Intracortical astrocyte subpopulations defined by astrocyte reporter Mice in the adult brain
Funding information: National Institutes of Health, Grant/Award Numbers: R21NS087391, R01MH106490, R01MH099554
Abstract
Although historically regarded as a homogeneous cell population, astrocytes in different brain regions exhibit differences in their physiological properties, such as gap-junction coupling, glutamate uptake dynamics, and intracellular Ca2+ response. Recent in vivo RNA profiles have further demonstrated the molecular heterogeneity of astrocytes in the adult CNS. Astrocyte heterogeneity exists not only inter-regionally but also intra-regionally. Despite the characteristic laminal organization of cortical layers and multiple sources of radial glia progenitors for (astro)gliogenesis, the molecular profile and functional properties of astroglial subpopulations in the adult cerebral cortex remain essentially undefined. Using two astrocyte reporter mouse lines: eaat2-tdTomato and Bac aldh1l1-eGFP, we identified tdT−eGFP+, tdTloweGFP+, and tdThigheGFP+ astroglial subpopulations (in an approximate 1:7:2 ratio) within the cortex. The tdT−eGFP+ astrocyte population is selectively localized at layers I–II and exhibits increased resting membrane potential and membrane resistance but reduced functional expression of the potassium channel Kir4.1. We also isolated individual astrocyte subpopulations through fluorescence activated cell sorting (FACS) and examined their transcriptome differences by RNA-seq. We found that the whole-genome transcriptional profiles of tdT−eGFP+ astrocytes are drastically different from that of tdTloweGFP+ and tdThigheGFP+ astrocytes. Particularly, elevated levels of several nonastrocyte genes that are typically specific to other glial cells, such as mog, mobp, Iba1, and pdgfrα, are observed in tdT−eGFP+ astrocytes, suggesting a less-specific molecular identity of these astrocytes. Overall, our study has unveiled molecular differences between adult cortical astroglial subpopulations, shedding new light on understanding their unique functions in the adult cortex.
1 INTRODUCTION
Astrocytes are an indispensable cell type in the CNS with many important and versatile functions, ranging from structural and metabolic support to modulation of synaptogenesis and synaptic transmission (Clarke & Barres, 2013). Astrocytes exhibit significant functional and molecular heterogeneity in vitro and in vivo (Chaboub & Deneen, 2012; Oberheim, Goldman, & Nedergaard, 2012), especially in a region-dependent manner (Fuentealba, Alvarez-Buylla, & Rowitch, 2015; Zhang & Barres, 2010). Recent in vivo profiling of astroglial mRNAs in major cortical and subcortical brain regions also found an interesting dorsal to ventral expression pattern of astroglial translating mRNAs (Morel et al., 2017). A recent cell-surface marker screening also defined several astrocyte subpopulations, with significantly distinct molecular signatures, in the brain of Bac aldh1l1-eGFP mice (John Lin et al., 2017). Functionally, regional astrocytes exhibit different levels of gap-junction coupling (Batter et al., 1992), glutamate uptake dynamics (Danbolt, 2001; Regan et al., 2007), and inward-rectifying K+ currents (Olsen, Campbell, & Sontheimer, 2007). In particular, brain stem astrocytes are able to uniquely modulate the inspiratory rhythm in response to environmental pH changes (Gourine et al., 2010).
Astrocyte heterogeneity exists not only inter-regionally but also intra-regionally. In the cerebellum, dorsal Bergmann glia and ventral velate astrocytes show opposite expression patterns for a number of synaptic proteins, such as GluA1, GluA4, GLAST, Kir4.1, and GLT1 (Farmer et al., 2016). Subsets of striatal astrocytes selectively respond to dopamine receptor D1 (DRD1) or D2 (DRD2) medium spiny neuronal (MSN) activity and subsequently activate NMDA receptors in homotypic but not heterotypic MSNs (Martin, Bajo-Graneras, Moratalla, Perea, & Araque, 2015). Cortical astrocytes are originated from radial glia (RG) in the ventricular or subventricular zone (VZ or SVZ) during the late embryonic or early postnatal phase when RG transition from GLAST+/Nestin+ to GLAST+/Nestin− progenitors (Siddiqi et al., 2014). Continuous local proliferation of newly generated protoplasmic astrocytes during the first 2–3 weeks postnatally is considered the major source of cortical astrocytes (Ge, Miyawaki, Gage, Jan, & Jan, 2012). Despite the characteristic laminal organization of cortical layers and VZ/SVZ sources of RG progenitors, intracortical astrocyte diversity has just begun to be understood. Cortical layer I astrocytes appear to strongly express glial fibrillary acidic protein (GFAP) with reduced cell density (Muralidhar, Wang, & Markram, 2013). Primate cerebral cortex layer I contains a large number of GFAP+/CD44+ interlaminar astrocytes (Colombo & Reisin, 2004), which extend long, straight, but less branched processes into deeper layers (Sosunov et al., 2014). There are also primate-specific varicose projection astrocytes which primarily reside in layers V–VI (Oberheim et al., 2009).
In the current study, we identified tdT−eGFP+, tdThigheGFP+, and tdTloweGFP+ astroglial subpopulations within the cerebral cortex using adult eaat2-tdT+ Bac aldh1l1-eGFP+ double reporter mice. The tdT−eGFP+ astrocyte population is selectively localized in layers I–II and exhibits distinct electrophysiological and unique molecular properties as compared to tdThigheGFP+ and tdTloweGFP+ astrocytes.
2 MATERIALS AND METHODS
2.1 Animals
The eaat2-tdT transgenic mice (C57Bl6/FVB mixed background) were generated as previously described (Yang et al., 2011). Bac aldh1l1-eGFP (Cahoy et al., 2008) transgenic mice (C57Bl6/FVB mixed background) were obtained from the GENSAT project through The Jackson Laboratory (stock#: 030247 Bar Harbor, ME, USA). Both male and female mice were randomized in all experiments. All mice were maintained on a 12 hr light/dark cycle with food and water ad libitum. Both male and female mice were used for all experiments. All procedures were in strict accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Tufts University Institutional Animal Care and Use Committee.
2.2 Tissue processing, immunohistochemistry, and confocal imaging
Bac aldh1l1-eGFP or eaat2-tdT reporter mice (P60–70) were deeply anesthetized with ketamine (100 mg/kg) plus xylazine (10 mg/kg) in saline by i.p. injection and perfused intracardially with 4% paraformaldehyde (PFA) in PBS. The brains were dissected and kept in 4% PFA overnight at 4 °C, then cryoprotected by immersion in 30% sucrose for 48 hr. Brains were embedded and frozen in Tissu-Tek OCT Compound (Sakura). Coronal sections (20 μm) were prepared with a cryostat (model HM525, Leica) and mounted on glass SuperFrost+ Slides (Fisher Scientific) with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific). For immunostaining, slides were rinsed three times in PBS, then treated with blocking buffer (1% BSA, 5% goat serum, and 0.2% Triton-X 100 in PBS) for 30 min at room temperature. Primary antibodies for Iba1 (Wako, 1:500) and APC (cc1 clone, Calbiochem, 1:50) were incubated overnight at 4 °C in blocking buffer. After washing slides three times in PBS, corresponding secondary antibody (1:2000) was added for 90 min at room temperature (RT). The sections were rinsed three times in PBS before mounting. For representative images of the large field cortex, automated stitching of fluorescence images was performed using the Keyence BZ-X700 Series Microscope at 20× magnification (numerical aperture, 0.7). Confocal images were taken using the Nikon A1R confocal laser scanning microscope (15 μm Z stack with 0.5 μm step) magnified with 40× (numerical aperture 0.8) or 60× (numerical aperture 1.0) objectives (Nikon Instruments Inc., Melville, NY). For images, we focused mostly somatosensory cortex.
2.3 Astroglial domain analysis
Reconstruction and measurement of cortical astroglial domain size were performed using Imaris (Bitplane, Zurich, Switzerland). All confocal images of tdT fluorescence from cortex of eaat2-tdT transgenic mice (P60) for Imaris analysis were taken with a 40× oil immersion objective lens. Images were taken under optimized settings to best illustrate the astroglial morphology. The 3D reconstruction images were first made using original confocal Z-stack images in Imaris software, as previously described (Morel, Higashimori, Tolman, & Yang, 2014). Briefly, the surface tool was used to build the astroglial domain. This function uses an automatic smoothing of the image with the Gaussian filter. The tdT fluorescence negative area in each of the confocal stack images was used as the internal control to determine the background fluorescence. The sensitivity threshold (absolute intensity) was manually adjusted so that the generated astroglial domain in the 3D image matches with that in the original confocal image. The cell somas were then detected based on size (≥12 μm in diameter) and used as seeding points to build the 3D domain. The quality (intensity) threshold was also manually adjusted to ensure that all cell somas were detected in a given image. The seeded watershed algorithm enables the Imaris software to recognize and split the domains of neighboring cells. The cells that were only partially included in confocal and 3D images were excluded from analysis. Overlapped domains were also carefully examined to ensure accuracy. The volume size of individual astroglia can be directly measured from generated 3D domains in Imaris. The expression levels of tdT have no effect on the labeling and quantification of astrocyte domain size, as previously shown (Morel et al., 2014) by dye fill of astrocytes.
2.4 Brain slice preparation
Cortical brain slices were prepared from young adult (P21–28) Bac aldh1l1-eGFP+ eaat2-tdT+ double transgenic mice. Animals were anesthetized with a ketamine/xylazine cocktail (110 mg/kg, 10 mg/kg); the cortex was quickly removed and 300 μm cortical slices were cut using a vibratome (Leica VT1200, Leica Microsystems) in ice-cold artificial cerebrospinal fluid (aCSF) (in mM): KCl 3, NaCl 125, MgCl2 1, NaHCO3 26, NaH2PO4 1.25, glucose 10, CaCl2 2, and 400 μM l-ascorbic acid, with osmolarity at 300–305 mOsm equilibrated with 95% O2–5% CO2. Slices were incubated at RT until needed.
2.5 Electrophysiology and dye-fill of astrocytes
For astrocyte recordings, cortical astroglial cells were identified by eGFP or tdT florescence and unique electrophysiological properties (−75 mV to −85 mV resting membrane potential and no action potential firing with −120 mV to +100 mV ramp sweep). For astrocyte patch-clamp, we focused mostly somatosensory cortex. Astroglial somatic whole-cell recordings were obtained following the procedure described above at a holding potential of −75 mV. For the application of BaCl2, BaCl2 (100 μM) was bath applied to cortical slices. Resting membrane potentials (RMPs) of three different cortical astrocyte populations were measured and manually recorded prior to and after BaCl2 application (5 min). I–V curves were generated by plotting capacitance-normalized current amplitudes as a function of applied voltage, from −100 mV to +250 mV, in 50 mV increments, as per voltage step protocol. For whole-cell recordings, input resistance and capacitance were measured in a voltage-clamp with 500 ms 5 mV step pulse from −75 mV holding potential. Cell capacitance was calculated by first obtaining the decay time constant of the transient current by the 5 mV step pulse and dividing this number by the series resistance. For the intracellular labeling of astrocytes, 5% Lucifer yellow (LY) potassium salt (Invitrogen) was added to the pipette solution. In voltage-clamp mode, the clamp potential was set at the resting membrane potential of astrocytes and the dye was injected into the cell by applying 0.5 s negative current pulse (1 Hz) until astroglial processes were completely filled. After the cells were filled, slices were immersed in 4% PFA overnight at 4C, rinsed in PBS, and mounted using Invitrogen ProLong Gold mounting medium.
2.6 Preparation of cortical cell suspension, fluorescence activated cell sorting of astrocytes, and RNA isolation
The total cortex of double Bac aldh1l1-eGFP+ eaat2-tdT+ transgenic mice (P60–90) was used for FAC sorting to increase yield. Animals were deeply anesthetized with Ketamine (100 mg/kg) + Xylazine (10 mg/kg) in saline by i.p. injection and perfused intracardially with Hanks Buffered Salt Solution (HBSS; Thermo Fisher Scientific). The total cortex was immediately dissected in cold Hanks buffer supplemented with glutamate receptor antagonists, 3 mM DNQX and 100 mM APV (Sigma, St. Louis, MO), and cut into small pieces. The cell suspension was prepared by following the manufacturer's instructions in the neural tissue dissociation kit (Miltenyi Biotech, Auburn, CA). Briefly, small pieces of tissue were treated with papain enzymatic mix (37 °C, 15 min) and then digested with DNase I (37 °C, 10 min), followed by careful trituration. Cell mixtures were then filtered through a cell strainer (40–70 mm) and resuspended in cold HBSS solution (5–10 × 106 cells/ml) for FACS. One mouse was used for preparing each FAC sorted sample. Cells were sorted using MoFlo MLS high-speed cell sorter (Beckman Coulter) with Summit version 4.3 software. The whole procedure for cell suspension preparation and FAC sorting process was completed within 2–3 hr. FAC sorted cells were spun down and RNA was isolated from the cell pellet using the standard TRIzol reagent.
2.7 RNA-Seq analysis
The 100 bp paired-end sequencing for all prepared sequencing libraries was carried out at the Tufts Genomics Core Facility using an Illumina HiSeq 2500 sequencer. RNA-sequencing libraries were assigned randomly to sequencing lanes to avoid lane bias. FastQC program was used to examine the quality of generated reads. For mouse samples, we usually obtained 25–50 million reads for each library with a Phred quality score > 32. The total number of mapped reads for each region was at least 50 million, sufficient for calculation of reliable Fragments Per Kilobase of Mapped Reads (FPKM) values and subsequent differential gene expression analysis. The Tuxedo suite of tools (Tophat/Cufflinks/Cuffdiff) was used to analyze RNA-Seq reads (Trapnell et al., 2012). Reads were aligned to the reference mouse genome (Jul 2007 NCBI37/mm9) using the Tophat (v 2.0.8) software. Greater than 50% of both read pairs were mapped for all samples. Cufflinks (v 2.1.1) was used to calculate FPKM values with –G option for all mapped transcripts.
Cuffdiff2 was used to identify significant changes in mRNA expression among different samples using the biological replicates with a default q-value (FDR-adjusted p value) cut-off of 0.05 (q < 0.05). The heatmaps were generated from the Qlucore software based on the output lists from Cuffdiff2 with a more stringent q value (q < 0.001). For generating the pie chart, genes that were differentially expressed between tdT−eGFP+ versus tdThigheGFP+ or between tdT−eGFP+ versus tdTloweGFP+ astrocytes were overlapped first, then upload to Ingenuity pathway analysis (Qiagen) for annotating their functions. The output gene functional categories are summarized in the pie chart.
2.8 qRT-PCR
For qRT-PCR of mouse genes, RNA isolated from FAC sorted cells was converted to cDNA using a high archive cDNA synthesis kit (Applied Biosystems, Foster City, CA). The relative expression levels of selected genes were measured using SYBR Green (Life Technologies, Carlsbad, CA) reagent and specific primers for analyzed genes were chosen from PrimerBank (https://pga.mgh.harvard.edu/primerbank/).
2.9 Experimental design and statistical analysis
Sample size and statistical approach used for each experiment are described in each method section and in figure legends. Briefly, for FACS and RNA-seq, two replicates were included from separate mice; for astrocyte recording, 16–20 astrocytes from 4 to 6 mice/group were used. The G*Power (version 3.1) was used to estimate the reasonable sample size. All statistical analyses were performed using GraphPad Prism6. All values were plotted as the mean ± SEM. For multiple groups (> 2), one-way ANOVA was used to analyze the variance, followed by a Tukey post hoc test to compare multiple groups. For two-group comparison, Student's t-test was used. Statistical significance was tested at a 95% (p < .05) confidence level and was denoted with an asterisk (*p < .05; **p < .01; ***p < .001).
3 RESULTS
3.1 Identification of astroglial subpopulations in the adult cortex
The majority of cortical astrocytes are produced in an unusual way by local proliferation from newly born and differentiating protoplasmic astrocytes during postnatal 2–3 weeks (Ge et al., 2012), although radial glia from the ventricular or subventricular zone are the initial (astro)glial progenitors during (astro)gliogenesis (Freeman, 2010). Previous observations have found that conventional GFAP immunostaining only labels a limited number of astrocytes (especially in layer I) in physiological conditions (Cahoy et al., 2008) even though astrocytes are abundantly present in the adult cortex, leading to speculation that there are astroglial subpopulations in the cortex. The laminal organization of the cerebral cortex from layers I–VI also makes it interesting to explore whether there are layer-based astrocyte subpopulations in the cortex.
We previously generated astrocyte reporter eaat2-tdTomato (tdT) mice in which the tdT reporter is widely expressed in cortical astrocytes (Morel et al., 2014; Yang et al., 2011), facilitating the identification of differentially labeled cortical astrocytes when bred with other astroglial reporter mice, such as Bac aldh1l1-eGFP mice (Cahoy et al., 2008). Based on somatic eGFP and tdT reporter expression in adult (P60–90) eaat2-tdT+ Bac aldh1l1-eGFP+ double positive mice, we characterized three distinct expression patterns of tdT/eGFP reporters in cortical astrocytes: tdT−eGFP+ (red arrows, Figure 1a), tdTloweGFP+ (yellow arrows, Figure 1a), and tdThigheGFP+ (white arrows, Figure 1a). Interestingly, tdT−eGFP+ astrocytes were selectively localized only at cortical layers I–II, while both tdTloweGFP+ and tdThigheGFP+ astrocytes were distributed across layers III–VI. Layer-specific distribution of astrocytes in the cortex was also recently observed using the cell surface antigen CD51 (John Lin et al., 2017). In addition, tdThigheGFP+ astrocytes (white arrows, Figure 1b) appeared to exhibit a more ramified astroglial domain with increased process arborization than tdTloweGFP+ astrocytes (yellow arrows, Figure 1b). We have previously developed a method to quantitatively measure astrocyte domain size, by converting the astrocytes into 3D surface images using Imaris software (Morel et al., 2014). We also showed that the expression levels of the tdT reporter have no significant effect in labeling the full morphology of astrocytes, following the comparison of dye-filled and tdT labeled astrocyte morphology (Morel et al., 2014). Using this approach, the ramification differences of tdTloweGFP+ and tdThigheGFP+ astrocytes were evidently illustrated (Figure 1b). Subsequent quantification of tdTloweGFP+ and tdThigheGFP+ astrocyte domains showed that the average domain size of tdThigheGFP+ astrocytes (19,326 ± 822, volume generally >10,000 μm3) is two times larger than that of tdTloweGFP+ astrocytes (9,987 ± 234, volume generally <10,000 μm3) (Figure 1c). To reveal the full morphology and quantitatively determine the domain size of tdT−eGFP+ astrocytes, we prepared cortical slices and dye-filled layer I tdT−eGFP+ astrocytes based on eGFP+ fluorescence. Although tdT−eGFP+ astrocytes show no detectable tdT fluorescence, their domain sizes are in a similar range (average domain size of 15,289 ± 1,430) as that of tdThigheGFP+ astrocytes. Our results are consistent with the idea that cortical astrocytes are morphologically diverse, and their morphology is not solely determined by their layer-specific localization (Lanjakornsiripan et al., 2018).
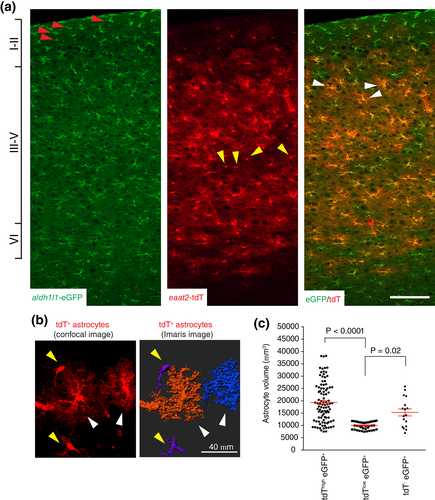
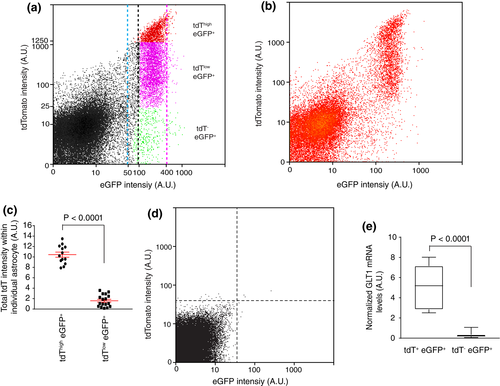
To quantitatively define cortical astrocyte subpopulations, we analyzed reporter expression and the relative distribution of these astrocyte subpopulations in the cortex of adult (P60–90) eaat2-tdT+ Bac aldh1l1-eGFP+ double transgenic mice using flow cytometry. The tdT−eGFP+ (in green, tdT A.U. < 25, Figure 2a), tdTloweGFP+ (in magenta, 25 < tdT A.U. < 1,250, Figure 2a), and tdThigheGFP+ (in red, tdT A.U. > 1,250, Figure 2a) cortical astrocytes were identified based on the drastic intensity differences of the tdT reporter. These subpopulations can be consistently gated based on the tdT intensity described above with minimal variability. These astrocyte subpopulations are distributed in an approximate 1:7:2 (tdT−eGFP+: tdTloweGFP+: tdThigheGFP+) ratio in the flow cytometry scatter plot (Figure 2a). We also analyzed a smaller number of cortical astrocytes to better illustrate the separated tdTloweGFP+ and tdThigheGFP+ subpopulations (Figure 2b). Meanwhile, the eGFP reporter fluorescence was generally constant with an intensity range of 100–400 A.U. (between black and magenta dashed lines, Figure 2a). We did observe a very small fraction of astrocytes with increased eGFP fluorescence (beyond the magenta dashed line, 400 < A.U. < 1,000), which were too few to be collected for analysis. It also appears that there might have a narrow band of cells with reduced eGFP levels (50 < A.U. < 100, between the blue and the black dashed lines, Figure 2a), however, these cells are intermingled with background debris fluorescence, making it less likely to be a distinct cell population. To confirm the differential expression levels of the tdT reporter in tdThigh and tdTlow astrocytes, we quantified the total tdT fluorescence intensity of typical tdThigh and tdTlow astrocytes (yellow and white arrows, Figure 1b) and indeed found drastically different total tdT intensity in tdThigh (mean intensity: 10.44 ± 0.49) and tdTlow astrocytes (mean intensity: 1.59 ± 0.28, Figure 2c). Importantly, the fold differences of the tdT fluorescence between these two populations of astrocytes quantified by either flow cytometry or Image J are comparable, further validating the intensity cut-off applied in the flow cytometry to define tdThigh and tdTlow astrocytes. In addition, only background auto-fluorescent signals from both tdT and eGFP channels (indicated by both dashed lines, generally with A.U. < 30) were observed in the cortex of wild-type control mice with the same gating cut-off, laser power, and equivalent number of cells analyzed (Figure 2d). It is intriguing that the expression of tdT is largely undetectable in the majority of layers I–II astrocytes as compared to cortical astrocytes in other layers, even though the tdT reporter is directed by the same human eaat2 promoter in all astrocytes. We then determined the GLT1 mRNA levels in tdT+eGFP+ and tdT−eGFP+ astrocytes isolated by FAC sorting from cortex of adult (P60–90) eaat2-tdT+ Bac aldh1l1-eGFP+ mice and found that GLT1 mRNA levels are drastically reduced (~5 times) in tdT−eGFP+ astrocytes as compared to tdT+eGFP+ (both tdThigh and tdTlow) astrocytes (Figure 2e). The significantly reduced GLT1 mRNA levels in tdT−eGFP+ astrocytes provide appealing evidence that the eaat2/glt1 promoter is less activated in these astrocytes than in tdT+eGFP+ astrocytes, suggesting that tdT−eGFP+ astrocytes represent a functionally distinct astrocyte subpopulation.
3.2 Whole-genome transcriptome analysis of cortical astroglial subpopulations
The observation that tdT− eGFP+ astrocytes are mainly localized in layer I–II and tdThigheGFP+ and tdTlow eGFP+ astrocytes are intermingled in other cortical layers makes it unfeasible to selectively isolate subpopulation-specific mRNAs using direct isolation approaches, such as translational ribosome affinity purification (TRAP) approach from these astrocytes. Although the cytoplasmic content can be extracted through the patching pipette for RNA-seq analysis after whole-cell patching using the “Patch-seq” technique (Cadwell et al., 2016), the much smaller astrocytic soma size (compared to neurons) makes it highly challenging to isolate the intracellular contents of astrocytic soma. A recent study physically dissected different cortical layers and then isolated layer-specific cortical astrocytes from Bac aldh1l1-eGFP mice (Lanjakornsiripan et al., 2018), which may introduce layer-specific bias due to the inconsistent separation of cortical layers. Alternatively, because these astrocyte subpopulations are differentially labeled with tdT or eGFP reporters, we isolated these subpopulations using fluorescence activated cell sorting (FACS) and then extracted total RNA for RNA-seq based transcriptome analysis. We first examined the specificity of RNA-seq datasets of astroglial subpopulations and found generally very low FPKM values (log2 FPKM <0) of representative genes from neurons, oligodendrocytes, microglia, and NG2 glial cells in these subpopulations, while FPKM values of known astrocyte genes, slc1a2, aqp4, aldh1l1, and gfap were generally high (Figure 3a) in all astroglial subpopulations, confirming that the RNA-seq datasets were specific to the astroglial transcriptome. We did observe that the expression levels of several nonastrocytic genes, such as mobp, iba1, cd11b, and pdgfrα, are elevated in tdT−eGFP+ astrocytes, indicated by higher FPKM values (Figure 3a), compared to tdT+eGFP+ astrocytes. We and others have previously demonstrated that the eGFP expression driven by the aldh1l promoter is specific to cortical astrocytes with no (neurons, microglia, and NG2 cells) or minimal (< 4%, oligodendrocytes) expression in other CNS cells (Cahoy et al., 2008; Yang et al., 2011). Therefore, the elevated expression levels of typical mRNA transcripts from multiple CNS cell types (microglia, oligodendrocytes, and NG2 cells) in tdT−eGFP+ astrocytes are unlikely to be a result of contamination during FACS. We further performed immunostaining of Iba1 and APC that specifically labels microglia and oligodendrocytes, respectively. Consistent with our previous results (Yang et al., 2011), very rare (< 2%) APC and no Iba1 staining signals (Figure 3b) were overlapped with eGFP+ cortical astrocytes.
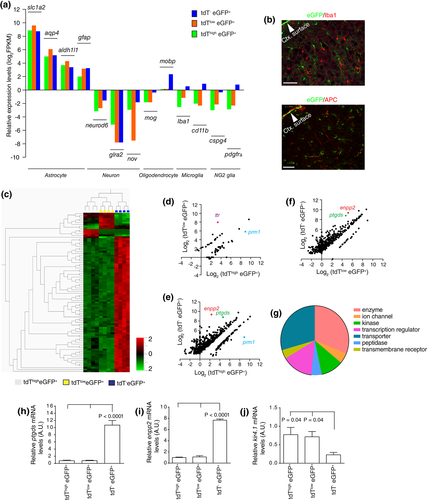
To identify unique mRNA expression profiles from different cortical astrocyte subpopulations, we performed multi-group cluster analysis with a stringent q value (FDR q < 0.001) and found that tdT− eGFP+ astrocytes showed a drastically different mRNA expression profile than that of tdThigh eGFP+ or tdTlow eGFP+ astrocytes (Figure 3c), clearly suggesting that this cortical subpopulation is molecularly unique. The representative top differentially expressed genes from each subpopulation are shown in Table 1. We next performed pairwise cuffdiff analysis of RNA-seq datasets to compare the transcriptome differences of any given two astroglial subpopulations. Consistently, we found only a small number (59) of genes that showed differential FPKM values (FC > 2, FDR q < 0.05) between tdTloweGFP+ and tdThigh eGFP+ astrocytes (Figure 3d, also in Supporting Information Table S1), among which only ttr and prm1 expression levels were dramatically different. In contrast, a large number of genes showed significantly different FPKM values (FC > 2, FDR q < 0.05) between tdT−eGFP+ and tdThigheGFP+ (528 genes, Figure 3e, also in Supporting Information Table S2), and between tdT−eGFP+ and tdTloweGFP+ astrocytes (480 genes, Figure 3f, also in Supporting Information Table S3), further suggesting that the molecular identity of tdTloweGFP+ and tdThigheGFP+ astrocytes is highly similar, but different from tdT−eGFP+ astrocytes. Interestingly, the vast majority (90%) of these genes show a reduced expression in either tdTloweGFP+ or tdThigheGFP+ astrocytes when compared to tdT−eGFP+ astrocytes. Particularly, expression levels of nonastrocyte glial marker genes, such as mog and mobp (oligodendrocytes), iba1 and cd11b (microglia), and pdgfrα (NG2 cells) in tdT−eGFP+ astrocytes are significantly higher than in tdTloweGFP+ and tdThigheGFP+ astrocytes (Figure 3a), suggesting a potential mixed glial lineage molecular signature in tdT−eGFP+ astrocytes.
| Gene symbol | tdT−eGFP+ | tdThigheGFP+ | Fold change |
|---|---|---|---|
| Cox8b | 119.386 | 0 | inf |
| Dynlrb2 | 25.3473 | 0 | inf |
| Fam183b | 15.106 | 0 | inf |
| Gm5480 | 43.1643 | 0 | inf |
| Tmem212 | 26.9697 | 0 | inf |
| Ttr | 5,413.74 | 10.2937 | 525.9275091 |
| Enpp2 | 659.926 | 5.01147 | 131.6831189 |
| Igf2 | 97.7155 | 1.24448 | 78.51914052 |
| Prm1 | 24.3464 | 488.855 | 0.049802907 |
| Gene symbol | tdT−eGFP+ | tdTloweGFP+ | Fold change |
| Eif2s3y | 24.2066 | 0 | inf |
| Gm5480 | 43.1643 | 0 | inf |
| Cd74 | 50.1016 | 2.20389 | 22.73325801 |
| Sostdc1 | 41.7345 | 1.84702 | 22.5955864 |
| Ttr | 5,413.74 | 248.832 | 21.75660687 |
| Igf2 | 97.7155 | 4.88869 | 19.98807451 |
| Enpp2 | 659.926 | 36.4721 | 18.09399514 |
| Apod | 108.46 | 6.09995 | 17.78047361 |
| Ptgds | 447.063 | 26.3646 | 16.95694226 |
| Rsph1 | 52.8741 | 3.15667 | 16.74996119 |
| Gene symbol | tdThigheGFP+ | tdTloweGFP+ | Fold change |
| Eif2s3y | 35.0507 | 0 | inf |
| Prm1 | 488.855 | 56.8842 | 8.593862619 |
| Ttr | 10.2937 | 248.832 | 0.041368072 |
- inf: infinity.
To determine functional pathways that are potentially distinct between tdT−eGFP+ and tdT+eGFP+ astrocytes, differentially expressed genes between tdT−eGFP+ and tdThigheGFP+ or between tdT−eGFP+ and tdTloweGFP+ were first overlapped and underwent functional pathway analysis using Ingenuity Pathway Analysis program (Qiagen). These genes are mostly clustered as transporters, enzymes, transcriptional regulators, and kinases (Figure 3g, detailed lists are shown in Supporting Information Table S4). We further performed qRT-PCR using RNA samples prepared from FAC sorted astrocyte subpopulation samples to specifically validate the expression differences of genes identified between tdT−eGFP+ and tdT+eGFP+, as tdThigheGFP+ and tdTloweGFP+ astrocytes share highly similar expression profiles (Figure 3c,d). The qRT-PCR confirmed that enpp2 and ptgds mRNA levels, two of the highly expressed genes in tdT−eGFP+ astrocytes and the most differentially expressed genes between tdT−eGFP+ and tdT+eGFP+ astrocytes based on RNA-seq data, are indeed highly elevated in tdT−eGFP+ astrocytes (Figure 3h,i). In addition, we also found that mRNA levels of the kir4.1 gene, one of the major inward-rectifying potassium channels that determine astrocyte membrane potential (Olsen et al., 2007), were significantly lower in FAC sorted tdT−eGFP+ astrocytes than in tdTloweGFP+ and tdThigh eGFP+ astrocytes (Figure 3j).
3.3 Physiological differences among cortical astroglial subpopulations
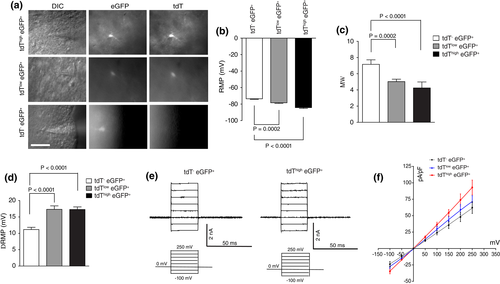
Encouraged by the molecular differences among cortical astroglial subpopulations, we next determined whether these astroglial subpopulations exhibit different physiological properties by performing astroglial recording on cortical slices (P21–28) of eaat2-tdT+ Bac aldh1l1-eGFP+ mice (Figure 4a). Interestingly, we found that the resting membrane potential (RMP) decreases gradually and significantly from tdT−eGFP+ (−73 ± 0.7 mV) to tdTloweGFP+ (−78.3 ± 0.8 mV, p < .001), and to tdThigheGFP+ astrocytes (−84.3 ± 0.9 mV, p < .001, Figure 4b). In addition, the membrane resistance of tdT−eGFP+ astrocytes (7.2 ± 0.6 MΩ) was significantly higher than that of tdTlow eGFP+ (5.0 ± 0.3 MΩ) and tdThigh eGFP+ astrocytes (4.3 ± 0.2 MΩ, Figure 4c). Because the resting membrane potential (RMP) of a mature astrocyte is primarily determined by the activity of inward-rectifying potassium channel 4.1 (Kir4.1; Olsen & Sontheimer, 2008), we examined the functional differences in Kir4.1 activity on these astrocytes. Interestingly, the application of BaCl2 (100 μM), a well-established inhibitor of Kir4.1, on cortical slices resulted in only a moderate depolarization of tdT−eGFP+ astrocytes (11.2 ± 0.7 mV) (Figure 4d). By comparison, BaCl2 application had a much stronger effect on both tdTloweGFP+ and tdThigh eGFP+ astrocytes (17.3 ± 1.1 or 0.8 mV, respectively, p < .001, Figure 4d), indicating that tdTloweGFP+ and tdThigh eGFP+ astrocytes express significantly larger inward Kir-mediated current, presumably as a result of higher Kir4.1 expression levels. Because the expression of Kir4.1 and K+ buffering is one of the central functions of mature astrocytes, the significantly smaller inward Kir-mediated current in tdT− eGFP+ astrocytes suggests that these astrocytes are likely to have a reduced buffering capacity for extracellular K+ than tdTlow eGFP+ and tdThigh eGFP+ astrocytes. The reduced inward Kir-mediated current results are also consistent with our observation above that kir4.1 mRNA levels are significantly lower in tdT−eGFP+ astrocytes than in tdTlow eGFP+ and tdThigh eGFP+ astrocytes. Moreover, we generated the I–V plot using capacitance (size)-normalized recorded currents as a function of voltage in Figure 4f. Representative current recording tracers for tdT−eGFP+ and tdThigheGFP+ astrocytes (tdTloweGFP+ astrocytes are similar to tdThigheGFP+ astrocytes) in response to stepwise increased voltage are shown in Figure 4e. Although current amplitude was slightly larger in tdThigh eGFP+ astrocytes than tdT−eGFP+ astrocytes, all three astroglial subpopulations demonstrated passive membrane currents typical of astrocytes with the reversal potential close to the K+ equilibrium potential (~ − 80 mV), confirming that they all carry typical astroglial membrane properties.
4 DISCUSSION
Our current study identified astrocyte subpopulations from the adult cortex based on the expression patterns of tdT and eGFP reporters from two distinct astrocyte reporter mouse lines and analyzed the functional and molecular differences among these cortical astrocyte subpopulations with a combined FACS/RNA-Seq approach. Particularly, the tdT−eGFP+ astrocytes appear to be mostly localized in cortical layers I–II with distinct physiological and molecular profiles from astrocytes in other cortical layers. Previous studies have shown that layer I cortical astrocytes tend to be better labeled with GFAP than other cortical astrocytes (Oberheim et al., 2009). However, it remains largely unexplored whether the physiological and molecular properties of layer I astrocytes are different from other cortical astrocytes. During the revision of this article, another study also investigated cortical astrocyte heterogeneity and found layer-specific molecular and morphological diversity of cortical astrocytes (Lanjakornsiripan et al., 2018).
Our results showed that tdT−eGFP+ astrocytes have significantly reduced functional expression of Kir4.1, accompanied by an increased resting membrane potential. The molecular profile of tdT−eGFP+, but not tdT+eGFP+ astrocytes, also showed elevated levels of a number of mRNAs including a few specific to multiple nonastrocyte glial cells (Figure 3a), suggesting a less-specific molecular identity for this subpopulation of adult cortical astrocytes. We previously showed that the eGFP reporter driven by the aldh1l1 promoter is selectively expressed in astrocytes with no or minimal expression in other glial cells (Yang et al., 2011). Additionally, all astrocyte subpopulations are sorted together from same cortical tissues. It is therefore unlikely that the elevated levels of other glial mRNAs—only observed in tdT−eGFP+ astrocytes—are a result of potential contamination from other glial cells during FACS; rather, the elevated levels of other glial gene mRNAs in tdT−eGFP+ astrocytes are likely to reflect the intrinsic molecular differences of tdT−eGFP+ astrocytes from tdT+eGFP+ astrocytes. Consistently, a number of genes that encode enzymes, transcriptional regulators, transporters, and kinases also show significantly increased expression in tdT−eGFP+ astrocytes (Supporting Information Table S4), also supporting the finding that the molecular profile of tdT−eGFP+ astrocytes is drastically different from that of tdT+eGFP+ astrocytes. Layer-specific molecular profiles of cortical astrocytes have also been observed in another recent study (Lanjakornsiripan et al., 2018). The high expression levels of these functional genes help shed new light on the unique functions of these layers I–II astrocytes in CNS development and physiology. Among these genes, enpp2 (encoding ectonucleotide pyrophosphatase) and ptgds (encoding Prostaglandin D2 Synthase) are two top genes with much higher FPKM values in tdT−eGFP+ astrocytes (660 and 447, respectively) than in tdThigh eGFP+ (5 and 25 respectively) and tdTloweGFP+ (36 and 26, respectively) astrocytes. ENPP2 (also named autotaxin) is a key secreted glycoprotein for the synthesis of lysophosphatidic acid (LPA), which promotes cell growth, survival, and differentiation for the development of the CNS (Moolenaar, Houben, Lee, & van Meeteren, 2013). PTGDS is the key enzyme for generating prostaglandin D2 (PGD2), a potent neuromodulator especially involved in sleep regulation (Urade & Hayaishi, 2011) and peripheral nervous system (PNS) myelination (Trimarco et al., 2014).
These molecular and functional differences between tdT−eGFP+ (mostly layers I–II) and tdT+eGFP+ astrocytes can be attributed to the cell fate timing and progenitor source differences during development. Layer I cortical astrocytes are generated as early as E12 in rodents (Tabata, 2015), significantly earlier than cortical astrocytes in other layers, which are typically born at E17 or early postnatally in rodents (Freeman, 2010). Because of their early generation at a time when the SVZ has not yet been formed, layer I astrocytes are generated at the basal lamina from VZ-derived progenitor cells (Tabata, 2015). Cortical astrocytes in other layers are mostly generated from initial SVZ-derived radial glia and massively produced later through a unique local proliferation from newly born immature astrocytes during early postnatal development (Ge et al., 2012). Notably, SVZ-derived radial glia have also been reported to contribute to the generation of layer I astrocytes in later embryogenesis (Tabata, 2015). In addition to the intrinsic differences of timing and progenitor source, localized external environment, especially localized neuronal signals, may differentially affect the development and functions of upper- and deeper-layer astrocytes. It is known that neuronal density is substantially lower in cortical layer I than in deep layers (Larkum, 2013; Muralidhar et al., 2013). The layer I neurons are also mostly GABAergic inhibitory neurons (Larkum, 2013; Muralidhar et al., 2013), unlike the predominantly glutamatergic neuronal populations in deep layers. Glutamatergic neuronal activity significantly regulates expression of functional peri-synaptic astroglial proteins (Morel et al., 2014; Yang et al., 2009) and the motility of astroglial processes (Theodosis, Poulain, & Oliet, 2008). We previously also showed that selective loss of VGluT1 neuronal glutamatergic signaling selectively impedes the morphological and molecular maturation of cortical astrocytes (Morel et al., 2014). Consistently, we observed that layer I adult cortical astrocytes exhibit properties of potentially immature astrocytes, such as reduced functional Kir4.1 with increased resting membrane potential and elevated expression of a number of genes including nonastrocytic glial genes, even though the cortical tissues from which astrocytes were molecularly profiled were from adult mice (P60). It is therefore conceivable that the lack of glutamatergic neuronal signaling in cortical layer I contributes to these unique immature properties of layer I astrocytes, even at the adult stage. The role of neuronal signals in directing localized astrocyte molecular and morphological properties is further demonstrated by the selective deletion of Dab1 in cortical neurons, which disrupts layer-specific neuronal migration during early development (Lanjakornsiripan et al., 2018). It is also likely that there are unique intracellular programs governing the molecular signature and functional properties of layer I astrocytes. Our datasets of the transcriptional regulators from tdT−eGFP+ and tdT+eGFP+ astrocytes may provide useful clues to understand potential intracellular programs and will be investigated in the future.
ACKNOWLEDGMENTS
We thank the Tufts Center for Neuroscience Research, Tufts FACS facility, and Tufts Genomics core facility for imaging, cell sorting, and sequencing support. We thank Rachel Jarvis for proofreading the manuscript. This work was supported by National Institutes of Health (R01MH099554, R01MH106490, and R21NS087391 to Y.Y.).
CONFLICT OF INTEREST
All authors declare no conflict of interest.




