+ T cells was studied (probes for LGALS1, CD3E, CD69, and CD30 were used). Two of the samples were selected from the patients of the scRNA-seq cohort while two were from cHL patients outside of our scRNA-seq cohort (Table 1). RNAscope was applied on entire slices of the cHL lymph nodes. For each patient, between four and eight representative regions with varying expression of the different markers were selected for further inspection (Supporting Information S1: Figure S7A–H). Interestingly, the total LAG3 expression per region correlated with CD8A expression (R2 ≥ 0.85), as did CD69 expression with CD3E (R2 ≥ 0.7 in each patient; Figure 5A,B). Second, CD3E was co-expressed in cells with CD69, but the enrichment over the background was low (Figure 5C, top panel). CD8A was more often co-expressed with LAG3 in the same cells compared with the background (Figure 5C, bottom panel). In summary, the RNAscope data confirm the frequent expression of LAG3 on CD8 T cells and the expression of CD69 on CD3 T cells, although at a lower frequency, which is in line with the scRNA-seq results (Figure 4B).
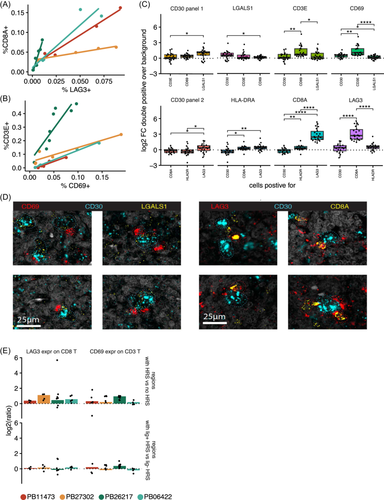
In terms of HRS ligand expression, CD30 and LGALS1 were co-expressed more often than background (Figure 5C, top panel), although this enrichment was not as high as the enrichment of the inhibitory receptors on the T cells (Figure 5C, bottom panel). HLA-DRA expression was the same on CD30-expressing cells as background. This means that HRS cells do express LGALS1 more than other cells but not HLA-DRA. Of note, large variability was observed in the expression of T and HRS markers between patients as well as between regions, with no consistent correlation between the two (Supporting Information S1: Figures S7A–H and S8). This suggests that HRS cells do not consistently induce expression of the inhibitory receptors on T cells across patients and tissue regions.
To see if there was an indication of cell–cell interactions, we analyzed the local enrichment of T cells around HRS cells. We found CD30+ cells closely surrounded by CD69+ and LAG3+ cells (Figure 5D). Therefore, we assessed whether T cells near HRS cells were more or less likely than other T cells to express CD69 or LAG3. In 40 out of 45 regions, T cells expressing CD69 or LAG3 were enriched near HRS cells. This indicates that this subset of T cells is either recruited toward HRS or the expression of the inhibitory receptor is induced near HRS cells. Still, the variation across different regions of the tumors was high in most patients (Figure 5E, top panel, and Supporting Information S1: Figure S9). The enrichment of CD69/LAG3-expressing T cells was compared between HRS cells that expressed the corresponding ligand and the other HRS cells. Interestingly, in patient PB26217 T cells near LGALS1+ HRS cells expressed CD69 more often in all tumor regions, but this was more variable for patients PB27302 and PB11473 and not the case for patient PB06422. The expression of LAG3 on T cells was not enriched near HLA-DRA+ HRS cells compared with other HRS cells in any patient (Figure 5E, bottom panel).
DISCUSSION
Here, we have applied scRNA-seq on pediatric cHL samples. In addition, while most previous cHL scRNA-seq studies only captured TME cells20, 21 and compared their data to publicly available bulk HRS cell profiles of another cohort,19 we managed to capture and sequence HRS cells using both plate- and chip-based methods. We validated the presence of HRS cells using multiple layers of evidence, including flow cytometry marker expression, CNVs, and WGS data. This unique data set allowed us to identify constitutively expressed HRS cell membrane protein genes and identify common HRS-immune cell interactions.
By capturing single HRS cells, new potential universal surface markers could be identified. Here, we identify 11 genes that are highly predicted to be expressed on the plasma membrane, expressed in only a few healthy tissues, and present on the HRS cells of at least ≥80% of the cHL patients. As expected, TNFRSF8 (CD30), which is already targeted by the clinically approved drug brentuximab vedotin, was one of the targets found in our study. Other candidate genes, such as TNF and NRXN3, could also be useful targets for (un)conjugated antibodies or CAR T cell therapy, and therefore warrant further research. In addition, in two patients with a high number of HRS cells sequenced, a clear continuum of the HRS-core transcriptional program could be identified. Extended cohorts capturing more HRS cells should validate this transcriptional heterogeneity and investigate its link with treatment response and prognosis, as this would, for example, reduce their usefulness as universal HRS markers in flow cytometry or as ubiquitous therapeutic targets. Combining scRNA with single-cell DNA or ATAC sequencing (assay for transposase-accessible chromatin using sequencing) might elucidate whether the transcriptional heterogeneity is due to genetic or epigenetic factors, like previously indicated for CNVs,51 or whether it is absent as was suggested for patients PB16107 and PB10130.
Next, we sought to explore which HRS–TME interactions could be potentially targetable. We identified the inhibitory interactions CD69/galectin-1 and LAG3/HLA-II between the HRS and T cells in most tumors based on scRNA-seq. Imaging analysis validated the expression of LAG3 and CD69 on CD8 and CD3 T cells, respectively, and indicated that their expression was enriched on T cells that surrounded HRS cells in all patients, although the amount of enrichment varied. However, when also studying the corresponding ligand, the CD69/galectin-1 interaction was present in one out of four investigated tumors, and the interaction was only identified in a subset of the tissue regions of the other tumors. The LAG3/HLA-II interaction was not observed in any patient. Possibly, other ligands on HRS cells are important for the interaction in the patient without enrichment, or the protein level of the ligands in HRS cells is different from transcript levels. In addition, previous research revealed that HRS cells were closely surrounded by frequent LAG3+ T cells in the subset of patients with cHL with loss of HLA-II expression on tumor cells.19, 22 The LAG3/HLA-II and CD69/galectin-1 interaction might thus not be universally targetable, but could pose a potential targetable interaction in a subset of patients. Experiments should validate the in vivo protein–protein binding of this ligand–receptor pair and should assess the effect of interfering with this interaction.
Our unique pediatric cohort allows us to compare the interactions predicted in our data set with those found in adult cHL. For example, we found a higher fraction of CD8 T cells expressing LAG3 compared with CD4 T cells, while the opposite was previously reported in adults.19 In both age cohorts, LAG3+ T cells seem to be surrounding HLA-II-deficient HRS cells. Although the galectin-1/CD69 axis was found in our cohort, it has yet to be described in adults, raising the question of whether this interaction is specific to pediatric cHL. On the contrary, in adult cohorts, galectin-9 expressed by HRS cells has been shown to interact with CD44+/CD45+ on macrophages, and galectin-3 with LAG3+ T cells.23 This suggests that, in both cohorts, galectins play an important role in the TME and thus need to be investigated further. In adults, the CXCR5/CXCL13 axis between HRS and macrophages has been associated with poor clinical outcomes and R/R HL.23 In our data set, this axis was only found between CXCR5+ HRS cells and CXCL13+ TFH cells in four patients, and the interaction was weak and inconsistent. This could hint that the CXCR5/CXCL13 axis might be less prominent in pediatric cHL.
Late complications of conventional chemotherapy and radiotherapy therapies can be reduced by the use of more targeted approaches, including interrupting important cell-to-cell interactions. In this study, we conclude that most HRS-immune cell interactions identified were only found in a subset of patients, and the strength of these interactions varied within the same tumor tissues. This highlights the importance of considering inter- and intra-patient heterogeneity of the cHL TME when investigating new targets for immunotherapies because heterogeneity can provide tumors with significant adaptability and can pose a challenge to accomplishing the goals of precision medicine.52 A possible way to tackle this issue could be the use of combination therapies, but further studies are required to prove their benefit in cHL. Our results suggest that the expression of multiple HRS cell surface markers is more consistent than the presence of tumor-immune cell interactions. This would suggest that future efforts into the development of new HL therapies should focus on targeting these surface markers, for example, by conjugated or unconjugated antibodies against these proteins.
METHODS
Patient material
All lymph nodes that were used for scRNA-seq were obtained as frozen single-cell suspensions from the biobank of the Princess Máxima Center for Pediatric Oncology, Utrecht, the Netherlands in accordance with the Declaration of Helsinki. The use of the material was approved by the Biobank and Data Access Committee under proposals PMCCRC2018016 (Hodgkin lymphoma lymph nodes) and PMCLAB2021-254 (RLNs).
Patients were selected who were diagnosed between 2019 and 2022 with cHL of any subtype and for whom frozen single-cell suspensions of lymph node material were available. After the initial two patients were processed by scRNA-seq (PB24752 and PB262127), the other samples were screened for having cells with HRS cell-like marker expression (see below). Only those samples that had those cells were selected.
IHC
IHC information was obtained from routine diagnostic IHC. IHC was performed on the Leila Bond III staining system. IHC stainings were analyzed by an experienced pathologist. Epstein–Barr virus status of HRS cells was determined by EBER in situ hybridization imaged on the same machine.
Single-cell suspension
Single-cell suspensions were made by the diagnostic technicians specialized in flow cytometry as follows. Wash medium (20% fetal calf serum, 80% RPMI-Glutamax) with 2% gentamycin was added to the lymph node biopsies, which were minced and pushed through a 100-μm cell strainer, spun down for 10 min at 300 g at room temperature and resuspended in washing medium and put on ice. Cells were divided over different ampuls, spun down at 469 g for 5 min, resuspended in 500 μL washing medium, 500 μL freezing medium was added in drops (80% washing medium, 20% dimethylsulfoxide), and cells were stored in liquid nitrogen freezers.
FACS
Samples were thawed and visible clumps were removed manually using a pipette. Cells were then stained for FACS. Sorting was performed on a Sony SH800S Cell Sorter. In all samples, all sorted events were DAPI-negative singlets, as determined by an FSC-H/FSC-A and an SSC-H/SSC-A gate. The Sony SH800S measures backward scatter, not side scatter, but as these are indicative of the same granularity/complexity “SSC” is used for clarity as the abbreviation throughout the manuscript. Except for the first two processed cHL lymph node samples (PB24752 and PB26217), the other nine samples were stained with fluorescently labeled antibodies against CD20, CD30, CD40, CD95, and CD15. The gating strategy for the HRS cell gate is depicted in Supporting Information S1: Figure S1. The BioLegend antibodies used were as follows: CD20-BV421 (clone 2H7, 302329, 1:50), CD15-FITC (clone HI98, 301903, 1:50), CD95-PE (clone DX2, 305607, 1:50), CD30-APC (BY88, 333909, 1:25), CD40-AF700 (clone 5C3, 334327, 1:50), and CD3-APC/Fire750 (clone SK7, 344839, 1:50). In addition, samples were stained with DAPI (Sigma-Aldrich, D9542-1MG, 500 mM, 1:250). RLNs were stained with CD20-FITC (clone 2H7, 302303) instead of CD20-BV421, DAPI, and DRAQ5 (50 μM, 1:100).
SORT-seq
Plate-based scRNA-seq was performed on 12 samples (PB24752, PB19568, PB09287, PB31727, PB26217, PB16107, PB11473, PB05088, PB09908, PB25394, PB32331, and PB32684) according to the SORT-seq protocol.24 For all samples, unbiased live singlets were sorted into the majority of the 384-well plates. In addition, for all samples, part of the wells was filled with SSC+ cells. Depending on the number of cells present after thawing and the fractions of cells that were part of these subsets, part of the wells were filled with SSC+CD20− cells (“tumor-lenient”) and SSC+CD20−CD30+CD40+CD95+CD15+ cells (“tumor-strict”). RLNs were stained with CD20 and part of the wells were filled with SSC+ and CD20+ cells. For a full overview of samples, cell numbers and sorting strategy, see Supporting Information S1: Table S1. The 384-well plates were filled with Sigma mineral oil (10 μL), RT primers (50 nL), and External RNA Controls Consortium (ERCC) spike-in transcripts. The first column was left empty to be able to control for background contamination after sequencing. Library preparation was done as previously described.24, 53 Paired-end 75 bp sequencing was performed on an Illumina NextSeq 500. Mapping to reference genome hg38, annotation using Gencode 26, and gene-level transcript quantification was done with the Sharq pipeline.54
Only wells in which the library was successfully constructed and sequenced, as judged from the ERCC transcripts, were considered. Then, DecontX was run, using all the successful wells from all plates, to remove ambient RNA.55
Singleron
Chip-based scRNA-seq was performed on four patients (PB16761, PB10130, PB05135, and PB20799) using the GEXSCOPE Single Cell RNAseq Library Kit LW (Singleron Biotechnologies, #4001091) following the manufacturer's instructions. Briefly, SSC+CD20−CD30+CD40+CD95+CD15+ cells (“tumor-strict”), SSC+CD20− cells (“tumor-lenient”), and unbiased live singlets were sorted in bulk. For a full overview of samples, cell numbers and sorting strategy, see Supporting Information S1: Table S2. The resulting single-cell suspension was adjusted to a concentration of 0.5 × 105 cells/mL with phosphate-buffered saline and loaded onto a microfluidic chip to capture 3000 cells. Of note, PB16761 (female) and PB10130 (male) were pooled before sorting and loaded on one microfluidic chip. These samples were later separated based on the expression of Y- and X-chromosome genes. Magnetic beads conjugated to oligo(dT) probes with unique molecular identifiers (UMIs) and barcodes were added, and the cells were lysed. The polyadenylated messenger RNA bound to the beads was extracted, reverse transcribed, and amplified by PCR. The resulting complementary DNA was fragmented and ligated to indexed Illumina adapters. The final amplified library fragment size distribution was analyzed using an Agilent Fragment Analyzer. Paired-end 150 bp sequencing was performed on an Illumina NextSeq 2000. The scRNA-seq data was preprocessed using the CeleScope software (v3.0.1), low-quality reads were discarded, and the remaining sequences were mapped to the human reference genome GRCh38 using STAR. Gene annotation was performed using Ensembl 99. The assignment of reads to genes was done using featureCount, resulting in a count matrix file that contained the number of UMIs for each gene within each cell.
scRNA-seq library integration, processing, and annotation
Count matrices from both platforms were integrated and further processed using the Seurat R package v5.0.3.56 Cells with less than 1000 transcripts, less than 200 measured genes, or with more than 50% mitochondrial reads were removed. Normalization to 10,000 transcripts, data scaling, and identification of the 2000 most variable features were then performed. PCA (100 principal components [PCs]) was performed, and the first 30 PCs were used for UMAP (Uniform Manifold Approximation and Projection) dimensionality reduction and shared nearest-neighbor clustering (resolution 0.05).
To identify cell types in the scRNA-seq data, first the HRS cells were identified. Then, the expression of HRS markers per cluster was compared with the HRS expression pattern based on the IHC of the pathology department. Then, SingleR package v2.5.228 was applied with the celldex v1.12.0 Monaco reference data. In addition, CHETAH v1.18.027 was run with the default tumor-immune reference. Based on these classifiers and canonical marker expression, cell types were assigned to each cluster. Subsequently, the clusters containing T cells were processed separately by Seurat as described above to better define the T cell subtypes. These subtypes were determined by T cell marker expression. The same procedure was performed for all myeloid cells.
Differential composition analysis was performed using the R package DCATS v1.0.042 with default settings to determine which cell types were depleted and enriched in cHL compared with RLN. The p values from the likelihood ratio test were false discovery rate (FDR) corrected.
CNV
The inferCNV package29 v1.18.1 was used to infer CNVs from the scRNA-seq data, using the standard settings “cutoff = 0.1, denoise = TRUE, cluster_by_groups = TRUE.”
Cell–cell interactions
The immune cell composition of the cHL lymph node from PB24752, in which no HRS cells could be detected, had a high fraction of GC-B and TFH cells, but almost no exhausted T cells. As this makes it likely that the part of the lymph node tissue that was analyzed had low or no HRS cells and was thus not representative of the tumor tissue, this sample was excluded in all subsequent immune cell analyses.
Receptor–ligand pairs were taken from the curated iCellNet interaction database.57 Only those interactions were selected for which all ligand and receptor genes were measured in the scRNA-seq data. First, the expression of each ligand and receptor was averaged per cell type per patient. For each patient, only cell types with five or more cells were used. Interaction scores were determined by multiplying the averaged ligand expression of one cell type with the averaged receptor expression of another cell type from the same patient. An interaction was considered to be active in a patient when the interaction score was 0.1 or higher. “Common” interactions between HRS and a specific cell type were those that were present in at least all but 2 patients (with a minimal of 3).
DEA was performed on T cells with CD69 expression compared with T cells expressing no CD69. The FindMarkers function from the Seurat package was used to find differentially expressed genes of CD69+ compared with CD69− T cells. Genes with log2 fold change (log2 FC) > 0.6 and p < 0.05 were set as significantly upregulated (“UP”), while genes with log2 FC < −0.6 and p < 0.05 were set as significantly downregulated (“DOWN”).
Bulk RNA-seq
Bulk RNA-seq data generated for routine diagnostics were obtained from the Princess Máxima Center biobank under proposals PMCLAB2021-205 and PMCLAB2021-254.
Cell-type deconvolution was performed with SCdeconR43 v1.0.0 using the OLS algorithm. Differential cell abundance was calculated with the Wilcoxon test and FDR corrected.
HRS markers
DEA was performed for the scRNA-seq data using the FindMarkers function from the Seurat package, comparing HRS cells to all other cell types, using the setting “logfc.threshold = 0, min.pct = 0, min.diff.pct = 0, min.cells.feature = 0, min.cells.group = 0.” Affymetrix data from Tiacci et al.13 and Steidl et al.31 were normalized using RMA (oligo package v1.60.058) and DEA was performed using limma v3.58.1 with standard settings.59 HRS cells from Steidl et al. were compared to bulk cHL, GC-B cells, and centroblasts. In the data from Tiacci et al. HRS were compared to naive B cells, memory B cells, centrocytes, and centroblasts. In the bulk-RNA-seq data, DEA was performed using the DESeq2 v1.42.1 package using standard settings and data of RLN samples as the control.60
For each of the four expression data sets, a gene was considered differentially expressed when the adjusted p value was lower than 0.01 and the log2 FC was higher than 0 for the bulk RNA-seq, or the average log2 FC in the scRNA-seq was higher than 0, or the minimal log2 FC of all comparisons with the normal B cell references was greater than 0 for Affymetrix data. HRS core genes were those that were identified in the two Affymetrix data sets and the scRNA-seq data.
Bulk RNA-seq data was also obtained from non-Hodgkin lymphoma samples from the Princess Máxima Center biobank. DESeq2 was applied to perform DEA between Hodgkin and non-Hodgkin lymphomas as described above.
GO term enrichment was performed using the enrichGO function from ClusterProfiler v4.10.1 using the “biological process” ontology.61 KEGG pathway enrichment was performed with the diffEnrich v0.1.2 package with the following setting, “N = 5.”62
SurfaceGenie63 was used to extract genes that express a protein that has a high likelihood of being present on the surface of the cell membrane. High likelihood was defined as being predicted as a membrane protein by at least four out of five methods. Then, HPAanalyze64 v1.20.0 was used to select genes that were expressed in fewer than 10 out of 127 normal cell types from 55 tissues from the Human Protein Atlas.
WGS, processing, SNV calling, and CNV
For patient PB16107, WGS was performed on 3500 bulk-sorted HRS cells. The sorting protocol was the same as described above for the scRNA-seq with cells sorted based on forward scatter/side scatter (FSC/SSC) characteristics and the following staining profile DAPI−SSC+CD20−CD30+CD40+CD95+CD15+. DNA was isolated using the NEBNext Ultra II FS DNA Library Prep kit. As a control, bulk T cells (CD20−CD3+) were sorted (~500,000 cells) and DNA was isolated with the Qiagen QIAamp DNA micro kit. DNA of HRS and T cells was sent for 30X WGS. FREEC was used to determine the CNV in this sample using a bin size of 2 Mb.65 The IAP pipeline was used for read alignment and variant calling and further filtering was performed using SMuRF v2.1.2 as described previously.66 For each SNV position in the WGS, all scRNA-seq reads from PB16107 that spanned the mutation sites were extracted. SNVs that had at least 80 reads spanning it were selected. From these reads, per cell, the number of alternative and reference reads was determined based on unique UMIs. Finally, per cell, the total number of UMIs that supported any of the alternative alleles was calculated.
RNAscope in situ hybridization
In situ hybridization assays were performed with RNAscope technology using the RNAscope Fluorescent Multiplex kit v2 (ACD, 323100) and 4-plex Ancillary Kit (ACD, 323120). Formalin-fixed, paraffin-embedded (FFPE) tissues from four cHL patients were cut into 6 μm sections using a microtome. Probes used included the following: Hs-TNFRSF8-C1 (ACD, 593451-C1), Hs-LGALS1-C2 (ACD, 486281-C2), Hs-HLA-DRA-C2 (ACD, 475891-C2), Hs-CD69-C3 (ACD, 494471-C3), Hs-CD8A-C3 (ACD, 560391-C3), Hs-LAG3-C4 (ACD, 553931-C4), and Hs-CD3E-C4 (ACD, 553971-C4). FFPE sections were deparaffinized in xylene and rehydrated in ethanol. RNAscope hydrogen peroxide was applied to block endogenous peroxidase activity before target retrieval was performed for 15 min in a preheated glass beaker (100°C) containing the target retrieval solution. Protein digestion was then carried out by applying RNAscope Protease Plus. Probes were hybridized for 2 h at 40°C followed by signal amplification. After amplification, probes were fluorescently labeled with Opal dyes: Opal 520 (Akoya Biosciences, FP1487001KT, 1:1500) was assigned to HLA-DRA and CD3E, Opal 570 (Akoya Biosciences, FP1488001KT, 1:1500) was assigned to LGALS1 and CD8A, Opal 620 (Akoya Biosciences, FP1495001KT, 1:1500) was assigned to CD69 and LAG3, and Opal 690 (Akoya Biosciences, FP1497001KT, 1:1500) was assigned to TNFRSF8. Finally, slides were incubated for 30 s in DAPI (4′,6-diamidino-2-phenylindole) and then coverslipped. Following staining, imaging was performed on a Leica STELLARIS 8 Confocal Microscope with a white light laser (tunable range 440–790 nm) using a ×20/0.75 NA multi-immersion objective set to oil. Tilescan images were acquired with 10% overlap at pixel size 0.142 × 0.142 µm in 16 bit. The tiles were merged in Leica LASX software.
RNAscope data processing
Upper and lower limits were set for each fluorescent label for each slide based on a manual inspection in ImarisViewer. ImarisViewer was also used to select five to seven regions of approximately equal sizes that encompassed all variability in the slides. In Python v3.9.16, the regions were isolated from each image, values outside the determined limits were capped, and the minimal values were restored to 0. Then, scikit-image (v0.19.2) was used to determine a threshold between true and false positive signals using Otsu's method. Values below this threshold were reduced to 0 to further reduce noise. Then, scikit-image was used to erode and dilute 1 px of the remaining values in order to remove the last noise. Finally, the signal of each block of 51 px (7.242 µm) was averaged. Subsequent analysis was done in R v4.2.1 and tidyverse 1.3.1. Mean intensities per block were converted to z-scores z = (intensityblock − min(intensityregion))/sd(intensityregion). Then, z-scores were normalized to 0–1. When investigating the z-scores, a bimodal distribution was observed with a minor peak near 0 and a major peak at 0.4. The dip between these peaks was found at 0.14 and values below 0.14 (3.3% of total positive values) were filtered out.
The expression of T cells expressing inhibitory markers was done by dividing the 7.242 µm blocks into three categories as follows: (1) those directly adjacent to blocks with CD30 expression and the interaction ligand (LGALS1/HLA-DRA), (2) those directly adjacent to blocks with CD30 expression, but not the expression of the ligand, and (3) those no adjacent to a block with CD30 expression. For the three categories, in each region in each tissue, the number of blocks with T cells was determined (based on CD3E/CD8A expression). Then, the fraction of these T cell blocks that co-expressed the inhibitory receptor (CD69/LAG3) was determined. To calculate enrichment, the fraction of T cells that expressed the inhibitory receptor was compared between categories 1/2 and 3 (general enrichment around HRS cells) or between categories 1 and 2 (enrichment around HRS cells that expressed the interaction ligand).
AUTHOR CONTRIBUTIONS
Friederike Meyer-Wentrup, Frank C. P. Holstege, and Thanasis Margaritis initiated the project. Jurrian K. de Kanter, Alexander S. Steemers, Arianne M. Brandsma, Thanasis Margaritis, Friederike Meyer-Wentrup, Niels Groenen, and Ruben van Boxtel were responsible for the experimental design. Jurrian K. de Kanter, Alexander S. Steemers, and Ruben van Boxtel drafted the manuscript. Arianne M. Brandsma, Alexander S. Steemers, Niels Groenen, Thanasis Margaritis, and Laurianne Trabut performed the experimental work for the scRNA-seq. Alexander S. Steemers and Ravian L. van Ineveld performed the RNAscope and imaging. Jurrian K. de Kanter and Diego Montiel González performed the data analysis. Marijn A. Scheijde-Vermeulen, Liset Westera, and Friederike Meyer-Wentrup provided clinical information. Frank C. P. Holstege, Anne C. Rios, Arianne M. Brandsma, Auke Beishuizen, Thanasis Margaritis, Ruben van Boxtel, and Friederike Meyer-Wentrup supervised the project.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
FUNDING
This work was funded by the Foundation Kids Cancer Free (KiKa; no. 424) to Ruben van Boxtel and Friederike Meyer-Wentrup. In addition, the work was funded by an ERC consolidator grant from the European Research Council (ERC; no. 864499) to Ruben van Boxtel and supported by a collaborative grant initiative between the Princess Máxima Center, Utrecht University and the University Medical Centre Utrecht. Additionally, this work was supported by the Oncode Institute, funding Jurrian K. de Kanter, Alexander S. Steemers, Niels Groenen, Ruben van Boxtel, and Friederike Meyer-Wentrup. We thank Single Cell Discoveries for their help with library preparation. We also thank the Princess Máxima Center Single Cell Facility for aiding in the single-cell RNA sequencing and performing data processing. Imaging services were performed in The Princess Máxima Imaging Center (Utrecht, the Netherlands). FACS was performed with the assistance of the Princess Máxima Center FACS facility. Data and material were provided by the Biobank Data Access Committee and the Princess Máxima Center genomics core. We thank all patients and parents for donating the tissue. Ruben van Boxtel is a New York Stem Cell Foundation—Robertson investigator. This research was supported by The New York Stem Cell Foundation.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.