Principal component analysis as an efficient method for capturing multivariate brain signatures of complex disorders—ENIGMA study in people with bipolar disorders and obesity
The complete author details for the ENIGMA bipolar disorder working group are available at https://enigma.ini.usc.edu/ongoing/enigma-bipolar-working-group/
The complete author details for the ENIGMA BMI-X working group are available at https://enigma.ini.usc.edu/ongoing/enigma-bmix/
Abstract
Multivariate techniques better fit the anatomy of complex neuropsychiatric disorders which are characterized not by alterations in a single region, but rather by variations across distributed brain networks. Here, we used principal component analysis (PCA) to identify patterns of covariance across brain regions and relate them to clinical and demographic variables in a large generalizable dataset of individuals with bipolar disorders and controls. We then compared performance of PCA and clustering on identical sample to identify which methodology was better in capturing links between brain and clinical measures. Using data from the ENIGMA-BD working group, we investigated T1-weighted structural MRI data from 2436 participants with BD and healthy controls, and applied PCA to cortical thickness and surface area measures. We then studied the association of principal components with clinical and demographic variables using mixed regression models. We compared the PCA model with our prior clustering analyses of the same data and also tested it in a replication sample of 327 participants with BD or schizophrenia and healthy controls. The first principal component, which indexed a greater cortical thickness across all 68 cortical regions, was negatively associated with BD, BMI, antipsychotic medications, and age and was positively associated with Li treatment. PCA demonstrated superior goodness of fit to clustering when predicting diagnosis and BMI. Moreover, applying the PCA model to the replication sample yielded significant differences in cortical thickness between healthy controls and individuals with BD or schizophrenia. Cortical thickness in the same widespread regional network as determined by PCA was negatively associated with different clinical and demographic variables, including diagnosis, age, BMI, and treatment with antipsychotic medications or lithium. PCA outperformed clustering and provided an easy-to-use and interpret method to study multivariate associations between brain structure and system-level variables.
Practitioner Points
- In this study of 2770 Individuals, we confirmed that cortical thickness in widespread regional networks as determined by principal component analysis (PCA) was negatively associated with relevant clinical and demographic variables, including diagnosis, age, BMI, and treatment with antipsychotic medications or lithium.
- Significant associations of many different system-level variables with the same brain network suggest a lack of one-to-one mapping of individual clinical and demographic factors to specific patterns of brain changes.
- PCA outperformed clustering analysis in the same data set when predicting group or BMI, providing a superior method for studying multivariate associations between brain structure and system-level variables.
1 INTRODUCTION
Large-scale, multisite brain imaging datasets are becoming more common through initiatives such as ENIGMA (McWhinney et al., 2023), ADNI (Cruciani et al., 2024), ABCD (Dahl et al., 2024), the human connectome project (Cohen et al., 2023), and others. Large datasets allow us to apply multivariate techniques of analyses, which model interplay between regions (Woo et al., 2017), but require larger, more ecologically valid samples to provide more replicable results (Marek et al., 2022). These techniques better fit the anatomy of complex neuropsychiatric disorders which are characterized not by alterations in a single region, but rather by variations across distributed brain networks (Hibar et al., 2018; Segal et al., 2023). However, there is little methodological clarity on which of the many available methods of multivariate data analyses are best suited to the task of relating brain structure to system-level variables. While development of new methods is one key aspect of the field, uncovering benefits and best-use scenarios for established methods is equally as important.
Analyzing brain imaging changes in BD is a suitable way to test multivariate techniques. Individuals with BD markedly vary in their clinical presentations and impact of the illness on their functioning. This clinical heterogeneity may reflect neurobiological heterogeneity, which can be studied by brain imaging. It is increasingly clear that brain alterations in severe mental illnesses (SMI) are multifactorial. Aside from the diagnosis, they also reflect the effects of additional clinical factors, including medications (Hajek et al., 2012; McWhinney, Abé, et al., 2022; Van Gestel et al., 2019), and comorbid psychiatric or physical conditions, such as obesity (McWhinney, Abé, et al., 2021; McWhinney, Brosch, et al., 2022; McWhinney, Kolenic, et al., 2021) and diabetes (Hajek et al., 2014, 2016). Understanding the brain changes in SMI and translating these findings into clinical settings requires sensitive and replicable methods that link patterns of brain alterations to system-level variables.
Broadly speaking, some methods, such as clustering, categorize participants into groups based on their brain structure, while others, such as principal component analyses, represent brain imaging data as a linear combination of features. While clustering has become a popular method for multivariate analyses of neuroimaging data (McWhinney, Abé, et al., 2022), we do not expect groups of individuals to fall neatly into distinct clusters (e.g. healthy vs. unhealthy). Also, external variables may not exhibit a binary effect on the brain, but rather a nuanced, continuous one. Indeed our previous study using clustering exemplified these issues. We found that there were no strictly separate clusters in brain imaging data and that the boundaries between BD and controls were not clear, i.e. many controls fell into the cluster together with BD individuals, while some BD individuals clustered with controls (McWhinney, Abé, et al., 2022). The cluster assignment of individuals in part depended on continuous variables including age and BMI and effectively resulted in categorizing of these continuous variables, which is not optimal.
Neuroimaging data are often strongly correlated naturally (i.e. brain networks) and due to preprocessing (i.e. coregistration, smoothing, etc.), which is a good reason for linear projection methods. Instead of categorization, such methods quantify degrees of variation and may be better suited to identifying sources of heterogeneity in brain imaging data, as many of these sources may in fact themselves be on a continuum. While machine learning (ML) techniques can overcome these challenges, such techniques require large training sets and out-of-sample validation, and results can be difficult to interpret and translate into practice. Principal component analysis (PCA) represents a potentially optimal middle ground between these approaches, as it can perform well using modest sample sizes while reliably reducing dimensionality across many variables (i.e. brain regions) and deriving robust low-dimensional data representations (Comrey & Lee, 2013). By identifying covariance across individuals in numerous regions simultaneously, PCA can identify patterns of distributed brain network changes that can subsequently be linked with clinical correlates, while maintaining interpretability (Behdinan et al., 2015; Maralakunte et al., 2023; Rehák Bučková et al., 2023; Yeh et al., 2010).
Our main goal here is to compare whether methods which represent brain imaging data as a linear combination of features are better in capturing associations with clinical variable than methods which categorize brain imaging data into clusters. To do that we selected the most established and representative examples of each approach, i.e. PCA vs K-means clustering. Specifically, we used PCA to identify patterns of covariance across regions of interest and related them to clinical and demographic variables. We then compared performance of this approach to our prior clustering study on identical sample. We expected that compared to clustering, patterns of covariance in brain imaging data, as identified by PCA, would show stronger associations with clinical and demographic variables.
2 MATERIALS AND METHODS
2.1 Participating sites
The ENIGMA-BD working group brings together researchers with brain imaging and clinical data from people with BD (Hibar et al., 2016, 2018; McWhinney, Abé, et al., 2021; McWhinney et al., 2023). Nineteen site members of this group from 13 countries on six continents contributed individual subject structural MRI data, medication information, and body mass index (BMI) values for a total of 2770 participants. We split this sample into the primary and replication samples. The primary sample (N = 2436) was identical to the one in our previous study (McWhinney, Abé, et al., 2022) and allowed us to directly compare the results of clustering and PCA on the same sample. An additional five sites contributed data to our replication sample for out-of-sample validation (n = 327). Two of the new sites also recruited individuals with schizophrenia (N = 107). We decided to include them for testing the diagnostic specificity of the findings. Table 1, as well as Supplementary Tables S1 and S2 list the demographic and clinical details for each cohort. Supplementary Table S3 provides the diagnostic instruments used to obtain diagnosis and clinical information. Supplementary Table S4 lists exclusion criteria for study enrollment. Briefly, all studies used standard diagnostic instruments, including SCID (N = 12 studies), MINI (N = 1), and DIGS (N = 1). Most studies (N = 8) included both bipolar I (BDI) and bipolar II (BDII) disorders, five studies included only BDI, and one study only BDII participants. At the time of scanning, most individuals with BD were euthymic (81%), with some depressed (15%), manic (2%), hypomanic (1%), or mixed (<1%). Substance abuse was an exclusion criterion in seven studies. Most studies did not exclude comorbidities, other than substance abuse. Consequently, the sample represents one of the broadest, ecologically most valid, and a generalizable representation of real-world BD studied to date. In order to test how well a method captures relevant clinical links, we need a broad representation of the diagnosis, which is not restricted to one subtype and ideally also includes representation of other diagnoses.
| Controls | Cases | Difference | ||
|---|---|---|---|---|
| Primary sample | Sample size (N) | 1600 | 836 | |
| Sex—N (%) male | 684 (42.8) | 353 (42.2) | χ2 = 0.25, p = .617 | |
| Age—mean (SD) | 35.47 (12.63) | 40.57 (12.81) | F(1,2433) = 49.64, p < .001* | |
| BMI—mean (SD) | 24.43 (4.12) | 27.10 (5.30) | F(1,2378) = 135.46, p < .001* | |
| BMI category—N (%) | ||||
| Normal | 1014 (63.4) | 331 (39.6) | χ2 = 158.55, p < .001* | |
| Overweight | 437 (27.3) | 298 (35.6) | ||
| Obese | 149 (9.3) | 207 (24.8) | ||
| Diagnosis in patients—N (%): BD-I/BD-III/BD-NOS | 572 (70.5)/234 (28.9)/5 (0.6) | |||
| Treatment at time of scan in patients—N (%): None/Lithium/Anticonvulsant/1st gen./2nd gen. antipsychotic/Antidepressant | 226 (27.0) / 373 (49.7) /244 (35.4) / 37 (5.4) /262 (37.4) / 248 (35.4) | |||
| Replication sample | Sample size | 136 | 191 | |
| Sex—N (%) male | 59 (43.4) | 115 (60.2) | χ2 = 2.85, p = .091 | |
| Age—mean (SD) | 38.54 (13.51) | 40.25 (12.87) | F(1,246) = 0.01, p = .986 | |
| BMI—mean (SD) | 24.60 (4.77) | 29.35 (6.63) | F(1,82) = 21.86, p < .001* | |
| BMI category—N (%) | ||||
| Normal | 89 (65.4) | 54 (27.7) | χ2 = 51.03, p < .001* | |
| Overweight | 30 (22.1) | 56 (29.3) | ||
| Obese | 17 (12.5) | 82 (42.9) | ||
| Diagnosis in patients—N (%): BD-I/BD-III/BD-NOS /Schizophrenia | 15 (7.8)/9 (4.7)/1 (0.1)/107 (56.0) | |||
| Treatment at time of scan in patients – N (%): None/Lithium/Anticonvulsant/1st gen./2nd gen. antipsychotic/Antidepressant | 57 (29.8)/24 (12.6)/38 (19.9)/35 (18.3)/112 (58.6)/60 (31.4) | |||
- Note: Asterisks indicate significant group differences (*p < .05).
All participating sites received approval from local ethics committees, and all participants provided written informed consent. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
2.2 MRI acquisition & processing
High-resolution T1-weighted brain anatomical MRI scans were acquired at each site, see Table S5. All groups used the same ENIGMA-standardized FreeSurfer protocol to derive region of interest (ROI) estimates of cortical thickness and surface area and performed standard visual and statistical quality assessment, as detailed at: http://enigma.ini.usc.edu/protocols/imaging-protocols/. These open-source protocols are standardized across the ENIGMA consortium, and available online to foster open science, replication, and better reproducibility. They were applied in prior publications by our group (Hibar et al., 2018; McWhinney et al., 2023), and more broadly in large-scale ENIGMA studies of major depression, schizophrenia, ADHD, OCD, PTSD, epilepsy, and autism (Thompson et al., 2020).
Briefly, FreeSurfer provides segmentations of 34 cortical regions per hemisphere, based on the Desikan–Killiany atlas, with estimates of cortical thickness and surface area for each region. Visual quality controls were performed on a ROI level aided by a visual inspection guide including pass/fail segmentation examples. We also generated diagnostic histogram plots for each site and outliers that deviated from the site mean for each structure by more than three standard deviations were flagged for further review. All observations failing quality inspection were withheld from subsequent analyses, see Table S6. Measurements were removed in 1.4% of participants per region on average, and missing values were imputed using the missForest algorithm (Stekhoven & Bühlmann, 2012). Prior analyses from the ENIGMA-BD working group showed that scanner field strength, voxel volume, and the version of FreeSurfer used for segmentation did not significantly influence the effect size estimates (Hibar et al., 2016).
2.3 Principal component analysis
We scaled each region to be zero-centered with a standard deviation of 1.0 and used PCA to obtain the loadings and scores of each principal component (PC) separately for cortical thickness and surface area, with each including all 68 cortical regions. Loadings indicated the contribution of each region in reduced-dimensional space for each component, and scores reflected the position of individuals in that component's space based on their cortical thickness or surface area weighted by each component's loadings. For each, we calculated the proportion of variance explained by each component and took the first component as the indicator of whole-brain structural covariance across individuals (Alexander-Bloch et al., 2013). The principal components are essentially anatomical patterns composed of highly correlated brain regions (Alexander-Bloch et al., 2013). We further focused on PCs that explained more than 10% of variance in either cortical measure.
The scores for these components provided each participant a single number to indicate their position on a continuous range of alterations across 68 ROIs throughout the whole cortical mantel. We plotted these scores to better visualize them. We tested for associations between the component score and clinical or demographic factors, as described below. First, this PCA model was completed in the primary sample which was identical to the sample we used for clustering in our previous work, in order to be able to directly compare the two methods (McWhinney, Abé, et al., 2022). Second, it was performed in our replication sample that included only newly contributing sites.
2.4 Statistical modeling
The list of all models we tested is included in the supplement. For each of cortical thickness and surface area, we used mixed linear regression modeling to test for associations between the individual component's score (and by proxy their associated patterns of brain structure) with group (BD or control), BMI, age, and sex. In previous studies, BMI has proven to be robustly related to cortical thickness (McWhinney et al., 2023; McWhinney, Brosch, et al., 2022). We tested for nonlinear effects of age (age squared), as well as for interactions between age, sex, and BMI. We additionally tested the inclusion of an interaction between group and BMI, including it if significant. BMI and age were each scaled to a range of 4.0 so that model estimates would equate to quartiles of their distributions.
This same procedure was performed among participants with BD using predictors of BMI, age, sex, diagnosis subtype (BD-I or BD-II), age of illness onset, history of psychosis (Y/N), and prescribed medications at the time of scanning (antidepressant, 1st or 2nd generation antipsychotic, anticonvulsant, and/or lithium), coded as yes or no for each medication class as separate predictors, as in prior ENIGMA BD analyses (McWhinney, Abé, et al., 2022). We tested for interactions between BMI and medications and included them if significant. All models included research site as a random effect. We checked for normality of residuals using QQ plots, and for multicollinearity by testing the variance inflation factor (VIF). All modeling was completed using the package lme4 (v1.1-21) and lmerTest (v3.1-3) in R version 4.1.1.
We compared the PCA with clustering analyses used in our previous paper (McWhinney, Abé, et al., 2022). We obtained the cluster number for each participant using the exact sample and procedure described in our previous study (McWhinney, Abé, et al., 2022). Both outcome measures (first component score for cortical thickness and cluster number) were tested as predictors of our two variables of interest, group (control or BD) and BMI, to determine which measure was a stronger predictor of each variable. We used mixed logistic regression modeling to test for associations between group and (1) covariates alone; (2) covariates with cluster number; (3) covariates with the first PC's score; and (4) all of the above. Covariates included BMI, age, sex as fixed effects, and research site as a random effect. We calculated the Bayesian Information Criterion (BIC), area under the curve (AUC) of the ROC curve, as well as predictor and model significance to compare the predictive power of each model in association with the participant group. We performed the same procedure using mixed linear regression modeling with BMI as the dependent variable but using the diagnostic group as a covariate instead of BMI and estimating model fit using R2 instead of AUC.
Lastly, we tested the fit of either average cortical thickness or the first component's score as dependent variables to BMI and group as predictors. We compared fit between these two models (R2, AIC, BIC). We additionally tested the Pearson correlation coefficient between average cortical thickness and the first component's score, and we further tested the significance of their association while adjusting for a random effect of research site.
2.5 Harmonization of between-site differences
The methods described above control for differences between sites using random effects in mixed regression modeling, identically to the preferred approach in most previous comparable studies (Hibar et al., 2016, 2018; McWhinney, Abé, et al., 2021; McWhinney, Abé, et al., 2022; McWhinney, Brosch, et al., 2022; McWhinney et al., 2023). As a sensitivity analysis, we additionally pre-processed the raw data using ComBat to mitigate between-site variability in the raw data (Johnson et al., 2007; Radua et al., 2020). We recompleted PCA in cortical thickness for our primary sample, calculated scores for the first component, and tested for associations with group, age, sex, and BMI as specified above. Estimates and significance of these effects were compared with and without the transformation of ComBat to test whether the combination of PCA and random effects adequately controlled for between-site variability.
2.6 Application in replication sample
The PCA model derived from cortical thickness of the primary sample was applied in the replication sample by applying the first PC's projections to the cortical thickness data for these new individuals. The first PC's score in the new data was tested for associations with group, BMI, age, sex, and a random effect of research site using linear mixed regression modeling. Group was first tested using two levels (healthy controls or patients), and second using three levels (healthy controls, individuals with BD, or schizophrenia). We included individuals with schizophrenia, as we wanted to test whether the method revealed something specific to BD or whether the results represented a general pattern across diagnoses. For comparison, a new PCA model was additionally run in the replication sample, resulting in each individual receiving a first component score from each of the two PCA models. These two scores were compared using the Pearson correlation coefficient.
3 RESULTS
3.1 Sample
Both the primary and replication samples are outlined in Table 1. In both samples, individuals with BD were significantly older and had significantly higher BMI relative to controls. All models that included both groups adjusted for both age and BMI.
3.2 Principal component analysis
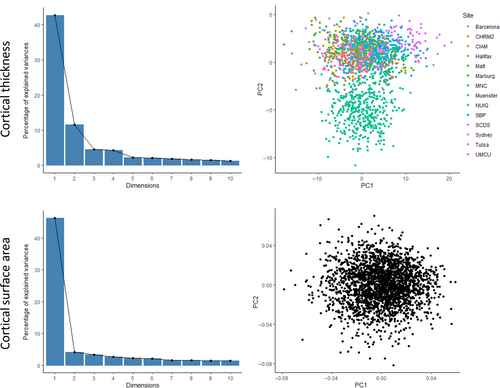
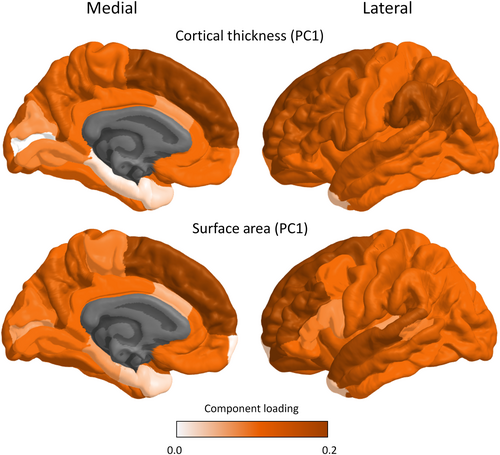
In the primary sample, the scores for each of the first PCs accounted for 42.7% of variance in cortical thickness, and 46.2% in surface area (see Figure 1). With exception of the second component for cortical thickness (11.5%, see Figure S1), all other components in both measures explained <5% of variance each. The first principal component scores were associated with higher cortical thickness and surface area in all studied regions, with some regional variations in the strength of association (see Figure 2). Consequently, if a clinical variable was associated with lower first component score, then it would be associated with lower cortical thickness across regions in a pattern reflecting the component's loadings across the regions.
For cortical thickness, the first component's score was negatively associated with diagnosis of BD, BMI, and age, indicating that BD, BMI, and age were independently associated with a diffuse pattern of thinner cortex, see Table 2. Among participants with BD, lithium and antipsychotic medications showed opposing associations with the first principal component, such that antipsychotics were negatively while lithium was positively associated with cortical thickness, see Table 2. The second component for cortical thickness was significantly associated with BMI, age, and sex, see Table S7.
| All participants | Participants with BD | ||||
|---|---|---|---|---|---|
| Estimate (SE) | Significance | Estimate (SE) | Significance | ||
| Cortical thickness | Group (BD) | −1.38 (0.17) | F(1,2421) = 67.80, p = .000* | n/a | n/a |
| BMI | −0.39 (0.13) | F(1,2419) = 8.79, p = .003* | 0.25 (0.29) | F(1,436) = 0.75, p = .387 | |
| Sex (F) | 0.16 (0.14) | F(1,2418) = 1.30, p = .254 | −0.01 (0.34) | F(1,435) = 0.00, p = .970 | |
| Age | −3.10 (0.10) | F(1,2422) = 1003.34, p = .000* | −3.37 (0.28) | F(1,439) = 143.67, p < .001 * | |
| Diagnosis BDII | n/a | n/a | −0.74 (0.57) | F(1,438) = 1.65, p = .200 | |
| Lithium | n/a | n/a | 0.80 (0.38) | F(1,437) = 4.50, p = .034 * | |
| Antipsychotic | n/a | n/a | −0.90 (0.38) | F(1,438) = 5.61, p = .018 * | |
| Anticonvulsant | n/a | n/a | −0.79 (0.40) | F(1,438) = 3.84, p = .051 | |
| Antidepressant | n/a | n/a | 0.20 (0.37) | F(1,436) = 0.28, p = .597 | |
| Age of onset | n/a | n/a | 0.03 (0.02) | F(1,437) = 2.29, p = .131 | |
| Psychosis | n/a | n/a | 0.44 (0.46) | F(1,439) = 0.92, p = .338 | |
| Cortical surface area | Group (BD) | −0.03 (0.22) | F(1,2431) = 0.02, p = .904 | n/a | n/a |
| BMI | −0.10 (0.18) | F(1,2424) = 0.35, p = .554 | 0.41 (0.38) | F(1,439) = 1.20, p = .274 | |
| Sex (F) | −5.77 (0.18) | F(1,2421) = 980.07, p = .000* | −6.40 (0.44) | F(1,436) = 210.13, p < .001* | |
| Age | −1.53 (0.13) | F(1,2430) = 139.82, p = .000* | −2.49 (0.36) | F(1,440) = 47.03, p < .001* | |
| Diagnosis BDII | n/a | n/a | −0.81 (0.74) | F(1,441) = 1.19, p = .276 | |
| Lithium | n/a | n/a | 1.55 (0.48) | F(1,440) = 10.23, p = .001* | |
| Antipsychotic | n/a | n/a | −0.36 (0.49) | F(1,441) = 0.54, p = .463 | |
| Anticonvulsant | n/a | n/a | 0.56 (0.52) | F(1,441) = 1.16, p = .282 | |
| Antidepressant | n/a | n/a | 0.33 (0.48) | F(1,437) = 0.47, p = .495 | |
| Age of onset | n/a | n/a | 0.03 (0.03) | F(1,441) = 1.01, p = .315 | |
| Psychosis | n/a | n/a | 0.34 (0.59) | F(1,440) = 0.33, p = .565 | |
- Note: Asterisks indicate significant associations (*p < .05).
For surface area, only sex, age, and Li treatment were associated with the first component scores for surface area, see Table 2. Females, relative to males, and older participants showed significantly smaller surface area, while those prescribed lithium showed larger surface area.
3.3 Comparing goodness of fit
Rankings of component loadings closely overlapped with the ranking based on clustering (Cohen et al., 2023) (Spearman ρ = 0.922, p < .001). We tested goodness of fit measures when using the covariates alone, relative to the addition of cluster number, the first PC score for cortical thickness, or both combined. Each of these sets of predictors was included in separate models for the prediction of group (control or BD) or BMI. Multicollinearity among the cluster number and the first component was acceptable in the combined model (VIF = 2.4). Results are shown in Table 3.
| Covariates | Covariates, cluster | Covariates, PCA | Covariates, cluster, PCA | |||
|---|---|---|---|---|---|---|
| Predicting group | Model fit | BIC | 2512 | 2501 | 2458 | 2465 |
| AUC | 0.811 | 0.815 | 0.823 | 0.823 | ||
| Predictor significance | Cluster number | – | χ2 = 18.52, p < .001* | – | χ2 = 0.19, p = 0.659 | |
| PCA first component | – | – | χ2 = 57.99, p < .001* | χ2 = 41.04, p < .001* | ||
| Model significance | VS Covariates | – | χ2 = 18.75, p < .001* | χ2 = 61.67, p < .001* | χ2 = 61.86, p < .001* | |
| VS Cluster model | – | – | – | χ2 = 43.11, p < .001* | ||
| VS PCA Model | – | – | – | χ2 = 0.19, p = .660 | ||
| Predicting BMI | Model fit | BIC | 3793 | 3803 | 3803 | 3816 |
| R2 | .159 | .162 | .167 | .167 | ||
| Predictor significance | Cluster number | – | χ2 = 3.95, p = .047* | – | χ2 = 0.16, p = .687 | |
| PCA first component | – | – | χ2 = 7.76, p = .005* | χ2 = 3.95, p = .047* | ||
| Model significance | VS Covariates | – | χ2 = 3.94, p = .047* | χ2 = 7.65, p = .006* | χ2 = 7.81, p = .020* | |
| VS Cluster model | – | – | – | χ2 = 3.86, p = .049* | ||
| VS PCA Model | – | – | – | χ2 = 0.16, p = .686 |
- Note: Asterisks indicate significant associations (*p < 0.05).
The BIC indicated that PCA offered the most accurate and parsimonious model over any other option when predicting the group (BD or controls). Both the clustering and PCA methods had a similar BIC when predicting BMI. However, goodness of fit (AUC for group, or R2 for BMI) was highest when using PCA to predict either group or BMI. While cluster number and the first component score were significant predictors of both group and BMI alone, when both were included as predictors in a single model, only the first PC was a significant predictor of BMI. Also, both the cluster and PCA models provided a significantly better fit than the covariate-only model. Critically, while the combined model performed significantly better than the cluster-based model, it was not a significant improvement over the PCA model (Table 3).
Lastly, when testing the association between the first component's score for cortical thickness with BMI and group, model fit (R2 = .065) was 28.6% higher than when using average cortical thickness (R2 = .050), with corresponding improvements in AIC and BIC, see Table 4. The first component score and average cortical thickness were highly correlated (r = .983, p < .001, see Figure S2), and average cortical thickness was significantly associated with the first component score of cortical thickness with adjustment for research site (t(2433) = 290.30, p < .001).
| Outcome | Predictors | R2 | AUC | AIC | BIC | Percent fit improvement |
|---|---|---|---|---|---|---|
| First component | BMI, Diagnosis | .065 | – | 6756 | 6780 | 28.6% |
| Avg. thickness | BMI, Diagnosis | .050 | – | 6793 | 6817 | – |
- Note: Fit is shown using R2 for linear models. Percent improvement in fit for using the first component relative to average thickness is shown in each model.
3.4 PCA predictions in replication sample
When applying the PCA model to the cortical thickness estimates in the replication sample on which the model was not trained, the first PC (i.e. thickness overall) was significantly smaller in patients (BD and schizophrenia) relative to controls (Difference estimate = 1.48, SE = 0.49, F(1,320) = 8.84, p = .003), and in older participants (F(1,321) = 193, p < .001), while there was no significant association with BMI or sex. These results are consistent with those for the original sample (see Table 2), except for the missing association with BMI, which may be due to lower statistical power in the smaller sample, which was 13.4% the size of the primary sample. When categorized using three diagnostic groups, significant group differences remained (F(1,318) = 7.21, p = .001), with the thickest cortex in controls, intermediate in BD (Est = −0.96, SE = 0.54), and the thinnest in schizophrenia (Est = −2.31, Est = 0.61).
Within the replication sample, the first PC derived from the training sample loadings was strongly correlated with the first PC from the new PCA, completed in this replication sample (r = .998, t(325) = 308.32, p < .001). These findings suggest that the PCA model is sensitive not only to generalizable differences seen in BD from other samples but also to similar variations seen in other SMIs.
3.5 Exploration of components
The distribution on the first component for cortical thickness was normal (W = 0.99, p = .422), whereas the second component showed a non-normal, bimodal distribution (Figure 1, W = 0.89, p < .001). The distinct cluster of lower scores consisted of data from a single research site; no other clinical or demographic data distinguished these clusters. The variance accounted for by each component is shown in Table S8.
3.6 Comparison with ComBat
Similar to the analyses without ComBat, the first PC of data which were preprocessed with ComBat remained significantly associated with age, BMI, and diagnosis of BD. Specifically, we found significantly thinner cortex in older participants (Est = −3.94, 95% CI [−4.05, −3.62]), those with higher BMI (Est = −0.50, 95% CI [−0.81, −0.18]), and those with BD (Est = −1.66, 95% CI [−2.03, −1.30]). In addition, the ranking of regional component loadings for the first PC with and without the application of ComBat using the Spearman rank order correlation coefficient was almost identical (ρ = 0.921, p < .001), see Table S9 for more details.
4 DISCUSSION
In this study, the first of 68 total PCs accounted for 42.7% of variance in cortical thickness. The first PC, which indexed a greater cortical thickness across all 68 cortical regions, was negatively associated with BD, BMI, antipsychotic medications, and age and positively associated with Li treatment. These associations between the first PC and cortical thickness closely mirrored the associations found when applying clustering to the same sample, where the cluster with lower cortical thickness was also associated with diagnosis of BD, higher BMI, and older age, and the cluster with higher cortical thickness was associated with Li treatment (McWhinney, Abé, et al., 2022). Only PCA, not clustering detected links with antipsychotic medications. Also, when directly compared, PCA outperformed clustering as predictors of clinical and demographic variables. When we applied the PCA to the previously unseen replication sample collected from additional ENIGMA-BD working group sites, on which the model was not trained, we found the same patterns of associations with diagnosis and age, even though the sample was almost 90% smaller. The same pattern of brain changes detected in BD was also associated with the diagnosis of schizophrenia. Similar to previous large-scale studies, surface area was not associated with diagnosis of BD or BMI (Hibar et al., 2018; McWhinney, Abé, et al., 2022; McWhinney, Brosch, et al., 2022; McWhinney et al., 2023). The different system-level correlates of CT and SA in our and previous studies further support the practice of keeping these measures separate.
We directly compared PCA and clustering in the same dataset. Both methods detected similar associations with system-level variables, including diagnosis, BMI, age, and Li exposure. However, PCA outperformed clustering in terms of model fit and sensitivity. We suspect there are systematic reasons for this. Even if there are no clearly defined discontinuities/clusters in the data, clustering would segment continuous distribution of findings into several parts. Similar categorization of a continuous range of values necessarily results in a loss of statistical power, as very alike individuals are considered distinct when on opposing sides of a clustering threshold. That is, clustering does not encode the distance between individuals in the multidimensional space where those assigned to the same cluster still differ from one another. Similarly, as there are no strict boundaries, individuals assigned to different clusters may be very similar to one another. In contrast, PCA does not need arbitrary criteria to delineate patterns and find orthogonal effects in the dataset. Within the same component, we were able to maintain the strength of the associations and distance between individuals. For both reasons, PCA should systematically outperform clustering when there are no clearly defined groups of individuals and indeed that was the case in our study.
While our study replicates previous findings of negative associations between cortical thickness and diagnosis of BD in mass univariate analyses (Hibar et al., 2018; McWhinney, Abé, et al., 2021; McWhinney, Brosch, et al., 2022; van Erp et al., 2018), the effect size for association between first PC (d = 0.33) and diagnosis of BD was stronger than associations between individual ROIs and the diagnosis of BD in previous ENIGMA studies (d = 0.015–0.29) (Hibar et al., 2018) and instead of running one model per region, we captured covariance across all regions by a single number. Even in mass univariate analyses associations between clinical variables and brain structure are evident across many regions and are not isolated to a single ROI (Hibar et al., 2018; McWhinney, Abé, et al., 2021; McWhinney, Brosch, et al., 2022; van Erp et al., 2018). The lesion model, where changes in a single region are necessary and sufficient to cause an illness, clearly does not apply to a complex disorder such as BD (Hibar et al., 2018; Reddan et al., 2017). Consequently, looking at distributed effects across groups of regions should be more informative than looking at individual regions. All in all, it is encouraging that our findings converge with these theoretical expectations and suggest that multivariate analyses are better suited to studying complex neuropsychiatric disorders than mass univariate ones.
While the second component for cortical thickness explained approximately 12% of variance, it was associated predominantly with research site; with the outlying site removed, the second component explained only 5% of the variance, see supplement. It is interesting that the first and second components, which are necessarily orthogonal to one another, were each predominantly associated with clinical/anthropometric variables, or research site, respectively. These differences need to be replicated in other studies, but they may represent a more generalizable pattern. After all, correlates of demographic and clinical variables are consistent in the same direction (i.e. negatively associated with cortical thickness). In contrast, the variations related to research sites may be less consistent and less predictable. Consequently, they may fall into separate components. Interestingly when we applied a different method of removing the site effects, i.e. ComBat, the associations between the first PC and clinical/demographic variables remained identical to the results obtained without ComBat. Even without removal of site effects from the raw data, by identifying orthogonal components, PCA may implicitly separate the site effects from the more predictable/systematic biological effects. If confirmed in future studies, this would be a major advantage of PCA.
The same distributed pattern of brain regions was associated with each of the system-level variables. When we applied the PCA projections to the replication sample, we found associations with similar system-level variables, consistent with observations by others (Cao et al., 2023). In fact, the same patterns were also associated with the diagnosis of schizophrenia. This is in keeping with other large-scale studies or meta-analyses, which have also demonstrated that there is a common, non-specific pattern of case-control differences across major psychiatric disorders (Goodkind et al., 2015; Hettwer et al., 2022; Matsumoto et al., 2023), with highly correlated neurostructural abnormalities between BD and schizophrenia (Opel et al., 2020) and with PCA detecting a profile of shared cortical thickness differences across 6 major psychiatric disorders, which explained 48% of variance (Writing Committee for the Attention-Deficit/Hyperactivity Disorder et al., 2021). Some have argued that there may be more cause-specific alterations after removing this non-specific pattern of the first component (Cao et al., 2023), though with exception of the second component for cortical thickness, subsequent components explained only a small fraction of variance (i.e. typically less than 5% each). All in all, this study contributed to the growing body of evidence for lack of specificity of associations between some key clinical and demographic factors and patterns of brain alterations, so-called neural P factor (Sprooten et al., 2022). Many different variables, including age, obesity, psychiatric diagnoses may be negatively associated with cortical thickness across a wide range of cortical regions.
When testing associations with BMI and group, the first PC explained 28.6% more variance than average cortical thickness. Principal component analysis, which accounts for the regional distribution of effects, was better than simple average cortical thickness, which collapses information across all regions. At the same time, the PC1 was highly correlated with average cortical thickness. While there are regional effects and accounting for these improves the fit of the model, the system-level variables are associated with some level of cortical thinning across most regions. This may further contribute to the lack of specificity of the system level to brain associations and is consistent with findings from another study indicating that accuracy of ML classifier comparing controls versus BD versus schizophrenia was strongly dependent on global grey matter measures (Schwarz et al., 2019).
Our findings confirmed that BD is characterized by diffuse regional structural brain alterations, specifically lower cortical thickness. The fact that these alterations are so diffuse as to resemble global atrophy is interesting. We can only speculate about why this would occur. First of all, most pathologies will result in atrophy, i.e. thinning of the cortex. Second of all, changes in one region are likely to propagate through the network, thus eventually involving more related regions. Thirdly, perturbations within one network are going to propagate to other networks, thus involving even more regions eventually to the point of resembling a global atrophy. These mechanisms could explain why so many regions are correlated across individuals, why the association with the first component is uniformly in one direction, and even why different predictors (age, BMI, BD, schizophrenia, medications) are all associated with the same global pattern.
The advantages of this study include the large sample size, the validation of the most prominent findings in a replication sample, and the multivariate approach which improved statistical sensitivity. Importantly, we were able to directly compare results between two unique multivariate analyses in the same sample: clustering and PCA.
The multi-site nature of the study is a limitation that complicates the data analyses and interpretation of the results. At the same time, our findings suggest that PCA may code site in separate PCs from the effects of clinical/anthropometric variables. Along similar lines, the sample contained a broad representation of BD, including individuals with BDII and we also included a sample of people with schizophrenia. In order to test how well a method captures relevant clinical links, we need a representative/generalizable snapshot of the disorder, which is not restricted to one subtype and ideally also includes representation of other diagnoses. More detailed clinical or biological markers beyond those analyzed were not broadly available throughout the ENIGMA-BD working group. While it is possible that associations with other clinical variables would show different patterns, the consistent and replicated nature of the direction of associations and spatial distribution of networks suggest this is unlikely. As these were independently collected datasets, not a centralized single study, we did not have access to raw, whole-brain data.
We performed these analyses on derived estimates of cortical thickness and surface area, but we cannot generalize our findings to other measures, which may show different patterns of associations. There are other methods within this broad category, which may be of specific use for example when attempting a multimodal data fusion, i.e. FLICA. However, comparing different methods within the broad group of linear representation of data was not our goal and would likely only lead to incremental gains, if any. We did not test other multivariate techniques including ML methods, which are difficult to interpret. Our interest was in applying traditional methods which may also be applied in smaller samples and in a wide range of situations, including clinical settings. Due to simplicity and straightforward application of the linear methods, they may be a method of choice, especially where the size/structure of the dataset would not allow for proper ML.
5 CONCLUSION
In this study, we confirmed that cortical thickness in widespread regional networks as determined by PCA is negatively associated with relevant clinical and demographic variables, including diagnosis, age, BMI, and treatment with antipsychotic medications or lithium. The action of these factors on a widespread network suggests that conceptualizing or studying their effects on individual regions may be misleading. In addition, significant associations of many different system-level variables with the same network suggest a lack of specificity to individual clinical and demographic factors. While there may be general agreement among multivariate techniques on these associations, PCA outperformed clustering and may better fit the nature of brain imaging data in SMI. More broadly, the results have demonstrated that representing data as a linear combination of features is superior to clustering when investigating links between brain and system measures in SMI. This work could help researchers make informed decisions about which methods to use and could save them from applying an ill-fitting method, when a simpler, more reproducible one is available.
ACKNOWLEDGEMENTS
We gratefully acknowledge the following contributions and research funding sources that made this study possible: PMT, CRKC, and SIT were supported by NIH grants U54 EB020403 from the Big Data to Knowledge (BD2K) Program, R56 AG058854 (The ENIGMA World Aging Center); R01 MH116147 (The ENIGMA Sex Differences Initiative), R01 MH129742-01 (The ENIGMA Bipolar Initiative), and the Baszucki Brain Research Fund & Milken Institute's Center for Strategic Philanthropy; CRKC also acknowledges, NIA T32AG058507. The St. Göran study was supported by grants from the Swedish Research Council (2022-01643), the Swedish Foundation for Strategic Research (KF10-0039), the Swedish Brain Foundation (FO2022-0217), and the Swedish Federal Government under the LUA/ALF agreement (ALF 20200036, ALFGBG-965444).
FUNDING INFORMATION
This work is also part of the German multicenter consortium “Neurobiology of Affective Disorders. A translational perspective on brain structure and function,” funded by the German Research Foundation (Deutsche Forschungsgemeinschaft DFG; Forschungsgruppe/Research Unit FOR2107). Principal investigators (PIs) with respective areas of responsibility in the FOR2107 consortium are as follows: Work Package WP1, FOR2107cohort and brain imaging: TK (speaker FOR2107; DFG grant numbers KI588/14-1, KI588/14-2, KI588/20-1, KI588/22-1), UD (co-speaker FOR2107; DA 1151/5-1, DA 1151/5-2), AK (KR 3822/5-1, KR 3822/7-2), IN (NE 2254/1-1, NE 2254/2-1, NE 2254/3-1, and NE 2254/4-1), BS (STR 1146/18-1), CK (KO 4291/3-1). Further support from the German sites was provided by MNC and FOR2107-Muenster: This work was funded by the German Research Foundation (SFB-TRR58, Project C09 to UD) and the Interdisciplinary Center for Clinical Research (IZKF) of the medical faculty of Münster (grant Dan3/012/17 to UD and grant SEED11/18 to NO); FOR2107-Muenster: This work was supported by grants from the Interdisciplinary Center for Clinical Research (IZKF) of the medical faculty of Münster (grant MzH 3/020/20 to TH) and the German Research Foundation (DFG grants HA7070/2-2, HA7070/3, HA7070/4 to TH, CRC1457/A7 to RN). The Medellin studies (GIPSI) were supported by the PRISMA UNION TEMPORAL (UNIVERSIDAD DE ANTIOQUIA/HOSPITAL SAN VICENTE FUNDACIÓN), Colciencias-INVITACIÓN 990 de 3 de agosto de 2017, Codigo 99059634. The San Raffaele site was supported by the Italian Ministry of Health RF-2018-12367789 project. The University of Galway research was supported by the Irish Research Council (IRC) Postgraduate Scholarship, Ireland awarded to LN and to GM, and by the Health Research Board (HRA-POR-324) awarded to DMC and (HRA_POR/2011/100) awarded to CMcD. We thank the participants and the support of the Welcome-Trust HRB Clinical Research Facility and the Centre for Advanced Medical Imaging, St. James Hospital, Dublin, Ireland. JS and RTK received support from the William K. Warren Foundation National Institute of Mental Health (R21MH113871); JS also acknowledges the National Institute of General Medical Sciences (P20GM121312). MB is supported by a NHMRC Senior Principal Research Fellowship and Leadership 3 Investigator grant (1156072 and 2017131). JH and EB were supported by the Czech Health Research Council Project No. NU21-08-00432 and by ERDF-Project Brain dynamics, No. CZ.02.01.01/00/22_008/0004643. IHG received support from the National Institute of Mental Health (R37MH101495). This study was also funded by EU-FP7-HEALTH-222963 “MOODIN-FLAME” and EU-FP7-PEOPLE-286334 “PSYCHAID.” The Barcelona group would like to thank CIBERSAM (EPC) and the Instituto de Salud Carlos III (PI18/00877 and PI19/00394) for their support. This work was supported by the Singapore Bioimaging Consortium (RP C009/2006) research grant awarded to K.S. The CIAM group (FMH—PI) was supported by the University Research Committee, University of Cape Town, and South African funding bodies National Research Foundation and Medical Research Council; DJS from CIAM was supported by the SAMRC. FS from CIAM was supported by a Brain-Behavior Unit Postdoctoral Research Fellowship. The Sydney studies were supported by the Australian National Health and Medical Research Council (NHMRC) Program Grant 1037196, Investigator Grants 1176716 (PRS) and 1177991 (PBM), Project Grants 1063960 and 1066177, the Lansdowne Foundation, Good Talk and Keith Pettigrew Family; as well as the Janette Mary O'Neil Research Fellowship to JMF. The study was also supported by NIMH grant number: R01 MH090553(to RAO). Funding for the Oslo-Malt cohort was provided by the South Eastern Norway Regional Health Authority (2015-078), the Ebbe Frøland Foundation, and a research grant from Mrs. Throne-Holst. EV acknowledges the support of the Spanish Ministry of Science and Innovation (PI15/00283, PI18/00805) integrated into the Plan Nacional de I + D + I and co-financed by the ISCIII-Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER); the Instituto de Salud Carlos III; the CIBER of Mental Health (CIBERSAM); the Secretaria d'Universitats i Recerca del Departament d'Economia i Coneixement (2017 SGR 1365), the CERCA Programme, and the Departament de Salut de la Generalitat de Catalunya for the PERIS grant SLT006/17/00357. Lastly, this study was supported by the Canadian Institutes of Health Research (103703, 106469, and 142255, 180449, 186254), Nova Scotia Health Research Foundation, Dalhousie Clinical Research Scholarship to TH, Brain & Behavior Research Foundation (formerly NARSAD); 2007 Young Investigator and 2015 Independent Investigator Awards to TH. The Minnesota sites were supported by the National Institutes of Health (U01MH108150 to SS; K01MH093621 to SU), the Center for Magnetic Resonance Research (P41 EB027061; 1S10OD017974), a Merit Review Award (#I01CX000227 to SS) from the United States (U.S.) Department of Veterans Affairs Clinical Science Research and Development Service, and Minneapolis VA Health Care System resources to SU and SS. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the United States government. LTE was supported by NIH MH083968 and Desert-Pacific Mental Illness Research Education and Clinical Center.
EV thanks the support by CIBER-Consorcio Centro de Investigación Biomédica en Red-(CB07/09/0004), Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation and grants PI18/00805 and PI21/00787, integrated into the Plan Nacional de I + D + I and co-financed by the ISCIII-Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER); the Instituto de Salud Carlos III; the Secretaria d'Universitats i Recerca del Departament d'Economia i Coneixement (2021 SGR 01358), the CERCA Programme, and the Departament de Salut de la Generalitat de Catalunya for the PERIS grant SLT006/17/00357. Thanks also for the support of the European Union Horizon 2020 research and innovation program (EU.3.1.1. Understanding health, wellbeing, and disease: Grant No 754907 and EU.3.1.3. Treating and managing disease: Grant No 945151).
JR thanks the support by the Spanish Ministry of Science and Innovation. Instituto de Salud Carlos III (PI19/00394 and PI22/00261), integrated into the Plan Nacional de I + D + I and co-financed by ERDF Funds from the European Commission (“A Way of Making Europe”), CIBERSAM, and the Secretaria d'Universitats i Recerca, Departament d'Economia i Coneixement and Departament de Salut (2021 SGR 01128).
CONFLICT OF INTEREST STATEMENT
PMT & CRKC received a grant from Biogen, Inc., for research unrelated to this manuscript. DJS has received research grants and/or consultancy honoraria from Lundbeck and Sun. LNY has received speaking/consulting fees and/or research grants from Abbvie, Alkermes, Allergan, AstraZeneca, CANMAT, CIHR, Dainippon Sumitomo Pharma, Janssen, Lundbeck, Otsuka, Sunovion, and Teva. TE received speaker's honoraria from Lundbeck and Janssen Cilag and is a consultant to Sumitomo Pharma America. Thanks also for the support of the European Union Horizon 2020 research and innovation program (EU.3.1.1. Understanding health, wellbeing, and disease: Grant No 754907 and EU.3.1.3. Treating and managing disease: Grant No 945151). EV has received grants and served as consultant, advisor, or CME speaker for the following entities (unrelated to the present work): AB-Biotics, Abbott, Allergan, Angelini, Dainippon Sumitomo Pharma, Ferrer, Gedeon Richter, Janssen, Lundbeck, Otsuka, Sage, Sanofi-Aventis, and Takeda. PMT and CRKC have received partial research support from Biogen, Inc. (Boston, USA) for work unrelated to the topic of this manuscript. EV has received grants and served as consultant, advisor, or CME speaker for the following entities: AB-Biotics, AbbVie, Adamed, Angelini, Biogen, Biohaven, Boehringer-Ingelheim, Celon Pharma, Compass, Dainippon Sumitomo Pharma, Ethypharm, Ferrer, Gedeon Richter, GH Research, Glaxo-Smith Kline, HMNC, Idorsia, Johnson & Johnson, Lundbeck, Medincell, Merck, Novartis, Orion Corporation, Organon, Otsuka, Roche, Rovi, Sage, Sanofi-Aventis, Sunovion, Takeda, and Viatris, outside the submitted work. Yatham reports grants from Abbvie and Dainippon Sumitomo, and served as an advisor or consultant or speaker to JAMA Pharma, Intracellular Therapies, Merck, Allergan, GSK, Gedeon Richter, Sanofi, Sunovion, and Alkermes, outside the submitted work.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




