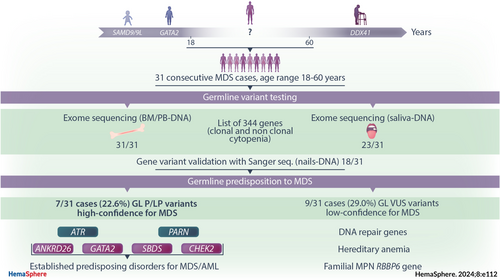
Prospective genetic germline evaluation in a consecutive group of adult patients aged <60 years with myelodysplastic syndromes
Graphical Abstract
Abstract
Relevance of germline (GL) predisposition in myelodysplastic syndromes (MDSs) was stressed in both 2022 WHO and International Consensus classifications, but its incidence is probably underestimated, especially in young adult patients. We selected a cohort of 31 consecutive de novo MDS patients with unusual young age (<60 years). We performed exome sequencing (ES) on DNA extracted from noninvasive sources (peripheral blood and saliva), filtering for a panel of 344 genes specifically tailored for detecting GL variants related to clonal and nonclonal cytopenia. We observed at least one high- or low-confidence GL MDS variant in 7/31 (22.6%) and 9/31 (29.0%) of cases, respectively. Four of 31 patients (12.9%) confirmed having established MDS/AML predisposing disorders. We found heterozygous variants in genes involved in DNA repair/cancer predisposition (ATM, ATR, FANCM, PARN, BRCA1, BRCA2, CHEK2, MSH2) in 9/31 (29.0%) cases and variants affecting ribosome biogenesis (SBDS), hematopoietic stem cell (GATA2), and megakaryocyte (ANKRD26) differentiation in single cases. Two cases had variants in RBBP6, a gene previously described exclusively in familial myeloproliferative neoplasms. Lastly, four cases had variants in genes related to inherited anemias (CUBN and PIEZO1 genes). Our results showed that “young” MDS patients aged 40–60 years carried reported and unreported GL variants with an unexpectedly high proportion, and these events co-occurred with somatic mutations recurrent in myeloid neoplasms. We explored the “no man's land” of the young adult MDS cases adopting a practical and scalable diagnostic tool, capable to detect GL variants avoiding invasive methods.
INTRODUCTION
Myelodysplastic syndromes (MDSs) are clonal hematopoietic diseases characterized by cytopenia, dysplasia, and propensity to evolve to acute myeloid leukemia (AML).1, 2 MDS affect elderly population, with a median age of 73 years in Italy (Italian FISiM Registry, www.fisimhematology.org) and with majority of patients older than 55 years.3 Senescence is considered an important determinant of MDS, as well as a factor influencing prognosis and therapeutical approach. Rarely, MDS patients are younger, and they more often present therapy-related MDS instead of primitive condition that occurs in the absence of a personal patient history of prior chemoradiotherapy for another cancer (de novo MDS).4
The increasing availability of large-scale genomic sequencing techniques in diagnostic routine has facilitated recognition of germline (GL) predisposing variants to myeloid neoplasms. The novel scientific knowledge led to the extension of the section of “Myeloid neoplasms associated with germline predisposition” in the latest 2022 WHO and International Consensus classifications. Although germline alterations may enhance the risk to develop therapy-related MDS,5, 6 younger patients with de novo MDS could be strongly suspected to carry inherited predisposition, and studies investigating genomic profile in this setting are increasing.7, 8 Young MDS patients are more frequently eligible for hematopoietic stem cell transplant, increasing the relevance for routine evaluation of GL predisposition.9, 10
However, systematic studies based on age at onset and aimed to establish the incidence of GL variants are scarce.11, 12 GL variants predisposing to MDS have been frequently found in children,13 where SAMD9/SAMD9L and GATA2 variants predominate,14-16 but also in elderly patients, where DDX41 can be associated with MDS with late adult onset.17 Variants of genes involved in DNA repair and telomere biology disorders prevail in the adolescent-young adults MDS patients.18 However, their incidence and relevance in contributing to occurrence of MDS at age ranging from 40 to 60 years are not known.
We decided to adopt different high-throughput sequencing techniques in order to optimally define the molecular assessment in a cohort of younger MDS patients (age range: 18–60 years) and to determine the frequency of GL variants in this setting. To address this point, we analyzed by next-generation sequencing (NGS) techniques the DNA extracted from MDS patients with scarce syndromic features or clinical signs of predisposing alterations to cover the widest panel of variants, beyond those known to be recurrent in myeloid neoplasms. We perform here a descriptive analysis, comparing the clinical characteristics with the genetic alterations.
MATERIALS AND METHODS
Study population
Thirty-one consecutive patients with a diagnosis of MDS and age <60 years were referred for second opinion.
Sample collection
Biological samples were obtained after informed consent in accordance with the Declaration of Helsinki Principles and local Ethics Committee's approval. Bone marrow (BM) samples were obtained by BM aspirate. BM and/or peripheral blood (PB) mononuclear cells were collected at diagnosis or re-evaluation, separated on Ficoll Hypaque density gradient (Lympholyte-H; Cedarlane Labs). DNA extraction from BM mononuclear cells was performed using the AllPrep DNA/RNA Mini Kit and the AllPrep DNA/RNA Micro Kit (Qiagen). Saliva was collected from all patients (Oragene OG-575; Genotek), and, after an incubation at 56°C overnight, DNA was extracted (S-DNA) using QIAsymphony® DSP DNA Midi Kit. Isolation of genomic DNA from nails was performed using the DNeasy® Blood & Tissue Kit (Qiagen). We avoided mucosal brushing to minimize blood contamination risk.
Cytogenetics
Cytogenetic analysis (conventional karyotyping) was performed using standard chromosomes banding techniques and documented according to ISCN recommendations.19 When metaphases were absent (one case, P29), fluorescent in situ hybridization (FISH) was performed to assess −7, del(5q), +8, del(17p) cytogenetic abnormalities.
Targeted next-generation sequencing (t-NGS) of somatic mutations
The presence of somatic mutations was evaluated by a t-NGS panel of recurrently mutated genes in myeloid neoplasms currently used in routine diagnostics of MDS in our Center. Custom panel comprising 24 myeloid-neoplasm-associated genes (ASXL1, CALR, CBL, KIT, CSF3R, DNMT3A, EZH2, IDH1, IDH2, IKZF1, JAK2, KRAS, MPL, NRAS, PTPN11, RUNX1, SETBP1, SF3B1, SH2B3, SRSF2, TET2, TP53, U2AF1, ZRSR2) performed on Ion Torrent S5 platform (kit Oncomine™ Myeloid Thermo Fisher, Thermo Fisher Scientific). NGS raw reads were aligned against the GRCh38/hg38 using NextGENe® software 2.4.2. Variant allele frequency (VAF) threshold was set to ≥0.02 in case of previously unreported mutations and ≥0.01 for known hotspots, with a median depth of coverage of 1000 × (minimum coverage of 95% of bases sequenced with minimum of 250 reads; SoftGenetics, LLC).
Exome sequencing (ES)
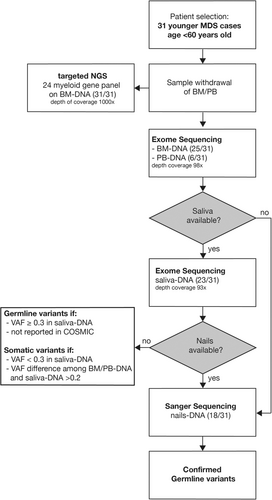
ES was performed on both BM/PB-DNA (25 and six cases, respectively) and for 23/31 also on S-DNA (Figure 1). Libraries were constructed with enzymatic fragmentation followed by end repair, A-tailing, adapter ligation, and library amplification (KAPA HyperPlus Kits; Roche). Libraries were hybridized to the whole-exome capture arrays (SeqCap EZ Exome v3, Nimblegen; Roche) and sequenced with NextSeq. 500/550 (Illumina Inc.). Commercially available kits were used for library preparation and sequencing. Reads were aligned to the reference genome Grch37 (hg19) using Burrows-Wheeler Aligner (BWA) tool,20 mapped and analyzed with the IGV software (Integrative Genome Viewer, 2013, Broad Institute).21 Variant calling was carried out using GATK caller,22 SAMtool,23 and Picard Tools (http://picard.sourceforge.net/), producing raw sequencing reads with a size of up to 300 bp, and the variant annotation was done using Annovar tool.24
Quality control of sequencing showed that 96% of the reads were mapped to the reference genome (hg19), and 97% of the targeted regions were covered by ≥30 × reads with average depth of 98× in BM/PB-DNA and 93× in S-DNA. ES assessed a list of 344 genes specifically selected by us on the basis of updated literature, giving evidence of gene involvement in predisposition or demonstrated pathogenetic for hematological cytopenia of clonal and nonclonal nature. Indeed, our curated gene list comprehends a first subgroup of genes involved in BM failure syndromes, inherited cytopenia(s), and those suspected to be involved in predisposition to myeloid neoplasms. Other subgroups refer to genes involved in inborn errors of immunity with signs of hemopathies, inherited thrombocytopenia, inherited anemia, and erythrocytosis (Supporting Information S1: Table 1).
Filtering of clinically relevant GL variants and pathogenicity assessment were performed according to the American College of Medical Genetics and Genomics (ACMG) guidelines.25 When gene-specific variant classification criteria were available, ClinGen recommendations were adopted for specific genes; otherwise, variants were assessed according to recent recommendations.26
- –
GnomAD MAF = 0, variant completely absent from gnomAD for dominant disease and present at a maximum of 10 alleles in recessive disease (leading to the application of the PM2 supporting);
- –
Prediction of in silico tools to discover pathogenic and benign of rare missense variants: revel score ≥0.52, or SpliceAI score ≥0.38 (leading to the application of the PP3 and ≤0.43, or SpliceAI score ≤0.2 leading to the application of the BP4);
- –
Variants located in mutational hotspot and/or critical and well-established functional domain without benign variation (leading to the application of the PM1 moderate/supporting);
- –
Case-control studies: an evaluation of the variant's prevalence in MDS patients was evaluated by pertinent revision of literature or by statistical analysis (by using 1600 adult non-MDS controls from local dataset) for some well-known and/or recurrent variants. If the variant identified was significantly increased in MDS cases compared to controls, PS4 criteria was applied (strong odd ratio, OD > 100, moderate OD > 30, supporting OD > 10);
- –
Patient phenotype or family history was highly specific for a disease with a single genetic etiology (leading to the application of the PP4).
Single heterozygous variants in genes associated with recessive MDS predisposing conditions were not considered because of their doubt clinical significance, and thus, they were not reported. Finally, variants were divided into two clusters: high-confidence (pathogenic, P and likely pathogenic, LP) and low-confidence GL variants (variants not primarily related to myeloid neoplasms and uncertain significance variants, VUS).
Sanger sequencing
Sanger sequencing of nails-DNA of 18/31 cases was performed for confirming the nature of the variants suspected GL (VAF ≥ 0.3) detected in BM/PB-DNA as well as in saliva DNA (where available). When nails samples were not available, we considered GL only those variants detected in saliva with VAF ≥ 0.3 and not previously reported as somatic in the COSMIC database (Figure 1).27
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8. For univariate comparison of potential differences, Fisher's exact test and Chi-squared test were used. A p ≤ 0.05 was considered statistically significant, when compared between groups.
RESULTS
Characteristics of the patients
MDS patients enrolled in this study presented a median age of 51 years (21–59 years), with male/female ratio 1.6 (Table 1). Anemia was the predominant cytopenia and a prolonged moderate cytopenia for >5 years before MDS diagnosis was observed in 9/31 patients (29.0%). Family history for onco-hematological disorders, unexplained cytopenia, or any other sign evocative of hereditary conditions was present in 12/31 (38.7%) of cases, while only one case presented with mild extra hematological syndromic features (Supporting Information S1: Table 2). Karyotype was altered in 10/31 cases. Patients were risk categorized according to the Revised International Prognostic Scoring System (IPSS-R):28 11/31 (35.4%) very low, 13/31 (41.9%) low, 2/31 (6.5%) 0ntermediate, 3/31 (9.7%) high, 2/31 (6.5%) very high.
| Characteristics | Patients (no. = 31) |
|---|---|
| Male/female | 19/12 |
| Median age (range) | 51 (21–59) |
| Age <50 years (%) | 13 (41.9) |
| Family history (%) | 12 (38.7) |
| ICUS >5 years before MDS diagnosis (%) | 9 (29.0) |
| MDS - post cytotoxic therapy (%) | 2 (6.5) |
| Number of cytopenia (%) | |
| 1 | 17 (54.8) |
| 2 | 8 (25.8) |
| 3 | 6 (19.4) |
| Type of cytopenia (%) | |
| Anemia | 23 (74.2) |
| Neutropenia | 14 (45.2) |
| Thrombocytopenia | 15 (48.4) |
| 2022 WHO classification (%) | |
| MDS-5q | 2 (6.5) |
| MDS-SF3B1 | 4 (12.9) |
| MDS-biTP53 | 1 (3.2) |
| MDS-LB | 15 (48.3) |
| MDS-h | 4 (12.9) |
| MDS-IB1 | 2 (6.5) |
| MDS-IB2 | 1 (3.2) |
| MDS-f | 2 (6.5) |
| IPSS-R (%) | |
| Very low | 11 (35.4) |
| Low | 13 (41.9) |
| Intermediate | 2 (6.5) |
| High | 3 (9.7) |
| Very high | 2 (6.5) |
| Number of somatic mutations (%) | |
| No somatic mutations | 10 (32.3) |
| 1 somatic mutation | 6 (19.4) |
| 2 or more somatic mutations | 15 (48.3) |
- Note: Characteristics of our cohort. Herein are reported demographic and clinical features, cytogenetic abnormalities and the number of somatic mutations, diagnosis according to 2022 edition of WHO classification, and prognostic score according to IPSS revised. One patient belonged to MDS-biTP53 category due to the detection of 2 TP53 somatic mutations along with loss of 17p.
- Abbreviations: IPSS-R, Revised International Prognostic Scoring System; MDS-f, MDS with fibrosis; MDS-h, MDS hypoplastic; MDS-LB, MDS with low blasts; MDS-5q, MDS with low blasts and isolated 5q deletion; no, number; pts, patients; yrs, years.
Somatic mutations
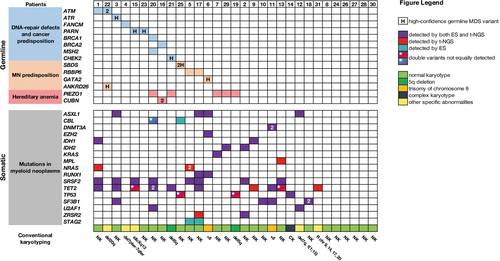
NGS analysis indicated that 67.7% of cases in our cohort carried at least one somatic mutation (Figure 2). In nine MDS patients who did not present GL variants, we observed at least one somatic alteration recurrently associated with MDS (ASXL1, DNMT3A, IDH1, IDH2, MPL, SRSF2, TET2, TP53, SF3B1, U2AF1, ZRSR2).29 MDS cases without any genetic alteration still presented a clear morphological evidence of myelodysplasia.
ES
We applied a diagnostic algorithm (Figure 1) to the ES data, and we identified a total of 24 GL variants distributed in 16/31 (51.6%) patients. Among these, 7/31 (22.6%) were classified as carriers of high-confidence variants for MDS and 9/31 (29.0%) of low-confidence variants (Table 2A–B). In MDS cases carrying high-confidence GL heterozygous variants, somatic genetic alterations were observed in all but one of them. In 9/31 (29.0%) cases, we found heterozygous GL variants that affected genes involved in mismatch DNA repair and telomere-structure maintenance, particularly ATM, ATR, FANCM, PARN, BRCA1, BRCA2, CHEK2, MSH2; 5/31 (16.1%) cases had GL variants in SBDS (P25), GATA2 (P6), ANKRD26 (P22), and RBBP6 (P5 and P17), gene related to myeloid neoplasms predisposition; and 6/31 (19.4%) cases had variants in genes related to nonclonal anemia (CUBN gene, in P16, and PIEZO1 in five patients, see Figure 2). Among these groups, four of 31 patients (12.9%) confirmed having established predisposing disorders for MDS/AML: GATA2 deficiency, Shwachman-Diamond syndrome (SDS), ANKRD26-related thrombocytopenia, and germline predisposition due to CHEK2 P/LP variants (P21).
| Pt | Diagnosis | Gender | Age (years) | Germline variants (ES) | Reference sequence (HGMD) | HGVS nomenclature | HGVS nomenclature | rs ID | ACMG score |
|---|---|---|---|---|---|---|---|---|---|
| c.DNA level | Protein level | ||||||||
| P3 | MDS-SF3B1 | M | 50 | ATR | NM_001 184.4 | c.2634-2A>G | / | / | LP (PVS1, PM2) |
| P22 | MDS-LB | M | 59 | ANKRD26 | NM_014 915.3 | c.−128G>A | / | rs1589393809 | P (PS4, PM2, PP4, PM1, PS3) |
| P15 | MDS-IB1 | F | 57 | PARN | NM_002 582.4 | c.482 A>G | p.Y161C | rs201990148 | LP (PS4, PP1, PP4) |
| P23 | MDS-LB | M | 51 | PARN | NM_002 582.4 | c.482 A>G | p.Y161C | rs201990148 | LP (PS4, PP1, PP4) |
| P21 | MDS-5q | F | 50 | CHEK2 | NM_007 194.4 | c.1100delC | p.T367fs | rs555607708 | LP (PVS1, PS3) |
| P25 | MDS-LB | M | 53 | SBDS | NM_016 038.4 | c.183_184delinsCT | p.K62* | rs120074160 | P (PVS1, PM2, PM3, PP4) |
| SBDS | NM_016 038.4 | c.258+2T>C | / | rs113993993 | P (PVS1, PM3, PP4) | ||||
| P6 | MDS-IB2 | M | 51 | GATA2 | NM_032 638.5 | c.372_373insT | p.P125Sfs*60 | / | P (PVS1, PM2, PP4) |
| B. List of low-confidence germline variants in “young” MDS patients. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| HGVS nomenclature | HGVS nomenclature | ||||||||
| Pt | Diagnosis | Gender | Age (yrs) | Germline variants (ES) | Reference sequence (HGMD) | c.DNA level | Protein level | rs ID | ACMG score |
| P1 | MDS-LB | M | 49 | ATM | NM_000 051.4 | c.5753G>C | p.R1918T | rs148064985 | VUS (BP4) |
| P22 | MDS-LB | M | 59 | ATM | NM_000 051.4 | c.346A>G | p.K116E | / | VUS (PM2, BP4) |
| ATM | NM_000 051.4 | c.4060C>A | p.P1354T | rs145119475 | VUS (BP4) | ||||
| P4 | MDS-h | M | 46 | FANCM | NM_020 937.4 | c.5832G>T | p.L1944F | rs201017015 | VUS (BP1, PP4, PM1) |
| P20 | MDS-LB | M | 59 | BRCA1 | NM_007 294.4 | c.1662G>C | p.E554D | rs876659028 | VUS (PM2) |
| MSH2 | NM_000 251.3 | c.509A>G | p.Q170R | rs1114167865 | VUS (PM2) | ||||
| PIEZO1# | NM_001 142 864.4 | c3667G>A | p.V1223I | rs185326407 | LP (PM1, PM7, PP1, PP2) | ||||
| P16 | MDS-LB | M | 45 | BRCA2 | NM_000 059.4 | c.10095delCins11 | p.S3366Nfs*4 | / | VUS (PM2) |
| CUBN# | NM_001 081.4 | c.7604A>C | p.E2535A | / | VUS (PM2, BP4) | ||||
| CUBN# | NM_001 081.4 | c.4864G>C | p.G1622R | / | VUS (PM2, BP4) | ||||
| P21 | MDS-5q | F | 50 | PIEZO1# | NM_001 142 864.4 | c.5863C>T | p.R1955C | rs547409918 | LP (PM1, PM2, PM7, PP3, BS2) |
| P5 | MDS-f | F | 57 | RBBP6 | NM_006 910.5 | c.3868C>T | p.R1290* | / | VUS (PM2, PVS1) |
| P17 | MDS-f | M | 56 | RBBP6 | NM_006 910.5 | c.4738A>G | p.K1580E | rs147630531 | VUS (BP4) |
| P7 | MDS-LB | M | 47 | PIEZO1# | NM_001 142 864.4 | c.6781A>G | p.S2261G | rs550322372 | LP (PM1, PM2, PM7, PP1, BS2) |
| P19 | MDS-5q | M | 58 | PIEZO1# | NM_001 142 864.4 | c.5863C>T | p.R1955C | rs547409918 | LP (PM1, PM2, PM7, PP3, BS2) |
| P29 | MDS-LB | F | 48 | PIEZO1# | NM_001 142 864.4 | c.6479C>T | p.P2160L | rs753068603 | LP (PM1, PM2, PM7, PP3) |
- Note: List of GL variants filtered in the cohort of 31 MDS patients <60 years. (A) Variants selected as high-confidence for MDS (pathogenic and likely pathogenic). (B) Variants selected as low-confidence for MDS (variants not primarily related to predisposition to myeloid neoplasms and uncertain significance variants). The symbol “#” refers to variants related to nonclonal anemia. rs ID:dbSNP Reference SNP (rs or RefSNP) number is a locus accession for a variant type assigned by dbSNP. Reference sequence according to HGMD transcript.
- Abbreviations: ES, exome sequencing; LP, likely pathogenic; MDS-f, MDS with fibrosis; MDS-h, MDS hypoplastic; MDS-IB, MDS with increased blasts; MDS-LB, MDS with low blasts; MDS-SF3B1, MDS with low blasts and SF3B1 mutation; MDS-5q, MDS with low blasts and isolated 5q deletion; no, number; P, pathogenic; pts, patients; VUS, variant of uncertain significance; yrs, years.
The patient affected by GATA2 deficiency (P6) presented classically HPV-anogenital warts, monocytopenia, and CD4 lymphocytopenia and developed high-risk MDS (MDS with increased blasts 2), which shortly progressed into secondary AML. The somatic genetic alterations found included trisomy of chr 8, along with mutations in ASXL1, EZH2, and RUNX1 (Figure 2). Patient P25 was referred to our center at age of 53 years for investigation of MDS. Clinical presentation included mild dysmorphic features and short stature. The medical history during childhood was unremarkable. Two years prior, a diagnosis of diffuse large B cell lymphoma was made, treated with conventional chemoradiotherapy as per standard practice. The prolonged cytopenia afterward prompted a BM evaluation, which showed multilineage dysplasia. Conventional karyotyping confirmed normal karyotype, while t-NGS analysis revealed 2 mutations in TP53 gene. ES analysis revealed the presence compound heterozygous variants of the SBDS gene (see Supporting Information Material), leading to an unusual late onset diagnosis of SDS. Patient P22 came to our center for a second opinion after a diagnosis of immune thrombocytopenia, not responding to prednisone. BM evaluation showed marked dysmegakaryopoiesis with micromegakaryocytes. The absence of syndromic features, the familial history of thrombocytopenia (Supporting Information S1: Table 2), and the normal platelet size prompted us to screen the 5′-UTR of ANKRD26 using ES, which led to the identification of a c.−128G>A heterozygous substitution.
We also identified in our cohort GL variants in two genes not clearly reported in literature related with MDS/AML occurrence: PARN (c.482A>G, p.Y161C, rs201990148—in P15 and P23) and RBBP6 (c.3868C>T, p.R1290*—in P5 and RBBP6 c.4738A>G, p.K1580E, rs147630531—in P17). The frequency of the variant for PARN was evaluated in 1600 adult non-MDS controls (internal dataset) and compared to that observed in our cohort. There was a significant difference (p < 0.001) indicating enrichment for this specific variant in our small cohort of MDS cases (Supporting Information S1: Table 4).
Variants in at least two genes were observed in 4/31 cases. In particular, in P22, we found a carrying heterozygous substitution in 5′ UTR ANKRD26 gene related to familial platelet disorder30 and two compound heterozygous variants in ATM gene.5, 31 A GL variant in BRCA2 and a compound heterozygous alteration in CUBN related to the autosomal recessive megaloblastic anemia (Imerslund-Grasbeck syndrome 1)32 were observed in another case (P16). In all MDS subjects (5/31) who presented GL variants in PIEZO1, these variants were demonstrated pathogenic for hereditary stomatocytosis by functional studies and segregation data.33 Ten of 12 patients with familial history in first- and second-degree relatives of oncological/hematological diseases presented at least one variant. This variable resulted the unique predictive parameter in GL predisposition for this selected cohort (p < 0.05).
DISCUSSION
MDSs are diseases correlated with senescence, and, in Western countries, median age of patients is >70 years. Occurrence of de novo MDS in younger population is extremely rare. While many studies focus on incidence of GL predisposition in children and/or adult MDS patients aged <40 years,8, 34 the frequency of GL variants across the age range of 40–60 years has not yet been fully investigated. We selected 31 consecutive MDS patients aged between 18 and 60 years, 29/31 patients of our cohort being aged 40–60 years. Somatic mutations were detected in 67.7% of cases, differently from the higher frequency currently observed for elderly MDS patients,35 but consistent with what was previously shown in youngsters.36 We anyhow could observe a significant high percentage of MDS cases with at least one high confidence (LP/P) GL variant (22.6%). Although the limited number of cases in our cohort, this occurrence appeared much higher compared to the frequency of the GL variants recently observed in a very large number of healthy individuals (≈9%).37 In this recent work, in line with our hypothesis, healthy individuals with GL variants (the most frequent being heterozygous variants in ATM and CHEK2 genes) were developing clonal hematopoiesis and predisposed to hematological malignancies.37 Most of GL heterozygous variants described in our study involved DNA repair genes, like ATM, ATR, BRCA1, BRCA2, and CHEK2. Therefore, this nonnegligible frequency of GL variants co-occurring with somatic mutations could constitute a contributing factor for early onset of disease in patients unusually “young” for MDS (<60 years).
Because saliva as control tissue is not optimal, we performed Sanger sequencing on nails-DNA. We could not perform skin biopsy and set up fibroblast culture.9 The difference in VAF between GL variants and somatic mutations in saliva is confirmed as a fairly reliable parameter for the segregation of variants according to their nature (Supporting Information S1: Figure 1). However, saliva could be reserved to well defined and limited number of cases.
We observed the presence of PARN in two MDS patients. Both cases presented the same PARN modification and had similar geographical origin but were unrelated (studying up to three generations). Autosomal dominant PARN variants are associated with aplastic anemia and MDS.8, 38-40 Patients P5 and P17 carried different RBBP6 GL variants, both presented BM fibrosis grade 1 and somatic mutations in SRSF2 and STAG2 genes, and both progressed rapidly to AML (Supporting Information S1: Table 5). RBBP6 variants identified are located next to the predicted p53 binding domain.41 Such variants are likely to increase the rate of mutagenesis, suggesting a possible role in accumulation of acquired somatic mutations as previously reported in other neoplasms for other genes.42 As variants in RBBP6 that predispose to myeloproliferative neoplasms have been previously reported,41 investigating their possible pathogenicity in MDS is warranted.
We hypothesize that together with ANKRD26 variant,43 a concomitant role of the two additional ATM variants could be responsible for accelerating MDS onset in P22. Similarly, P16 presented multilocus inheritance with two variants in CUBN (justifying low B12 levels not responding to oral B12 supplement but only to intramuscular injections) and one in BRCA1.
Our results indicate that an extended diagnostic work-up could be relevant in younger MDS cases. For instance, our cohort have included adult cases diagnosed with SDS and GATA2 deficiency, not otherwise clinically evident. The importance of scalable diagnostic tools becomes clearer when analyzing somatic profiles can help diagnose specific genetic predispositions. In this context, clonal hematopoiesis in predisposed individuals (CHIPI) often arises as a compensatory response to counteract harmful GL mutations and maintain hematopoietic cell fitness.44 However, distinguishing between adaptive and maladaptive clonal responses is complex. Certain somatic alterations that improve cellular fitness in GL disorders carry a significant risk of leukemic transformation. TP53 mutant clones are frequently observed in SDS, and their presence, while not exclusive to this disorder, can inform the diagnostic process.45 Similarly, the mutational profile observed in patient P6—including trisomy of chromosome 8 and somatic mutations in RUNX1 and ASXL1—is indicative of GATA2 deficiency, although in the absence of the classical monosomy 7.46-49 Early identification of such inherited BM failure syndromes is crucial for tailoring surveillance strategies and detecting early signs of leukemic progression. Patients could benefit from specific therapeutic strategies in case of demonstration of GL variants clearly predisposing/determining MDS. On the same line, although with completely different outcome and management, inheritance like that of CUBN can reveal pathologic conditions that are worsening MDS conditions and that can be treated efficiently. We may affirm that ES may help in identifying actionable variants within genes linked to both clonal and nonclonal cytopenia.
On another level, the biological and conceptual relevance of these observations in a small group of MDS cases is intriguing. The co-presence of GL variants and somatic mutations was observed in almost all cases. In specific, GL variants and SRSF2 mutations have been identified in 6/16 cases. As the synergy between heterozygous GL and somatic mutations as concurring causes of tumor onset has already been observed in solid neoplasms, we may assume that the same mechanism of transformation occurs in adult MDS (Figure 2).42, 50, 51 This phenomenon was observed recently37 and could indeed indicate a different dynamic respect to the pathogenesis of inherited BM failure syndromes and childhood MDS determined by homozygous variants.
Onset of MDS at an unusually young age should always prompt suspect of a predisposing condition. An integrated diagnostic approach, defined as the convergence of diagnostic techniques (ES, t-NGS, Sanger sequencing) and clinical evaluation, allowed us to characterize the genetic asset of 31 nonsyndromic MDS cases with “young” age (median age: 51 years). Our locally designed panel used for ES analysis has the advantage of identifying genetic alterations not included in the currently proposed t-NGS panels for GL evaluation in MDS. It equally gave us the possibility to provide a proof of concept, uncovering a conspicuous number of patients with GL variants, although few of them clinically actionable and immediately relevant for management. Our data support the relevance of genetic screening for MDS patients aged 40–60 years, both to reach a certain diagnosis of MDS and clarify atypical cases of cytopenia, as well as modify management in specific, although rare instances.
AUTHOR CONTRIBUTIONS
Enrico Attardi and Valeria Santini was involved in contribution, identified and managed MDS patients, designed the study, collected and interpreted the data, wrote the manuscript. Lucia Tiberi and Daniela Formicola performed NGS experiments and participated in the analysis interpretation. Giorgio Mattiuz performed statistical analysis, edited the manuscript and gave helpful intellectual insights during the study. Elia Dirupo performed bioinformatics data extrapolation and analysis. Marco G. Raddi and Angela Consagra helped in sample and data collection. Debora Vergani and Rosangela Artuso helped in NGS experiments; Valeria Santini took responsibility for the integrity and the accuracy of the data presented; and all authors reviewed and approved the final version of this manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
FUNDING
This research was funded by Associazione Italiana per la Ricerca sul Cancro (AIRC) IG-26537-2021 Investigator Research Grant to V.S.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.