RNA binding protein-directed control of leukemic stem cell evolution and function
Abstract
Strict control over hematopoietic stem cell decision making is essential for healthy life-long blood production and underpins the origins of hematopoietic diseases. Acute myeloid leukemia (AML) in particular is a devastating hematopoietic malignancy that arises from the clonal evolution of disease-initiating primitive cells which acquire compounding genetic changes over time and culminate in the generation of leukemic stem cells (LSCs). Understanding the molecular underpinnings of these driver cells throughout their development will be instrumental in the interception of leukemia, the enabling of effective treatment of pre-leukemic conditions, as well as the development of strategies to target frank AML disease. To this point, a number of precancerous myeloid disorders and age-related alterations are proving as instructive models to gain insights into the initiation of LSCs. Here, we explore this myeloid dysregulation at the level of post–transcriptional control, where RNA-binding proteins (RBPs) function as core effectors. Through regulating the interplay of a myriad of RNA metabolic processes, RBPs orchestrate transcript fates to govern gene expression in health and disease. We describe the expanding appreciation of the role of RBPs and their post–transcriptional networks in sustaining healthy hematopoiesis and their dysregulation in the pathogenesis of clonal myeloid disorders and AML, with a particular emphasis on findings described in human stem cells. Lastly, we discuss key breakthroughs that highlight RBPs and post–transcriptional control as actionable targets for precision therapy of AML.
INTRODUCTION
Hematopoietic stem cells (HSCs) are essential for the life-long production of healthy blood, as they differentiate into all short-lived mature hematopoietic cell types found in blood through a series of transient progenitor states, while stably renewing themselves to sustain the stem cell reservoir.1 The tightly controlled balance between self-renewal and differentiation is essential for proper HSC function and blood production, and its dysregulation underlies many disease states including anemias, bone marrow failures, and malignancies such as acute myeloid leukemia (AML). AML is characterized by the rapid, clonal expansion of immature cells of the myeloid lineage, which are either stalled in their differentiation or have differentiated into abnormal, nonfunctional cells.2 AML accounts for over 30% of adult leukemias, and despite significant efforts toward therapy improvement, patients continue to suffer from chemoresistance, high relapse rates, and a less than 25% 5-year overall survival, indicating that novel AML therapeutics continue to be an urgent, unmet need.2, 3
Seminal findings validating the cancer stem cell model confirm that AML is sustained by a small population of leukemic stem cells (LSCs). LSCs originate from hematopoietic stem or progenitor cells (HSPCs) that acquire genetic alterations and dysregulation of their cellular and molecular organization which impart hyper-competitive self-renewal properties and abnormal differentiation profiles.4, 5 These alterations endow transformed HSPCs the capacity to durably sustain the leukemia in a manner analogous to that by which HSCs support the healthy blood system.6, 7 In addition to underlying AML initiation and maintenance, evidence suggests LSCs uniquely evade standard chemotherapy regimens and seed disease relapse, worsening AML outcomes.4, 5, 8 Thus, we believe deeper insights into the molecular framework of LSC evolution, maintenance, and chemoresistance can be harnessed to develop targeted anti–leukemic agents that effectively eradicate the disease from its roots.
AML incidents increase with age where age-related clonal hematopoiesis is typical.3, 9 Additionally, AML secondary to myeloid disorders (sAML), including myelodysplastic syndrome (MDS),4, 10, 11 Shwachman–Diamond syndrome (SDS) and Diamond Blackfan Anemia (DBA),12 occurs at substantially higher rates than the general population (Figure 1A). These conditions can in their own right impose significant disease burdens that will benefit from enhanced therapeutic options. For example, MDS manifests as cytopenia in one or more myeloid lineages with median overall survival of approximately 3 years and up to 40% transformation to sAML with more dismal outcomes.11 Moreover, as clear leukemia-sensitized states that harbor pre-leukemic clones, these conditions represent important stages for early disease intervention and provide a window into identifying key drivers of the evolution of the LSC-precursor cells (pre-LSC) that sustain them into bona fide LSCs.4, 13

A significant body of evidence is emerging that dysregulated post–transcriptional regulation underlies critical aspects of malignant myeloid development including propagation at the stem cell level. RNA is regulated post–transcriptionally through a combination of modifications, such as constitutive or alternative splicing, polyadenylation, methylation, and RNA fate decisions such as localization, stability, and translation (Figure 1B,C). Through these processes, post–transcriptional regulators diversify and fine-tune the genetic code with influence over all cellular processes. RNA-binding proteins (RBPs) are core effectors of post–transcriptional control, where they form dynamic ribonucleoprotein (RNP) complexes to spatially and temporally regulate their RNA targets. RBPs can also cooperate with or directly regulate noncoding RNAs, which themselves are post–transcriptional regulatory agents.14 RBPs function through their RNA-binding domains (RBDs), where the composition and arrangement of these domains within any one RBP creates a combinatorial effect that not only dictates the function of that RBP but also the specificity of its binding to RNA.15, 16 RBPs can also co-ordinately regulate classes of functionally related RNAs, to induce rapid pathway level alterations, and thus significant changes in cell function in response to both cell intrinsic and extrinsic stimuli.14 Recent technologies that couple RNA capture with mass spectrometry have revealed that RBPs number in the thousands and that many have not been explored for their functions in post–transcriptional control with a subset lacking conventional RBDs. Interestingly some of these newly identified RBPs have additional functionalities beyond RNA-association, such as DNA binding or metabolic regulation, highlighting the possibility that certain RBPs may participate in orchestrating the crosstalk of multiple layers of cellular programming.17, 18 Overall, the widespread and coordinated function of RBPs can classify them as master regulators of cellular identity and fate.
Given their powerful role as influencers of essential cellular processes, it is perhaps unsurprising that alterations in expression of, or mutations in, RBPs have often been implicated in carcinogenesis.19, 20 With methodological advances having only recently provided the ability to globally assess properties of the transcriptome and proteome with high precision at low inputs or single-cell resolution, there exists significant scope for future research into the cancer and/or stem cell-driving aspects of RBPs.21-23 In this review, we highlight RBP-driven processes that drive pathogenic myeloid behavior with an emphasis on regulation at the stem cell level across disease evolution, by describing RNA modifications, mRNA translation, stability, and splicing as the major post–transcriptional axes at play. We describe RBPs that are required for healthy HSC function, but whose activity becomes maladapted in the promotion of pre-leukemic and AML states (Table 1). In addition, we also highlight instances where RBPs are preferentially required for certain stages of disease, highlighting the complex networks of RBPs that drive disease evolution. Finally, we explore therapeutic strategies that have been used and are being developed to target RBPs in the treatment of AML.
| RBP | RBP function | Role in healthy hematopoiesis | Role in preleukemia | Role in AML | Citations |
|---|---|---|---|---|---|
| METTL3 | RNA modifications - m6A writer | Mettl3 KO in murine HSCs results in impaired differentiation, while catalytic Mettl3 loss causes an increase in neutrophil progenitors and reduced erythroid progenitors. METTL3 KD in human HSPCs causes increased differentiation and loss of primitive cells. | METTL3 is enriched in AML, where METTL3 loss induces differentiation, apoptosis, reduced cell cycle, and delayed in vivo reconstitution. | Healthy,24-26, 90 AML26, 27 | |
| METTL14 | RNA modifications - m6A writer | METTL14 KD in human CB CD34+ HSPCs induces monocyte/macrophage differentiation. | METTL14 is elevated in AML and promotes mRNA stability and translation to support AML maintenance and LSC function. | Healthy and AML28 | |
| MSI2 | Regulatory RBP – translational regulation | Enriched in murine and human HSCs. Msi2 influences the translation of transcripts involved in TGF-β signaling. Overexpression of MSI2 in CB HSPCs enhances self-renewal through translational inhibition of the AHR pathway. | MSI2 is elevated in HSPCs from MDS patients, where its loss reduced malignant engraftment in the NUP98-HOXD13 murine model of MDS. | MSI2 is upregulated in AML, where it impairs AML and LSC survival by regulating a variety of oncogenic pathways, as well as suppressing AHR signaling. | Healthy,29, 30: Preleukemia,31: AML,32-39, 148 |
| 4EBP1/2 | Translational regulator | Murine HSCs have hypophosphorylated 4E-BPs, where mice with double 4E-BP1/2 deletion have impaired secondary reconstitution. | Healthy40 | ||
| LIN28b | Regulatory RBP – miRNA regulation | Lin28b is preferentially expressed in mouse fetal HSCs compared to BM HSCs and inhibits biogenesis of let-7 miRNAs, leading to maintained expression of Hmga2. As human fetal HSCs mature into a neonatal state, HMGA2 undergoes alternative splicing mediated by CLK3 and SRSF1 to remove let-7 binding sites and escape degradation. | Aberrant elevation of LIN28/LIN28B facilitates transformation through the repression of let-7 microRNA maturation and subsequent de-repression of its targets. | Healthy:46, 95 AML,42, 43, 149 | |
| CLK3 | Splicing factor | As human fetal HSCs transition into the intermediate neonatal state, let-7 binding sites in the HMGA2 3′ UTR are spliced out, in part mediated by CLK3, to escape repression. | Healthy41 | ||
| YTHDF2 | RNA modifications - m6A reader | Conditional deletion of Ythdf2 results in upregulation of an inflammatory state, expansion of the HSC compartment and myeloid skewed primary reconstitution with secondary reconstitution severely impaired. | YTHDF2 is increased in AML, where its genetic inhibition impairs cell survival, clonogenic potential, and engraftment capacity in primary human AML through decreasing the half-life of pro-apoptotic mRNAs. | Healthy:96, AML44 | |
| RBM15 | Splicing factor | Rbm15 KO results in the upregulation of a shortened c-Mpl receptor isoform that in turn impairs thrombopoietin signaling and HSC engraftment. | Recruited to promoter-bound RNAs through RBFOX2 activity, and subsequently recruits the m6A methyltransferase complex to further modify the RNA. | Healthy,45: AML46 | |
| U2AF1 | Splicing factor | U2af1 knockout results in pancytopenia, lethality, and a reduction in immunophenotypic HSCs with impaired reconstitution capacity, attributed to altered exon inclusion and increased DNA damage. | Found to be mutated in MDS, typically as a heterozygous non-synonymous mutation. Mutations confer altered 3′ splice site recognition. Promotes the gain-of-function of IRAK4 through the inclusion of exon 4. | Recurrently mutated in AML, with a higher frequency in secondary AML. | Healthy,47: Preleukemia,48-51: AML52 |
| SF3B1 | Splicing factor | Heterozygous deletion of Sf3b1 in murine HSCs results in hypo proliferative HSCs, with reduced HSC frequency and impaired competitive reconstitution. | Found to be mutated in MDS, typically as a heterozygous non-synonymous mutation, and associated with the presence of ring-sideroblasts. Mutations confer altered 3′ splice site recognition. Promotes IRAK4 gain-of-function through inclusion of exon 6. | Recurrently mutated in AML, with a higher frequency in secondary AML. | Healthy,53, 54: Preleukemia,48, 50, 51, 55-57: AML52 |
| SRSF2 | Splicing factor | Homozygous Srsf2 deletion in murine HSCs results in leukopenia, anemia, and impaired reconstitution in competitive transplantation experiments. | Found to be mutated in MDS, typically as a heterozygous non-synonymous mutation. Mutations confer altered recognition of exonic splicing enhancers. The SRSF2 P95H mutation promotes inclusion of PTCs. This mutation also results in the inclusion of the EZH2 poison exon. | Recurrently mutated in AML, with a higher frequency in secondary AML. | Healthy,58: Preleukemia,48, 50, 51, 58, 59: AML52 |
| ZRSR2 | Splicing factor | Zrsr2 deletion in mouse HSCs results in their increased competitive self-renewal and clonal advantage. | Found to be recurrently mutated in MDS, where mutations encode for premature termination codons or loss-of-function inducing frameshifts. Typically results in the retention of conserved minor U12-type introns. | Recurrently mutated in AML, with a higher frequency in secondary AML. | Healthy,60: Preleukemia,48, 50, 102 AML52 |
| Ribosomal subunits: RPS19, RPL5, RPS26, RPL11, RPS14 | Ribosome component | Mutations in ribosomal subunits are prevalent in DBA, where reduced ribosome activity can selectively impair the translation of select transcripts. | Preleukemia61 | ||
| SBDS | Translational regulator | SBDS mutations, found in patients with SDS, results in defects in ribosome biogenesis through impaired loading of the large ribosomal subunit onto the small subunit. | Preleukemia61 | ||
| DNAJC21 | Translational regulator | DNAJC21 binds ribosomal rRNA and aids in the maturation of the 60 S ribosomal subunit. Patients with DNAJC21 mutations present with symptoms highly similar to SDS. | Preleukemia61 | ||
| PUS7 | RNA modification - pseudouridine writer | Loss of PUS7 impairs the differentiation of human HSPCs. | PUS7 is decreased in MDS with monosomy 7 or del(7q), and re-introduction of modified 5-tRNA fragments into high-risk MDS dampens protein synthesis to drive MDS differentiation and enhances lineage balanced engraftment in vivo. | Healthy,62: Preleukemia63 | |
| DDX41 | Regulation of splicing | Germline loss-of-function mutations in DDX41 induces MDS or AML, where shRNA mediated loss-of-function assays in human healthy HSPCs or primary MDS cells imparts hyperproliferative features. | Preleukemia64 | ||
| RBM39 | Splicing factor | Upregulated in AML, where it is needed for the efficient splicing of HOXA9 target mRNAs. | AML65 | ||
| RBM17 | Splicing factor | RBM17 is upregulated in LSC containing fractions of human AML, where its inhibition leads to myeloid differentiation, impaired colony output and engraftment. RBM17 repression leads to the inclusion of poison exons and subsequent NMD of pro-leukemogenic transcripts. | AML:133 | ||
| FBL | RNA modification − 2′O-Me writer | FBL deposits 2′-O-Me marks on rRNA to facilitate ribosome biogenesis. Depletion of FBL in patient AML samples is associated with reduced colony formation, and elevation of FBL increases leukemogenesis and confers LSC properties to blast cells. | AML66 | ||
| eIF4E | Translational regulator | eIF4E is often elevated in AML, where its chemical inhibition reduces expression of multiple oncogenic proteins. | AML67 | ||
| ZFP36L2 | Regulatory RBP | ZFP36L2 helps to maintain the undifferentiated state of AML by promoting mRNA degradation of myeloid maturation genes through 3′-UTR binding. | AML68 | ||
| ELAVL1 | Regulatory RBP | ELAVL1 is elevated in functionally validated LSC containing fractions of human AML. ELAVL1 loss induces myeloid differentiation of human AML and impaired engraftment, through suppression of RNAs related to mitochondrial function. | AML69 | ||
| ADAR1 | RNA modification - Base editor | ADAR1 is highly expressed in leukemic BM, where a splicing switch leads to overexpression of a hyper-editing form of ADAR1, ADAR1p150, within an inflammatory niche. ADAR1p150 leads to LSC generation and treatment resistance. | AML,70-72, 168 | ||
| FTO | RNA modification - m6A eraser | FTO is elevated in AML where its forced expression promotes leukemic transformation and impairs ATRA-induced AML differentiation. | AML73, 74 | ||
| ALKBH5 | RNA modification - m6A eraser | ALKBH5 is highly expressed in AML and is associated with poor prognosis, where it is nessessary for primary human AML engraftment. ALKBH5 functions through the stabilization of pro-leukemic transcripts. | AML75, 76 | ||
| YTHDC1 | RNA modification - m6A reader | YTHDC1 has increased expression in AML. YTHDC1 undergoes liquid-liquid phase separation upon binding m6A, and forms condensates that prevent mRNA degradation. Additionally recruited to promoter-bound RNAs by RBFOX2 activity. | AML:46, 152 | ||
| RBFOX2 | RNA modification - m6A reader | RBFOX2 recognizes m6A-modified noncoding promoter-associated RNAs, where it then recruits RBM15. RBFOX2 knockdown induces differentiation and reduces leukemic engraftment. | AML46 | ||
| IGF2BP1 | RNA modification - m6A reader | IGFBP1 supports leukemic activity by maintaining levels of HOXB4, MYB, and ALDH1A1. | AML77 | ||
| IGF2BP3 | RNA modification - m6A reader | Knockdown of IGF2BP3 impairs AML progression by destabilizing RCC2 in an m6A-dependent manner. | AML78 | ||
| eIF4A | Translational regulator | Inhibition of the RNA helicase eIF4A using rohinitib, shows antileukemic activity against FLT3-ITD mutated AML. | AML79 | ||
| RBM25 | Splicing factor | Clonal dominance of expanding Tet2 knockout HSC clones can partly be explained by reduced expression of Rbm25, potentially due to hypermethylation of its promoter. | In the context of Cebpa mutant murine leukemia, RBM25 knockdown enhances leukemia through decreased apoptosis and increased proliferation. As such, RBM25 functions as a tumor suppressor by regulating the splicing of apoptosis and MYC regulators. | Preleukemia,80: AML81 |
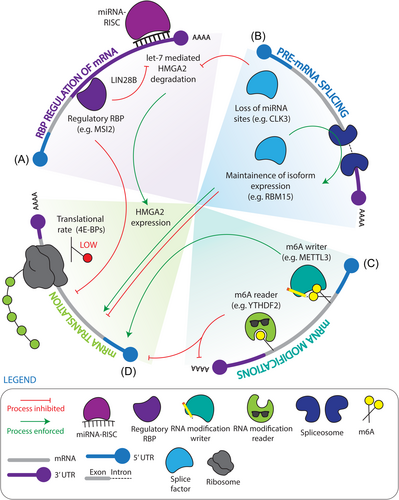
RBP-guided control of healthy hematopoietic stem cells
HSC fate decisions, including to remain quiescent, self-renew, differentiate, or induce apoptosis are critical to the life-long maintenance of a healthy blood supply. Transgenic murine models importantly accelerated the identification of determinants of HSC fate decisions.82 To extend this knowledge specifically in the human context, where there exists key differences, human hematopoietic dysregulation at the stem cell level is now best modelled by xenotransplantation into permissive niches in immunodeficient mice.83 The vast implications of fundamental insights derived from these assays have included forwarding paradigms in regenerative medicine, disease modelling, preclinical evaluation of experimental therapies, and the discovery of novel pathogenic factors. With regard to insights into the post–transcriptional level, it is clear that similar to other hierarchical tissues, HSCs and highly primitive progenitors exhibit significant de-coupling of proteomic and transcriptomic profiles compared to mature cells, implicating a role for post–transcriptional control within the primitive hematopoietic compartment.84, 85 Accordingly, seminal research has also demonstrated that murine HSCs and multipotent progenitors uniquely rely upon tightly controlled, low rates of protein synthesis to sustain their functional integrity compared to more mature cells, and this holds true independent of total mRNA abundance or quiescence status.40, 86 Considered together, these studies suggest that post–transcriptional regulation plays an essential role in dictating HSC fate and that its dissection may offer critical insights with translational impact.
Multimodal enforcement of the healthy HSC translatome
Beyond global translation rates, dynamic regulation of mRNA translation into protein products is influenced by abundant N6-methyladenosine (m6A) mRNA modifications.87 m6A deposition is mediated by a methyltransferase complex, including methyltransferase-like 3 and 14 (METTL3 and METTL14), along with Wilms tumor-associated protein (WTAP).87 Intriguingly, mouse long-term (LT)-HSCs have higher total RNA m6A levels compared to progenitors.88 Accordingly, knockout of Mettl3, the major catalytic unit of the m6A methyltransferase complex, in murine bone marrow results in an accumulation of non-functional HSC/progenitor-like cells, caused by an inability of HSCs to differentiate.24, 25 Moreover, Mettl3 knockout mouse HSCs stimulated to differentiate using cytokines were found to be unable to upregulate Myc protein,24, 25 a well-established enforcer of HSC activation and commitment.89 Since Myc mRNA levels remain unchanged, these findings indicate that decreased m6A levels may reduce Myc mRNA translation, stalling differentiation. Intriguingly, scRNA-seq analysis of murine Lin− and c-kit+ cells treated with the Mettl3 small-molecule inhibitor STM2457 to specifically block the catalytic activity of Mettl3, as opposed to full protein loss, showed a specific bias toward neutrophil progenitors at the expense of erythroid progenitors rather than the more multilineage impairment in maturation seen with Mettl3 knockout. This suggests that certain hematopoietic defects resulting from the loss of Mettl3 protein could be attributed to m6A-catalysis-independent functions.90 However, the role of METTL3 in healthy human hematopoietic stem and progenitor cells (HSPCs) remains paradoxical, as in contrast to the mouse context, in vitro culture of human cord blood CD34+ HSPCs upon METTL3 short hairpin RNA (shRNA) knockdown results in enhanced differentiation and loss of the primitive CD34 cell surface marker, while METTL3 overexpression shows the inverse.26 While differences observed across these studies could be due to differing levels of Mettl3 loss achieved by the distinct experimental methodologies used, it is also possible that the species, cell-type, or microenvironment may differentially influence the physiological role of deposited m6A. Of note, METTL14 has also been shown to play a role in human hematopoiesis, where METTL14 shRNA knockdown in cord blood CD34+ HSPCs induces monocyte/macrophage differentiation, suggesting that METTL14 is additionally required to control HSC fate.28
In addition to RNA modifications, translation of mRNA can also be influenced by the involvement of individual, sometimes cell-type specific, RBPs with the translation initiation machinery. The 5′-cap binding eukaryotic initiation factor (EIF) 4E binding proteins (4E-BP1-3) are ubiquitous inhibitors of translation initiation by binding and segregating EIF4E from EIF4G, thereby preventing the assembly of the EIF4F translation initiation complex. 4E-BPs are phosphorylated upon mTORC1 signalling, which reduces their binding to EIF4E and thus increases translation.91 In the mouse system, 4E-BP1 and 4E-BP2 are expressed in HSCs, where the 4E-BP pool is hypo-phosphorylated concordant with dampened translation rates.40 While mice with 4E-BP1/2 deletion have higher frequencies of immunophenotypic HSCs, these HSCs exhibited impaired reconstitution upon secondary transplantation, suggesting that low translation mediated by 4E-BPs may be necessary for long-term HSC maintenance or recovery through transplantation stress.40 Musashi-2 (MSI2) is an example of an RBP that can act to influence the translation of a select set of targets in a cell-type-specific manner to influence HSC fate. One mechanism by which MSI proteins are thought to repress translation is through competitive association with poly(A) binding proteins (PABP) to prevent complex formation with EIF4G that is required for ribosome assembly on target transcripts.92 In both the murine and human contexts, MSI2 is highly localized to the HSC compartment and in line with this expression profile, MSI2 has been shown to be essential for both murine and human HSC self-renewal.29, 36, 93 This has been linked to Msi2 binding and influencing the protein output of transcripts involved in pro-renewal TGF-β signaling in mouse HSCs such that Msi2 knockout HSCs exhibit reduced flux through this pathway.94 Supraphysiological MSI2 levels can also enforce human cord blood HSC expansion by inhibiting the translation of components of the aryl hydrocarbon receptor (AHR) signalling pathway.30
Post–transcriptional gene silencing is also mediated through microRNAs (miRNAs) which interfere with translation or direct the degradation of target transcripts. As miRNA interactors, specific RBPs can exert direct effects over these processes through miRNA intermediaries. A well-known HSC-associated RBP-miRNA axis is Lin28b–let-7–HMGA2. In the murine hematopoietic system, the Lin28 homolog B (Lin28b) RBP is preferentially expressed in highly renewing fetal HSCs, where it antagonizes the pro-differentiation let-7 class of miRNAs. This downregulates let-7-mediated mRNA degradation of the transcriptional regulator High mobility group AT-hook 2 (Hmga2), which enables high HSC renewal.95 Interestingly, let-7 levels rise as human HSCs undergo a developmental switch from fetal to neonatal states, and thus to ensure HSC renewal through this switch, a unique alternative splicing axis involving CLK3 and SRSF1 mediates excision of the let-7 target sequence from the HMGA2 3′-UTR.41 This strategy of insulating HMGA2 from miRNA-mediated repression illustrates the use of a choreographed interplay of post–transcriptional regulation of stability and splicing in governing a transcriptional master regulator of HSC fate and renewal decisions. Recently, the m6A reader YTH domain-containing family protein 2 (YTHDF2) has been shown to critically regulate mRNA stability through m6A recognition and promotion of de-adenylation. Conditional Ythdf2 deletion using the hematopoietic-specific Vav-iCre initially demonstrated an expansion of the HSC compartment, with no overt hematopoietic defects, along with reconstitution of primary recipients with myeloid skewing.44 However, reconstitution of secondary recipients was severely reduced, demonstrating an impairment of HSC self-renewal.96 Interestingly, transcriptomic profiling of Ythdf2-deficient HSC-enriched fractions demonstrated upregulation of an inflammatory signature. Since inflammation can drive HSC cycling and differentiation at the expense of self-renewal, Ythdf2 may thus maintain an pro-HSC state by post–transcriptionally suppressing HSC-depleting inflammatory responses.96, 97
mRNA isoform-level regulation of HSC activity
Eukaryotic precursor mRNAs (pre-mRNAs) harbor introns, which are non-coding regions that undergo combinatorial excision to produce a variety of mature mRNA isoforms. Studies of alternative splicing (AS) within the human hematopoietic system have intriguingly highlighted a sharp increase in AS and differential isoform abundance as HSCs differentiate into more mature populations.98 This may indicate that AS is required for effective blood production in the hematopoietic system by allowing HSCs to adopt diverse lineage fates. By extension, it also indicates the potential for dysregulation of this axis to seed impaired hematopoiesis. In contrast to more global analyses of splicing landscapes, several targeted approaches have in addition highlighted specific splicing factors critical to HSC function. Rbm15 for instance has been shown to regulate the thrombopoietin receptor, c-Mpl, which is necessary for megakaryocytic development and HSC renewal. Rbm15 knockout long-term (LT)- and short-term (ST)-HSCs showed an increased expression of a short c-Mpl isoform, termed Mpl-TR, which is known to have a dominant-negative effect on c-Mpl/thrombopoietin signalling and thus inhibits HSC engraftment in transplantation assays.45 SF3B1 is a critical component of the U2 spliceosome RNP complex that binds intronic regions upstream of the branch point, facilitating the recognition of 3′ splice sites (3′SS) by interacting with the U2AF1/U2AF2 heterodimer.99, 100 Deletion of one allele of the splicing regulator Sf3b1 in mouse HSCs had minimal effect on differentiation capacity or in situ hematopoiesis, however, HSCs were hypoproliferative, reduced in frequency, and impaired in competitive reconstitution capacity.53, 54 Mice with conditional HSC knockout of the splicing factor U2af1 demonstrated severe pancytopenia, with reductions in immunophenotypic HSCs that were also impaired in their repopulating capacity. Importantly, this was associated with an altered profile of exon inclusion in HSPC fractions and linked to increased DNA damage, potentially a consequence of mis-splicing of genes associated with HSC survival.47 Unlike SF3B1 and U2AF1, SRSF2 regulates alternative splicing by binding to exonic splicing enhancer sequences promoting exon recognition.101 Homozygous deletion of Srsf2 in mouse HSCs led to leukopenia and anemia, and importantly impaired HSC self-renewal when assessed by competitive transplantation.58 ZRSF2, found on the X-chromosome, is involved in the recognition of 3′SS of introns alternatively spliced by the minor U12-spliceosome.102 Surprisingly, Zrsr2 deletion in murine HSCs is associated with enhanced competitive self-renewal and clonal advantage.60 Together these findings exemplify splicing control as a key regulatory layer that acts on core HSC renewal factors.
In summary, there is an emerging appreciation that post–transcriptional control, a highly interconnected system, is essential to HSC fate determination (Figure 2), and the presence of RBPs at this key nexus implicates them as factors that may become dysregulated in malignancy. In the next sections, we will explore how a number of these same regulators become maladapted for disease and the mechanisms that allow them to do so. We will also describe regulators that become newly acquired in malignancy, and thus represent attractive therapeutic targets for their healthy HSC-sparing nature.
RBPs in leukemia predisposition
Leukemia-associated mutations can be found in healthy patients long before clinical diagnosis, including from birth.103, 104 Their presence in a heterogeneous pool of HSPCs promotes clonal fitness and the opportune setting for accumulation of further genetic or intracellular dysregulation to give rise to frank AML, supporting the existence of pre-LSCs that drive pre-leukemic states.4, 13 Clonal hematopoiesis (CH) occurs when dominant HSC clones contribute more prominently to hematopoietic output. CH is virtually guaranteed with increasing age,104, 105 and while it can be asymptomatic, it is associated with elevated risk of AML transformation.9, 106, 107 MDS is also an age-related stem cell-driven hematopoietic premalignancy with approximately 30% of patients progressing to highly refractory secondary AML.4, 13 In contrast to CH and MDS, Schwachman–Diamond syndrome (SDS) and Diamond Blackfan anemia (DBA) are congenital bone marrow disorders characterized by neutropenia and anemia, respectively, and associated with elevated AML predisposition.12 A striking overrepresentation of RBP mutations, as well as burgeoning evidence of molecular-level disruption of RBP-directed processes, in the stem cells of these conditions underscores a critical contribution of post–transcriptional dysregulation in the genesis of pre-LSCs (Figure 1, 3). The sections below explore this paradigm using these intermediate states as instructive models of the process of leukemia initiation and evolution.
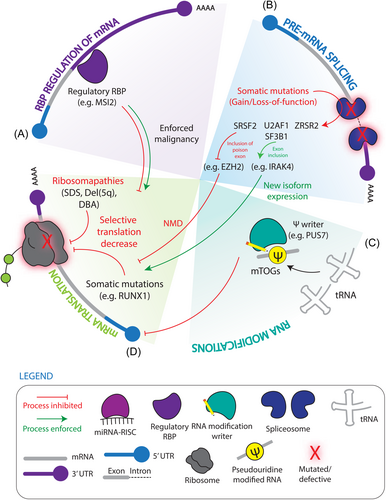
Translational regulation in preleukemia
Ribosomopathies are underpinned by mutations in ribosome proteins (e.g., DBA: RPS19, RPL5, RPS26, RPL11, del(5q): RPS14) or defects in ribosome biogenesis (e.g., SDS: SBDS and DNAJC21 mutations). While inherited SDS and DBA affect a number of tissues, the prominent manifestation of hematopoietic dysfunction underscores a unique reliance on ribosome homeostasis and post–transcriptional control for healthy blood production and seeding transformation in HSCs.12, 61, 108, 109 The molecular basis of tissue specificity of ribosomopathies is in part related to altered translation of select subsets of transcripts. A chief example with clinical implications is the selective reduction in translation efficiency of GATA1 mRNA encoding the erythroid-lineage master regulator in ribosome protein (RPS19 and RPL5) haploinsufficient human HSPC models of DBA.110 Two mutually inexclusive models to explain this have been proposed, one being that defective ribosome components result in the formation of heterogeneous or specialized ribosomes with unique propensity for translating a subset of mRNA targets. An alternative concentration hypothesis posits that defective ribosome components results in fewer translationally competent ribosomes having an outsized effect on reducing translation on the subset of transcripts.108 Importantly, ribosome deficiency also triggers a p53-dependent nucleolar stress response to induce cell cycle arrest and apoptosis, and one proposal explaining the tissue-specificity of ribosomopathies is the contribution of compensatory mechanisms to quell p53 signaling.111 Lastly, the apparently paradoxical predisposition of these hypo-proliferative disorders to secondary leukemia speaks to the role of translation-level dysregulation acting as a priming agent in HSC transformation.
Indeed, it is largely believed that regulated low translation rates in healthy HSCs confers protection during HSC-depleting stress, which may serve as a mechanism of clonal selection. The transcription factor RUNX1 is commonly mutated in a variety of hematological malignancies and MDS, often presenting as an early disease driver.112 Intriguingly, Runx1-deficent murine HSPCs demonstrate reduced translation rates and ribosome biogenesis, which was associated with resistance to endogenous or genotoxic stress.113 Additionally, in a mouse model of pre-leukemia driven by expression of the exon 9a variant of the AML1-ETO fusion protein, the progression from a pre-leukemic to leukemic state can be accelerated through the loss of Kat2a. In this model, loss of Kat2a can supress protein synthesis and ribosome biogenesis to drive this transition, a finding replicated by pharmacological repression of translation in Kat2a expressing AML1-ETO9a HSPCs.114 Interestingly, high-risk MDS patients show increased rates of protein synthesis,115 suggesting that while low translation rates may be exploited in seeding a dominant pathological clone, as disease aggression increases, demands for enhanced translation follow.
RNA pseudouridylation (Ψ) is a highly abundant modification present on a diverse range of RNA species including transfer RNA (tRNA), ribosomal RNA (rRNA), and mRNA, and is synthesized by pseudouridine synthase (PUS) enzymes which can catalyse the uridine to Ψ conversion.116 PUS7 was originally identified as enriched in human embryonic stem cells, where it modifies small 5′-tRNA fragments derived from tRNA containing 5′-oligoguanine (mTOGs).62 mTOGs were found to be inhibitors of cap-dependent translation initiation by associating with polyadenylate binding protein 1 (PABPC1) and reducing its cap-association.62, 63 Intriguingly, PUS7 and mTOG levels are reduced in MDS with monosomy 7 or deletion 7q, and concordantly shRNA knockdown of PUS7 in healthy human HSPCs resulted in impaired differentiation.62 Importantly, lipid-based delivery of Ψ-mTOGs into high-risk MDS samples selectively dampened protein synthesis compared to healthy controls and enhanced in vitro differentiation.63 Xenotransplantation experiments revealed that the addition of Ψ-mTOG enhanced engraftment of high-risk MDS cells, while decreasing malignant stem and progenitor cells defined by CD123 expression and promoting more balanced lymphoid-myeloid output.63 These findings demonstrate the potential for modulating RNA modifications as clinical tools to treat pre-malignant hematopoietic states through ultimately influencing global translation. An example of potentially more selectively rewired translation dysregulation in the pre-leukemic context has been provided by MSI2, which was found to be elevated in the stem and progenitor-enriched fraction of high-risk MDS bone marrow and associated with poor survival.31 Msi2 knockout in the NUP98-HOXD13 transgenic mouse model of MDS significantly impaired competitive clonogenicity in vivo allowing for recovery of the host hematopoietic system to reverse the MDS-like disease state. Inversely, Msi2 overexpression exacerbated disease burden induced by NUP98-HOXD13 MDS HSCs while activating a number of disease-associated transcriptional patterns including NRAS-activated signature, reduced quiescence, and increased progenitor phenotypes.31 Altogether these findings showcase the dysregulation of translational control in pre-leukemic states and also demonstrates that essential players in healthy HSCs can be pathologically repurposed underscoring the need for careful dissection of their context-dependent targets and effectors.
Contributions of spliceosomal mutations to pre-leukemic states
More than 60% of MDS patients harbor early driver mutations in the splicing factors SF3B1, SRSF2, U2AF1, or ZSRS2,117 which are also present in and strongly predispose CH cases to leukemic transformation.48, 50, 51 Accordingly, MDS and AML patient transcriptomes exhibit profoundly altered splicing landscapes, suggesting that global splicing dysregulation is a hallmark of myeloid pre- and frank malignancies.118-120 Together, this emphasizes the importance of appropriate regulation of proteome diversity in preventing malignant states, while conversely demonstrates that transcriptome-level lesions through aberrant changes in isoform presence are essential for promoting pathogenesis. Moreover, as the majority of these alternative splicing events have yet to be assigned roles in MDS/AML genesis, this spotlights a vast platform for discovery of therapeutic targets.
Mutations in SF3B1, SRSF2, and U2AF1 typically occur as heterozygous nonsynonymous substitutions concentrated in hotspots located within or surrounding protein domains responsible for protein–RNA and protein–protein interactions.51, 57 These result in aberrant recognition of 3′ splice sites by SF3B1 and U2AF1, or exonic splicing enhancers by SRSF2.49, 56, 58, 59 The “change-of-function” nature of these mutations is exemplified by experiments with paired murine models of Srsf2 knockout or the Srsf2 P95H mutation which, despite imparting some similar hematopoetic defects, were associated with differences in RNA recognition and splicing patterns. Differential RNA binding preferences of SRSF2 P95H promotes the inclusion of premature termination codons (PTCs) in target transcripts, and together with its enhanced capacity over SRSF2 WT to recruit NMD machinery downstream of PTCs, results in a widely reshaped transcriptional landscape through AS-coupled NMD.58, 121 A chief example of this regulatory mechanism is in the context of both murine HSPCs and primary patient myeloid malignancy where the SRSF2 P95H mutation, but not murine Srsf2 knockout, enforces preferential inclusion of a “poison exon” in the enhancer of zeste 2 (EZH2) transcript to introduce a PTC and induce NMD. In SRSF2 P95H expressing human leukemia cell lines, EZH2 protein levels and the H3K27me3 chromosome modifications it installs are reduced, consistent with the NMD event. Introduction of canonical EZH2 transcript partially rescued hematopoiesis, with cells expressing the canonical transcripts forming ~50% more colony-forming units compared to control. Notably, EZH2 loss-of-function mutations, common in MDS, are mutually exclusive with SRSF2 mutations, emphasizing the profound impact post-transcriptional players can have through their hold over master regulators of cell fate and pathogenesis.58 In a further example of this, another well characterized aberrantly spliced transcript in MDS is the interleukin-1 receptor-associated kinase 4 (IRAK4) which undergoes isoform switching in the context of both U2AF1 and SF3B1 mutations. While mutated U2AF1 promoted inclusion of exon 4 and mutated SF3B1 promoted inclusion of exon 6, both long IRAK4 isoforms appear to act as gain-of-function splice variants that increased toll-like receptor-mediated NF-kB signaling. Treatment of both U2AF1- and SF3B1- mutated patient MDS samples with IRAK4 shRNAs or small molecule inhibitor increased differentiation assessed by colony formation and reduced pre-LSC driven MDS burden as measured by reduced primary patient-derived xenotransplantation efficiency and impaired secondary engraftment.55, 122 These findings indicate that while there exists splice-factor mutant subgroups in MDS that often possess unique features, convergent splicing dysregulation, as exemplified by IRAK4, may enable a more global clinical targeting regardless of mutational subgroup and thus allowing therapies to benefit a larger patient population. In contrast to change-of-function variants, ZRSR2 mutations encode premature stop codons or frameshifts conferring a loss-of-function, causing the retention of highly conserved minor U12-type introns102 and to this point, as discussed above, ZRSR2 knockout mice exhibit highly MDS-like phenotypes.60
Additionally, germline frameshift or nonsense mutations of the RNA helicase DDX41 induces a hereditary MDS/AML with long latency but high penetrance. Hypomorphic somatic mutations in the helicase domain of DDX41 are also common in myeloid malignancy, but their frequent co-occurrence in germline mutated patients suggests a correlation between leukemogenesis and DDX41 inactivation. Indeed loss-of-function modelling with shRNAs in CD34+ healthy HSPCS, primary MDS, and various cell lines indicated DDX41 depletion imparted hyperproliferative features. As DDX41 mutations are associated with normal karyotypes and the absence of other known driver mutations they are postulated as a “first hit” that imparts a bona fide pre-leukemic state. While RNA helicases can operate in a number of molecular functions, mass spectrometry-based profiling of DDX41 protein interactors in HEK293 cells uncovered an enrichment of splice factors that is disrupted by the common somatic mutation (R525H), and deep sequencing of blast cells from 5 DDX41-mutated patients revealed substantially altered exon inclusion, suggesting that perturbed splice regulation at least in part underpins DDX41-driven leukemogenesis.64
Beyond overt mutations in splice-regulating RBPs, evidence also supports that inappropriate molecular-level control over post–transcriptional regulators contributes to the development of pre-leukemic clones. For example, loss-of-function mutations in the epigenetic modifier ten-eleven translocation 2 (TET2) are common drivers of CH and Tet2 knockout in murine HSCs also induces clonal dominance.123 Importantly loss of Tet2 alone cannot ubiquitously enforce clonal expansion and for the select set of clones that do become dominant there was a diminished expression of RNA splicing regulators as a class. The differential expansion capacities can in part be explained by reduced levels of the splice regulator Rbm25, likely due to hypermethylation of its promoter.80 Altogether, genetic and molecular features of these leukemia-sensitizing states and their stem cells points strongly to the role of post–transcriptional dysregulation as leukemia-priming agents.
RBP regulation of the AML state and leukemic stem cell function
As a stem cell-driven cancer,124 the future of effective AML management demands a paradigm shift that prioritizes the elucidation of LSC vulnerabilities which can be translated into therapies to eradicate AML at its roots. Importantly, the function-based definition of LSCs requires an in vivo transplantation approach to conclusively and specifically assess altered LSC function through defective serial reconstitution potential.6, 7, 125 To these points, our group identified a significant elevation of RBPs as a class in expression profiles of fractions containing primitive leukemic cells from AML patients and engineered a two-step in vivo screen validating RBPs as pervasive dependencies in LSCs.69 Moreover, a vast and growing arena of investigation is assigning pro-leukemic roles for specific RBPs, including in some cases as LSC dependencies, highlighting promising therapeutic opportunities, which we discuss in the sections below (Figure 4).
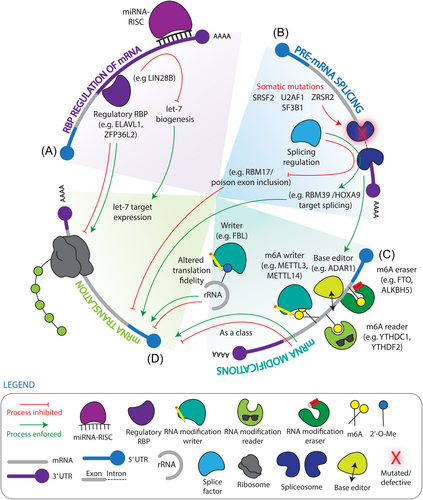
Dysregulation of RBPs involved in mRNA splicing
Approximately 29% of expressed genes are differentially spliced in primitive CD34+ bone marrow cells in AML patients compared to healthy donors.126 Importantly, splicing signatures of AML patients improved the accuracy of the existing three-group risk classification, suggesting that altered splicing is linked to disease outcomes and could serve as prognostic clinical signatures.127-129 Intriguingly, recurrent differential AS exists between patients with poor versus good prognosis independent of splicing factor mutations, indicating that altered splicing may be a global and essential pathogenic process in AML, whose targeting can have broad benefits across the patient population.127
Somatic mutations in SRSF2, SF3B1, U2AF1, and ZRSR2 are present in approximately 10% of AML patients130, 131 and are more frequent in older patients and in AML secondary to MDS, concordant with the descent of these leukemias from pre-leukemic MDS stem cells.52 Abnormal splicing factor expression is also found in AML samples compared to healthy controls.65, 132 RBM39, upregulated in AML compared to normal human CD34+ HSPCs, is required for efficient splicing of mRNAs encoding HOXA9 targets.65 Diminished RBM25, which was previously discussed as a mediator of Tet2-induced CH, is a tumour suppressor in AML cell lines that negatively regulates MYC through splice control of its post–translational regulator BIN1.81 More recently, our study focusing on dysregulated expression of splicing factors in human AML demonstrated that upregulation of the splicing factor RBM17 preferentially marks LSC-containing fractions, sustains patient LSCs in vivo, and directly correlates with shortened AML patient survival. RBM17 inhibition in primary AML cells leads to myeloid differentiation, impaired colony formation and in vivo engraftment. Integrative omics analyses showed that RBM17 repression leads to inclusion of poison exons and production of nonsense-mediated decay (NMD)-sensitive transcripts for pro-leukemic factors EIF4A2, RBM39, EZH2 and HNRNPDL,133 presenting another example of AS-NMD in the molecular pathology of leukemia and suggesting that RBM17 and its downstream leukemic AS events represent potential therapeutic targets for AML treatment.
Intriguingly, aberrant AS can also be linked to other mutation types within AML. For instance, in the 10%–15% of AML patients with the t(8,21)(q22;q22) AML1-ETO translocation, the resulting fusion protein retains the DNA-binding specificity of AML1 and the ability to recruit ETO-associated regulators that support self-renewal of the hematopoietic progenitor cells.134, 135 While AML1-ETO expression can induce myeloproliferative disorders in mice, expression of this fusion protein alone is not sufficient for leukemogenesis.134, 136-142 What was not tested however is expression of AML1-ETO9a, an alternatively spliced isoform of AML1-ETO known as AE9a that lacks the c-terminal NHR3/4 domains in ETO and is expressed in 70% of t(8,21) + AML patients.143 In contrast to AML1-ETO, when the shorter AE9a was similarly interrogated in later work by overexpression in mouse fetal liver cells it was found that introduction of this isoform drives AML initiation, highlighting a direct role for post–transcriptional regulation of splicing in leukemogenesis.144, 145 Overall, these studies demonstrate that genetic- and molecular-level dysregulation of splicing is a pro-leukemic pathogenic feature that is also shared in LSCs and likely carried forward from pre-LSCs.
Dysregulation of RBPs controlling the translational landscape
As is typical for cancer cells, leukemia mRNA translation landscapes are altered at the level of the translational machinery with the observation that eukaryotic translation initiation factor eIF4E is often elevated. Chemical inhibition of eIF4E/eIF4G assembly using 4EGI-1 was found to reduce ribosome-association and protein expression of multiple oncogenic transcripts including c-Myc, Cyclin D1, and Bcl-xL while inducing leukemic cell death.67 More recently, the paradigm of translation inhibiting anti-leukemic therapies has been explored at the stem cell level, whereby the protein synthesis inhibitor flavagline rocaglamide impaired in vivo primary AML leukemic repopulating cells while comparatively sparing normal HSCs.146 Additionally, MSI2 expression is upregulated in human AML and LSC-enriched fractions and is strongly associated with poor prognosis.32, 36-38 Validating it as an AML dependency, knockdown of MSI2 in leukemic cells results in increased apoptosis, decreased proliferation, enhanced chemosensitivity and reductions in LSC frequency.33, 35-37, 94, 147 MSI2 regulates a large proportion of genes integrally linked to oncogenesis, including Tspan3 and FLT3, which maintains LSC self-renewal and disease progression.34, 147 Interestingly, global mapping of MSI2 RNA targets in enriched fractions of mouse HSCs and LSCs using an adapted HyperTRIBE technique showed MSI2 binding was significantly increased in LSCs, leading to selective regulation of MSI2's oncogenic targets, providing a possible indication of differential dependencies on MSI2 targets in LSCs and HSCs.148 Here, the translational regulation by MSI2 was demonstrated through decoupling of mRNA and protein expression of MSI2 bound targets, where despite mRNA levels of targets remaining unchanged, protein levels were substantially reduced demonstrating translational repression.148 Indeed, while MSI2 also translationally represses AHR signaling in human HSCs to promote self-renewal, we have further demonstrated that AHR signalling is suppressed in human LSCs, where activation of AHR signaling was sufficient to impair LSC activity but not normal human HSPCs. This indicates that divergent molecular landscapes between leukemic and normal cells can be co-opted by RBPs in directing distinct malignant vs healthy fate decisions and enable the selective targeting of LSCs.37
Aberrant re-activation of fetal-specific LIN28/LIN28B in the adult context facilitates hematopoietic cell transformation42 and its inhibition results in cell cycle arrest, cell growth inhibition, and metabolism impairment.149 Since LIN28B is downregulated in healthy adult HSCs it represents a potentially selective anti–leukemic target which may spare adult HSCs co-existing within the bone marrow.41, 95 AU-rich elements present in mRNA 3′-UTRs are also central hubs for regulation of mRNA stability.150 A recent study using surface antigen-guided CRISPR screening identified ZFP36L2 as a critical regulator of maintaining the undifferentiated state and survival of AML cells through promoting mRNA degradation of key myeloid maturation genes through association with AU-rich elements in 3′UTRs.68 Recently, through the above mentioned in vivo CRISPR screens focusing on functionally assessing RBPs necessary for LSC activity, we identified the AU-rich associated RBP ELAVL1 to be a cross-species requirement for LSC activity.69, 151 ELAVL1 is elevated in functionally validated LSC-containing fractions of human AML, with relatively reduced expression in healthy LT-HSCs compared to more mature progenitors, suggesting selective activity within LSCs. Indeed, shRNA-mediated depletion of ELAVL1 induced myeloid differentiation of primary AML cells and impaired their overall in vivo engraftment upon xenotransplantation, underscoring an impairment of stem cell function. Multiomic mapping of ELAVL1 RNA targets in AML cells revealed a suppression of RNAs related to mitochondrial function,69 an emerging critical determinant of LSC function, further demonstrating important layers of oversight and crosstalk between post–transcriptional and metabolic regulation of LSCs.
Dysregulation of RBPs controlling mRNA modifications
Aberrant regulation of m6A mRNA modifications has been described in leukemia and LSC promotion in considerably greater detail than in HSC or pre-LSC contexts. As a class, m6A mRNA modification regulators, including writers (RNA methytransferases), readers, and erasers (RNA demethylases), appear elevated in AML. While this appears paradoxical in the promotion of disease, one explanation is that their co-expression does not necessitate equal-and-opposite functions, but rather reflects a global disruption of m6A use in leukemia. The highly complex, intertwined, and transcript-dependent actions of readers, writers, and erasers has also complicated the identification of global patterns of dysregulation, and may suggest that identification of the direct targets of m6A regulators holds the greatest potential to reveal tractable leukemic vulnerabilities. The m6A writer METTL3 is more abundant in primary AML patient samples compared to normal CD34+ HSPCs and correlates with higher global m6A levels.26 Depletion of METTL3 in human AML induces myeloid differentiation, apoptosis, cell cycle arrest, and delayed in vivo leukemia progression.26, 27 Through an integrative omics approach, Vu et al. showed that m6A promotes translation of oncogenic transcripts such as MYC, BCL2, and PTEN, and loss of METTL3 results in activated PI3K/AKT and apoptotic pathways which contributes to myeloid differentiation and apoptosis.26 With regards to other m6A writers, METTL14 has also been found elevated in human AML compared to healthy controls, and is required for AML development, maintenance, and LSC self-renewal, by promoting the stability and translation of MYB and MYC.28
The m6A eraser, obesity-associated protein (FTO), is highly expressed in AML with t(11q23)/MLL-rearrangements, t(15;17)/PML-RARA, RLT3-ITD and/or NPM1 mutations. Forced expression of FTO promotes leukemic transformation and inhibits all-trans-retinoic (ATRA)-induced AML cell differentiation.73 Transcriptomic m6A and RNA-seq analysis of human MONOMAC-6 AML cells with or without overexpression of FTO demonstrated that FTO enforces reduced RNA levels of its downstream targets, such as ASB2, RARA, and immune checkpoint genes of the LILRB4 family by reducing m6A levels.73, 74 ALKBH5, another m6A demethylase, is highly expressed in AML, is associated with poor prognosis, and is essential for primary AML LSC function as measured through xenotransplantation. Interestingly, ALKBH5 was found to increase mRNA stability of the receptor tyrosine kinase AXL and mitotic spindle stabilization factor TACC3 in an m6A-dependent manner.75, 76 These studies demonstrate that depending on the effector and transcript, m6A modifications can be stabilizing (of pro-leukemic targets) or destabilizing (of anti-leukemic targets), supporting the concept that m6A regulation is highly nuanced.
The YTH family of proteins are readers of m6A, which recognize and bind to m6A-modified transcripts and regulate their expression by controlling mRNA splicing, stability, structure, export, and translation.70 YTHDC1 was found to be highly expressed in different AML subtypes compared with normal controls.152 Through binding to m6A, YTHDC1 undergoes liquid–liquid phase separation and forms nuclear YTHDC1-m6A condensates (nYACs). Importantly, nYACs are more abundant in AML cells compared with normal HSPCs and are essential for the maintenance of AML cell survival and blocked differentiation. Mechanistically, nYACs protect m6A-mRNAs, in particular MYC, from the nuclear exosome and polyA tail exosome-associated RNA degradation, thus promoting mRNA stability and translation.152 Another family member, YTHDF2, is highly expressed across human AML with diverse cytogenetic abnormalities, where its expression correlates with LSC activity and its genetic inhibition impairs cell survival, clonogenic potential, and engraftment capacity of primary AML cells.44 Gene expression and transcriptome-wide mRNA m6A profiling of YTHDF2 knockout mouse cells demonstrated that YTHDF2 decreases the half-life of diverse m6A transcripts, including the pro-apoptotic factor TNFR2, that contributes to the overall integrity of LSC function.44 YTHDC1 and YTHDF2 provide a further example that while readers may have the same directive of recognizing m6A, they can enforce different consequences to the marked transcripts that endows them their diverse function. Recently, RBFOX2 was identified as a novel m6A reader, where it intriguingly links m6A-based RNA regulation directly with chromatin state and gene transcription as opposed to the more common mRNA regulation explored previously in this review. RBFOX2 was found to recognize m6A-modified non-coding promoter-associated RNAs (paRNAs), where it recruits RBM15 and subsequently the METTL3-METTL14 containing m6A methyltransferase complex to additionally methylate paRNAs in a suggested positive-feedback loop. The deposited m6A then recruits YTHDC1 which in turn commandeers the polycomb repressive complex 2 (PRC2) and subsequent transcriptional repression. Interestingly, RBFOX2 is elevated in AML patient samples compared to healthy controls and associated with reduced survival, where its knockdown reduced leukemic engraftment and increased differentiation in AML xenografts. Thus, RBFOX2 represents an example of crosstalk that can occur between transcriptional and post-transcriptional control to regulate leukemic cells.46 Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) have recently been reported as a distinct family of m6A readers that protect m6A-modified mRNAs in P-bodies and stress granules from degradation through interacting with various RBPs to promote mRNA stability and translation.153 IGF2BP3 is upregulated in AML patients and correlated with poor prognosis, and knockdown of IGF2BP3 significantly impairs AML progression both in vitro and in vivo through destabilizing regulator of chromosome condensation (RCC2) mRNA in an m6A-dependent manner.78 While IGF2BP1- and IGF2BP2-mediated m6A regulation has not been linked to their roles in leukemia, IGF2BP1 in particular is required for supporting LSCs by maintaining levels of HOXB4, MYB, and ALDH1A177 and IGF2BP2 is upregulated in AML patients and are correlated with poor prognosis.154
In addition to m6A modification of mRNA, 2′-O-methylation (2′-O-Me) of ribosomal RNA (rRNA) by the methyltransferase FBL impacts ribosome biogenesis and translation. Intriguingly, rRNA global 2′-O-Me patterns correlate with cellular identities in the human hematopoietic hierarchy and patient AML. A particular set of 2′-O-Me events is strongly correlated with LSC gene signatures and is selectively impaired with a concomitant reduction in progenitor activity by FBL depletion in patient AML. Conversely in this setting, enforced FBL expression elevated immunophenotypic LSCs and imparted leukemia-initiating capacity to leukemic blast cells in xenotransplant assays. Mechanistically, the FBL-induced rRNA ribomethylome promotes utilization of optimized codons to ensure the translational fidelity of amino acid transporters, and thus acts as a post–transcriptional master regulator of amino acid metabolism homeostasis required to sustain human LSCs.66
RNA editing also occurs at the level of RNA base conversions, among which adenosine-to-inosine (A to I) editing within double-stranded RNA (dsRNA) mediated by adenosine deaminases (ADARs), such as ADAR1, is the most prevalent. The resultant inosine bases are recognized as guanosine, which can alter the transcript's subsequent splicing and translation.155 ADAR1 was found highly expressed in AML-infiltrated bone marrow compared to cells from AML patients at complete remission (CR) and healthy controls at both mRNA and protein levels. Moreover, a splicing switch leading to the overexpression of a hyperediting form of ADAR1 (ADAR1p150) in response to an inflammatory niche has been shown to promote LSC generation and confer secondary AML (sAML) treatment resistance.71, 156 Depletion of ADAR1 in the K562 cell line resulted in reduced proliferation, cell cycle arrest, and the repression of Wnt pathway effectors β-catenin, c-Myc, TCF4, and CCND2168 while ADAR1 knockdown in primary sAML cells reduced expression of LSC-associated transcripts including CD44v3 and MCL1-L,72 indicating it has important functions in maintaining leukemic cell function. Altogether these studies underscore diverse epitranscriptomic alterations and their dysregulated effectors as critical determinants of AML, including in the LSCs seeding the disease.
Therapeutic approaches to target pre-leukemic/leukemic stem cell RBP-driven regulons
The identification of many post-transcriptional determinants of pre-LSC and LSC function, or AML in general, provides the impetus to explore these factors as targets for a new class of anti-leukemic therapeutics. One of the key pillars of any therapeutic is ensuring specificity of the agent to the leukemic compartment and ideally also the LSCs, while sparing healthy HSCs co-existing within the bone marrow that are essential for the regeneration of a healthy blood system following clearance of the leukemia. Recently, new methods to deliver a variety of bio-active molecules and compounds have emerged, broadening the toolset available to target LSCs. To investigate potential efficacy in the human context, these methods can be paired with preclinical xenotransplantation strategies that model the coexistence of AML and healthy hematopoietic cells within an in vivo microenvironment, thus enabling a more clinically relevant evaluation of RBP targeting therapies as a novel means to halt leukemia growth and relapse. Below we will touch on preclinical evaluation and clinical trials of RBP inhibitors as potent and selective AML- and LSC-targeting therapies (Table 2).
Elevated dependence on protein biosynthesis in cancer cells compared to healthy counterparts makes translation inhibition a coveted goal in cancer and AML therapy, and previously described experiments of translation inhibition using 4EGI-1 support its anti-leukemic effects.67 However, broad-acting translation inhibitors have been challenging to translate into the clinic due to high multiorgan toxicity, necessitating the elucidation of agents with higher leukemia-selectivity/specificity. Rohinitib, which targets the RNA helicase eIF4A and dampens the translation of oncogenic transcripts, showed enhanced anti-leukemic activity against AMLs with FLT3-ITD mutations. Rohintib treatment of primary AMLs was performed ex vivo and the effect on LSCs was measured by xenotransplantation, therefore it would be of great interest to evaluate its in vivo efficacy on established patient-derived xenotransplants.79 Beyond these preclinical studies, in strong support of their potential, translational inhibitors have progressed past the preclinical stage. For example, omecetaxine was tested in a phase 2 trial in combination with sorafenib for treatment of patients with relapsed/refractory or newly diagnosed FLT3-ITD mutated AML (NCT03170895), and was shown to be safe and effective.157, 162
| Clinical intervention | Malignant state | Preclinical studies | Clinical trials | Citations |
|---|---|---|---|---|
| Lipid-based delivery of pseudouridine-modified mTOGs | Preleukemia: high-risk MDS | Pseudouridine-modified 5′-tRNA fragments from tRNA containing 5′-oligoguanine (mTOGs) selectively dampen translation in high-risk MDS and promotes more balanced myelo-lymphoid engraftment in vivo. | [62, 63] | |
| Rocaglamide | AML | Global reduction in nascent translation using the flavagline rocaglamide results in the loss of AML-repopulating cells while sparing healthy HSPCs. | [146] | |
| FICZ | AML | FICZ is an AHR agonist, where in vivo treatment of prior established AML xenografts reduces leukemic burden in primary mice and reduced secondary engraftment. | [37] | |
| Dihydrotanshinone-I (DHTS) | AML | DHTS inhibition of ELAVL1 increases levels of myeloid antigens, increases apoptosis, and decreases CFU output in primary AMLs in vitro, as well as primary AML reconstitution of cells pre-treated with DHTS. | [69] | |
| MS-444 | AML | Impairment of ELAVL1 by MS-444 increases differentiation and cell death in vitro of primary AML samples. In vivo treatment of established AML xenografts reduces leukemic engraftment. Secondary transplant at limiting dilution revealed a quantitative loss of functional LSCs. | [69] | |
| 4EGI-1 | AML | 4EGI-1 is a 4E-BP1 mimic, thus inhibiting translation complex assembly. 4EGI-1 treatment results in a reduction of translation in primary AML cells, particularly of genes known to be related to oncogenesis. | [67] | |
| Rohinitib | AML | Rohinitib is an inhibitor of the RNA helicase eIF4A and shows anti-leukemic activity particularly against AMLs with FLT3-ITD mutations. Xenotransplantation of AMLs pre-treated with rohinitib shows impaired engraftment. | [79] | |
| Omecetaxine | AML | In vitro drug screening identified the global protein synthesis inhibitor omecetaxine (or homoharringtonine) as a particularly effective growth inhibitor of FLT3-ITD mutated primary AML samples. | Omecetaxine was safe and effective in the treatment of FLT3-ITD mutated AML when used with sorafenib in a Phase II clinical trial. NCT03170895 | [157, 162] |
| Ro 08-2750 | AML | Ro 80-2750 binds the RRM1 RNA binding domain of MSI2 to block MSI2-RNA binding, and thus represses abnormally stabilized targets and promotes leukemic toxicity. | [39] | |
| STM2457 | AML | STM2457 is a catalytic inhibitor of METTL3. Treatment of AML cell lines showed reduced growth, increased apoptosis, myeloid differentiation and cell cycle arrest. Treatment of MLL-rearranged xenografts reduced leukemic burden and secondary engraftment. | [90, 158] | |
| STC-15 | AML | STC-15 is a METTL3 inhibitor, where STC-15 mediated m6A depletion causes the formation of double-stranded RNA which induces interferon production and immune responses from the tumor microenvironment. | STC-15 was the first inhibitor of RNA modifications to enter phase I clinical trials for adults with advanced cancers. NCT05584111 | [159] |
| CS1 and CS2 | AML | CS1 and CS2 are inhibitors of FTO, where in vitro treatment of primary AML with CS1 and CS2 reduced cell viability, and xenograft assays showed impaired engraftment and greater survival. | [74] | |
| CA-4948 (Emavusertib) | Pre-leukemia: high-risk MDS, AML | CA-4948 is an IRAK4 inhibitor, as IRAK4 is a prevalent alternatively spliced transcript in MDS and AML. | CA-4948 is currently being evaluated in a Phase 1/2a clinical trial for treatment of patients with AML or high risk MDS (NCT04278768), or for anemia in patients with very-low to intermediate risk MDS (NCT05178342). | [160] |
| Indisulam (E7070) | AML | Indisulam degrades RBM39 through ubiquitin ligase activity, where treatment of leukemia cell lines induced cell cycle arrest and apoptosis. In vivo treatment of pre-established AML xenografts reduced leukemic burden. | Evaluated in a Phase 2 clinical trial in combination with cytarabine, idarubicin and dexamethasone for the treatment of relapsed AML or high-risk MDS. NCT01692197 | [65] |
| 17S-FD-895 | AML - with enhanced efficacy in secondary AML | 17S-FD-895 inhibits the SF3B spliceosome component. Treatment of primary AML with 17S-FD-895 reduces survival and clonogenic potential. Secondary transplants of treated primary xenografts showed reduced engraftment. | [161, 164] |
Toward enhancing drug selectivity, strategies that seek to target-specific pro-LSC translation axes over global translational inhibition may provide a more precise alternative. To this point, small molecule-based interference of LIN28 domains needed for let-7 association has emerged as a strategy to de-repress let7 microRNAs, and thus enable the de-stabilization of oncogenic transcripts whose levels are aberrantly elevated in the presence of high LIN28.43 Similarly, small molecule inhibitors of MSI2 found via small molecule screening can repress abnormally stabilized targets and promote leukemic apoptosis.39
Given the large network of dysregulation that promotes pro-leukemic m6A RNA modifications, inhibition of RNA modifying enzymes is emerging as a promising area of inhibitor development. The small molecule STM2457 for instance was recently found to be a highly specific and potent inhibitor of METTL3, capable of reducing the catalytic activity of the METTL3-METTL14 complex and consequently m6A levels, as well as the translation of key pathogenic targets.158 Treatment of human AML cell lines in vitro with STM2457 demonstrated reduced cell growth, increased apoptosis, myeloid differentiation, and cell cycle arrest, whereas mouse primary AML demonstrated reduced progenitor output and increased apoptosis. Effective in vivo targeting of primary AML by STM2457 treatment was demonstrated using MLL-rearranged AML xenografts, a result re-capitulated with mouse models. This study also reported a reduced population of immunophenotypic CD93+ LSCs and markedly reduced engraftment of primary STM2457-treated grafts challenged in secondary transplantation experiments, demonstrating that LSCs within the primary graft were effectively targeted. Promisingly, treatment of mice with an efficacious anti-leukemic dose of STM2457 had minimal effects on the size of endogenous murine HSPC or HSC compartments, and scRNA-seq confirmed that while a neutrophil lineage bias is stimulated, as described previously, the abundance of the most primitive murine HSCs is unaffected, suggesting that specific impairment of Mettl3 catalytic activity, rather than full genetic knockout, may represent a tolerable therapeutic strategy.90, 158 In the human hematopoietic context, STM2457 was shown to have minimal effect on the colony-forming potential of cord blood CD34+ HSPCs.158 Additionally, the METTL3 inhibitor STC-15, developed by STORM Therapeutics, was the first inhibitor of RNA modifications to enter clinical trials in November 2022 (NCT05584111). Intriguingly, m6A depletion by STC-15 induces the formation of double-stranded RNA that in turn activates interferon production and subsequent immune responses from the tumor microenvironment, intriguingly linking cell-intrinsic changes to extrinsic responses. While STC-15 is pre-dominantly being assessed in solid cancers in its phase I clinical trial, it has shown promising results against AML in vivo.159
In other studies, through screening on the MONOMAC-6 cell line, CS1 and CS2 were found to be potent inhibitors of the m6A eraser FTO by binding and blocking its catalytic pocket.74 Primary AML samples showed reduced viability upon CS1 and CS2 treatment in vitro compared to healthy controls, whereas cell lines showed reduced cycling. Importantly, quantification of LSCs in mouse AML treated with CS1 and CS2 using transplantation at limiting dilution showed reducing LSC frequency, and human xenografts showed impaired engraftment and greater survival of the treated mouse cohort.74 In the same study, Su and colleagues demonstrate that, intriguingly, treatment of primary AML with decitabine and azacytidine showed increased levels of the checkpoint inhibitors PD-L1, PD-L2, PD-1, and LILRB4 along with elevated FTO levels and decreased m6A, suggesting that increased FTO levels upon treatment with hypomethylating agents can drive immune evasion and reduce the efficacy of the treatment.74 Indeed this study went on to confirm that CS1 or CS2 treatment enhanced T cell cytotoxicity by reducing LILRB4 expression, thus synergizing with hypomethylating agents74 and highlighting a potential for exploiting RBP-directed therapies as combinatorial agents.
Finally, in MDS, small molecule inhibitors of the spliceosome are intriguing therapeutic agents to target mis-splicing due to spliceosome mutations, as these cells are uniquely sensitive to further splicing disruptions compared to healthy cells given their already altered splicing state.163 Similarly to translation inhibition, this broad action is not without consequence to healthy cells, thus encouraging the identification and targeting of specific pathogenic splice variants or more selective splice-regulating RBP axes driving MDS and/or AML. For example, IRAK4 inhibitors have entered clinical trials, including CA-4948 which is currently being tested for treatment of high-risk MDS, AML, and lymphoma (NCT05178342, NCT04278768).160 Within AML, small molecule inhibition of RBM39 was shown to have therapeutic effects,65 in addition to the splice modulator 17S-FD-895, both of which have also been shown to reverse AML-specific pro-survival splicing events.161, 164 Lastly, antisense oligonucleotides (ASOs), which are short, single-stranded DNA molecules, can hybridize to pre-mRNA to interfere with splicing, and are therefore anticipated to be impactful clinical tools for targeting specific oncogenic splicing events driving disease.164, 165 ASOs capable of inducing gene-level transcript depletion in AML, or pathogenic splice forms in Duchenne Muscular Dystrophy have reached clinical trials, demonstrating that ASOs have reached an advanced stage within the therapeutic pipeline for translation to a greater number of diseases and targets.166, 167 Thus, while a body of work is accumulating to support that indeed RBPs can be effectively targeted through traditional small molecule methods and achieve potent preclinical therapeutic benefits, targeted interference of specific RNA interactions and processes themselves hold promise in their potential to offer more nuanced specificity in inhibiting pro-leukemic pathways. Regardless of the therapeutic modality, given the stem-cell-driven nature of both MDS and AML, long-term evaluation of efficacy represent important experiments that would be advantageous to build into future preclinical assessments of candidate inhibitors as they will define their capacity to effectively target the disease-driving cells.
CONCLUSION
In this review, we have showcased the essential nature of RBP-directed post-transcriptional control in both driving the evolution of LSCs and maintaining their pathogenic function. As master regulators of RNA fate, RBPs hold incredible potential as targets for LSC-selective therapies. Future investigation into the wider RNA interactome of individual RBPs and how these dynamic RNP complexes change over the course of leukemic development and in response to primary therapy will bring forth a critical layer of understanding that will aid in understanding the specific vulnerabilities capitalized over time and during treatment and relapse. Together, this will lay the ground work for development of novel therapeutic approaches for targeting malignant stem cells at all stages of AML disease progression in order to ultimately establish a curative treatment program for patients both at-risk of, or who have already developed, AML.
AUTHOR CONTRIBUTIONS
Pratik Joshi, Ava Keyvani Chahi, Lina Liu, Steven Moreira, and Ana Vujovic wrote the manuscript. Kristin J. Hope conceived the project and wrote the manuscript. All authors reviewed the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
FUNDING
Pratik Joshi and Ana Vujovic were supported by an Ontario Graduate Scholarship. Lina Liu and Steven Moreira were supported by a Princess Margaret Cancer Centre Postdoctoral Award and Kristin J. Hope was supported by an Ontario Institute for Cancer Research Investigator Award and funds from the Princess Margaret Cancer Foundation.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in GEO at https://www.ncbi.nlm.nih.gov/geo/.