Comparison of genetic risk prediction models to improve prediction of coronary heart disease in two large cohorts of the MONICA/KORA study
Abstract
It is still unclear how genetic information, provided as single-nucleotide polymorphisms (SNPs), can be most effectively integrated into risk prediction models for coronary heart disease (CHD) to add significant predictive value beyond clinical risk models. For the present study, a population-based case-cohort was used as a trainingset (451 incident cases, 1488 noncases) and an independent cohort as testset (160 incident cases, 2749 noncases). The following strategies to quantify genetic information were compared: A weighted genetic risk score including Metabochip SNPs associated with CHD in the literature (GRSMetabo); selection of the most predictive SNPs among these literature-confirmed variants using priority-Lasso (PLMetabo); validation of two comprehensive polygenic risk scores: GRSGola based on Metabochip data, and GRSKhera (available in the testset only) based on cross-validated genome-wide genotyping data. We used Cox regression to assess associations with incident CHD. C-index, category-free net reclassification index (cfNRI) and relative integrated discrimination improvement (IDIrel) were used to quantify the predictive performance of genetic information beyond Framingham risk score variables. In contrast to GRSMetabo and PLMetabo, GRSGola significantly improved the prediction (delta C-index [95% confidence interval]: 0.0087 [0.0044, 0.0130]; IDIrel: 0.0509 [0.0131, 0.0894]; cfNRI improved only in cases: 0.1761 [0.0253, 0.3219]). GRSKhera yielded slightly worse prediction results than GRSGola.
1 INTRODUCTION
Coronary heart disease (CHD) is the leading cause of death worldwide (Global Health Estimates 2016, 2018). People at high risk for CHD can benefit from lifestyle or pharmacologic interventions, even if the risk is of genetic origin (Abraham et al., 2016; Damask et al., 2020; Khera et al., 2016; Mega et al., 2015). Therefore, reliable prediction models are needed. Genetic information may add predictive value to established clinical risk models like the Framingham risk score (FRS) (D'Agostino et al., 2008). In the last few years, high-throughput genotyping has become more and more cost-effective (Schwarze et al., 2018; Wetterstrand, 2020). Genetic prediction modeling has several advantages compared to prediction based on blood biomarkers, such as proteins and metabolites (Herder, Karakas, et al., 2011). Genotyping needs to be carried out only once in a lifetime and its results can be used to determine the risk of various diseases. Moreover, risk assessment can be performed early in life, when lifestyle changes are likely to have the greatest impact.
The field is evolving rapidly and there is an ongoing debate on how genetic information, provided as single-nucleotide polymorphisms (SNPs), can be most effectively integrated into predictive models for complex diseases like CHD. The genetic information can be integrated in the form of (weighted) genetic risk scores (GRS) consisting of preselected variants (Antiochos et al., 2016; Beaney et al., 2017; Elliott et al., 2020; Inouye et al., 2018; Iribarren et al., 2016; R. W. Morris et al., 2016; Paynter et al., 2010; Said et al., 2018; Tada et al., 2016; de Vries et al., 2015). Machine Learning approaches without score formation are alternatively applied (Boulesteix et al., 2020; Dogan et al., 2018; Gola et al., 2020; Okser et al., 2014). Apart from the method of inclusion, studies vary regarding the utilized genotyping tool. Predictive SNPs may stem from genome-wide SNP array data or specialized genotyping arrays such as the Metabochip, which focuses on SNPs associated with cardiometabolic outcomes (Voight et al., 2012).
Until recently, genetic risk prediction studies on complex diseases focused on variants identified as significantly associated in genome-wide association studies (GWAS) or candidate gene association studies (Müller et al., 2016). Studies on cardiovascular disease-related events included only a few literature-based confirmed SNPs in their analyses to investigate the predictive value of genetic information in addition to established risk prediction models (Antiochos et al., 2016; Beaney et al., 2017; Iribarren et al., 2016; R. W. Morris et al., 2016; Paynter et al., 2010; Said et al., 2018; Tada et al., 2016; de Vries et al., 2015). More recent studies showed that taking into consideration a larger number of SNPs tends to give more accurate prediction results beyond established risk factors than including only genome-wide significant SNPs (Abraham et al., 2016; Elliott et al., 2020; Inouye et al., 2018; Mars et al., 2020). Khera et al. (2018) derived a polygenic risk score for coronary artery disease, based on genome-wide SNP array data. It has been repeatedly evaluated in several different studies and can therefore be regarded as a benchmark in coronary artery disease risk prediction (Aragam et al., 2020; Dikilitas et al., 2020; Hindy et al., 2020; Mosley et al., 2020; Wünnemann et al., 2019). Another comprehensive polygenic risk score previously derived by Gola et al. (2020) based on Metabochip data, was reported to yield even better prediction results. However, the Gola et al. (2020) results lack sufficient external investigation and comparison.
The aim of the present study is to compare the ability of different genetic risk quantification approaches to improve the accuracy of CHD prediction beyond the established FRS variables, including the recently published Gola score.
2 METHODS
2.1 Study populations
We used data from a prospective case-cohort study incorporated within the population-based Monitoring of Trends and Determinants in Cardiovascular Diseases/Cooperative Health Research in the Region of Augsburg (MONICA/KORA) study as a training data set. This study was initiated in the early 1980s by the World Health Organization. Details of design and procedures of this study have been described elsewhere (Herder, Baumert, et al., 2011; Koenig et al., 2006; Thorand et al., 2005; Tunstall-Pedoe & WHO MONICA Project Principal Investigators, 1988). Briefly, for the original cohort study, three independent cross-sectional population-based surveys were conducted in the years 1984/1985 (Survey S1), 1989/1990 (S2), and 1994/1995 (S3) in Augsburg and two adjacent counties in Southern Germany. In total 13,427 individuals (6725 men and 6702 women) aged 25–64 (S1) or 25–74 years (S2, S3) participated at baseline.
As test data set, we used data from the population-based KORA S4 cohort study. The cross-sectional baseline survey was performed in 1999–2001 in the same study region as the MONICA/KORA S1–S3 cohort study. In total 4261 individuals (2090 men and 2171 women) aged 25–74 years participated at baseline (Ward-Caviness et al., 2017).
Data on sociodemographic and lifestyle variables were collected through standardized interviews in both studies. Data on medication use was assessed based on the packaging of all medication, which the study participants had taken during the last week before the investigation and brought along due to request in advance. In addition, standardized medical examinations were carried out and blood samples were collected (Holle et al., 2005). All participants were prospectively followed within the KORA framework.
Because of the low incidence of CHD in those younger than 35 years, we restricted the source populations of the present studies to persons between 35 and 74 years at baseline. Further exclusion criteria consisted of missing blood samples, withdrawn consent, double examinations and prevalent CHD, yielding two cohorts of interest comprising 9299 persons in the MONICA/KORA S1–S3 cohort study and 3289 persons in the KORA S4 cohort study. Because participants in S3 were followed for a maximum of 14–15 years, we limited the follow-up time of both studies to 14 years. After the restriction of follow-up time, a case-cohort study was formed in the surveys S1–S3. The case-cohort design of the present study has been described in detail before (Huth et al., 2020; Thorand et al., 2020). Briefly, a random sample subcohort of 2163 participants (1154 men and 1009 women) was drawn from the cohort of interest, stratified by sex and survey. The subcohort was extended by all further cases occurring within 14 years in the cohort of interest. In the following, we excluded persons with missing values, persons without genotyping informed consent and persons who failed preimputation genotype quality control (QC) in both studies. The finally analyzed MONICA/KORA S1–S3 case-cohort study consisted of 451 incident CHD cases and 1488 noncases (“MONICA/KORA trainingset”; median follow-up time: 14.0, 25th percentile: 10.3, 75th percentile: 14.0 years). The KORA S4 cohort study included 160 incident CHD cases and 2749 noncases (“KORA testset”; median follow-up time: 14.0, 25th percentile: 14.0, 75th percentile: 14.0). All study individuals are German residents and carry a German passport. For details on the selection of participants see Figure 1.
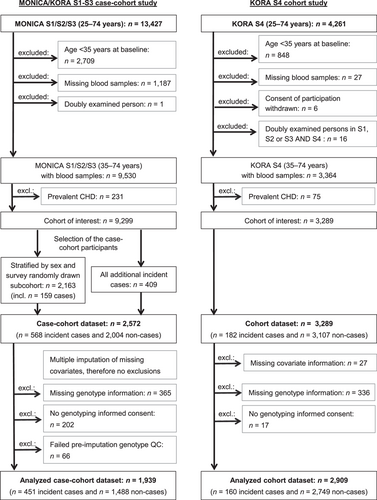
Written informed consent was given by all study participants, the study protocols were approved by the Ethics Committee of the Bavarian Chamber of Physicians. The studies were carried out in accordance with the Declaration of Helsinki.
2.2 Assessment of prevalent and incident CHD
The outcome CHD was a combined endpoint of nonfatal myocardial infarction as well as coronary death and sudden death (International Classification of Disease 9th Revision: 410–414 and 798).
Until December 2000, the diagnosis of a major, nonfatal myocardial infarction and coronary death was based on the MONICA algorithm (H. Tunstall-Pedoe et al., 1994) in which a diagnosis of a major CHD event was based on symptoms, cardiac enzymes (creatine kinase, aspartate aminotransferase, and lactate dehydrogenase), serial changes from 12-lead electrocardiograms (ECGs) evaluated by Minnesota coding (Prineas et al., 1982), necropsy results and history of CHD in fatal cases. Since January 1, 2001, the diagnosis of myocardial infarction was based on the European Society of Cardiology and American College of Cardiology criteria (Alpert et al., 2000).
Incident events were identified through follow-up questionnaires or through the MONICA/KORA myocardial infarction registry, which monitors the occurrence of all in- and out of-hospital fatal and nonfatal myocardial infarctions among the 25–74-year-old inhabitants of the study region (Löwel et al., 1991). Follow-up questionnaires were sent to all MONICA/KORA S1–S3 study participants in 1997/1998, 2002/2003, and 2008/2009 and to the KORA S4 study participants in 2008/2009 and 2016. The MONICA/KORA myocardial infarction registry covers the study region only. Therefore, subjects who had moved outside the study area reported incident events through follow-up questionnaires sent to their new addresses, which were traced by address research. Additionally, all participants from S1 were invited to participate in a follow-up examination in 1987/1988. For the KORA S4 study participants, two follow-up examinations were conducted in 2006–2008 and 2013/2014. Initially identified self-reported incident cases and the self-reported date of diagnosis not covered by the MONICA/KORA myocardial infarction registry, were validated by hospital records or by contacting the patient's treating physician. Deaths from myocardial infarction were validated by death certificates, autopsy reports, chart reviews, or information from the last treating physician.
2.3 Genotyping and assessment of risk score variables used for benchmarking
Details on genotyping, genetic QC, and genetic imputation can be found in Supporting Information Methods, Table S1, and Figure S1.
Genetic information was added to two reference clinical models: (1) age, sex, and survey; (2) sex, survey, and the FRS variables according to D'Agostino et al. (2008) with a minor modification. The original FRS contains the variables age, total cholesterol, high-density lipoprotein (HDL) cholesterol, systolic blood pressure, antihypertensive medication use, diabetes status, and current smoking. All metric variables, except for age, were naturally logarithmically (ln) transformed and (0,1)-standardized. Prevalent diabetes at baseline was defined as any type of self-reported diabetes that could be validated by the responsible physician or medical chart review or as current self-reported use of glucose-lowering medication. As a minor modification, we included former smokers in addition to current smokers as a separate category and used never smokers as the reference category. As we determined the combined hazard for men and women, we additionally included sex as a predictor variable, as well as a survey indicator, to account for the study design. During baseline examinations, a venous blood sample was collected while sitting. Samples were centrifuged within 120 min, refrigerated at 4–8°C and shipped on refrigerant packaging within 2–4 h to the laboratory of the Augsburg Central Hospital (now university hospital of Augsburg) for measurement of serum HDL cholesterol and total cholesterol (routine enzymatic method cholesterol oxidase phenol 4-aminoantipyrine peroxidase; Boehringer Mannheim [Roche from 1997 on]). HDL cholesterol, total cholesterol as well as several other collected biomarkers and variables (not used in the present analyses) had some missing values in the MONICA/KORA S1–S3 case-cohort study (HDL and total cholesterol: 0.04%). Therefore, to enable unbiased analyses and to use the available data as efficiently as possible, we replaced missing values using 20-fold multiple imputations by chained equations (MICE) (R version 3.2.3 and R package MICE version 2.25 [van Buuren & Groothuis-Oudshoorn, 2011; van Buuren et al., 1999]). As there was only a small number of missing values in the KORA S4 cohort study, we excluded persons with missing values from the KORA testset (Figure 1).
2.4 Compared genetic models—Main analyses
The prediction performance of all main analysis models was evaluated in the KORA testset based on regression coefficients (ln-transformed hazard ratios) estimated in the MONICA/KORA trainingset (Figure 2). We compared three different approaches to take genetic information into account. The first two were based on all Metabochip SNPs published to be genome-wide significantly (p < 5 × 10−8) associated with CHD. These were extracted from the literature received by a systematic Pubmed literature search on January 21, 2019, using the terms (metabochip OR cardiochip OR cardiometabochip). In the first approach, we used these SNPs to build a weighted GRS (GRSMetabo). In the second approach, we selected the most predictive SNPs from the identified genome-wide significant variants using a newly developed method called priority-Lasso, where reference model variables were prioritized over the SNP variables (PLMetabo) (Klau et al., 2017, 2018). Prediction included the unpenalized regression coefficients from the reference variables together with the shrunk regression coefficients from the selected SNPs. In the third approach, we evaluated a weighted GRS published by Gola et al. (2020) (GRSGola). Briefly, Gola et al. (2020) performed a random forest approach to preselect variants with corrected Gini importance greater 0. This was followed by a hyperparameter tuning to evaluate the optimal number of SNPs and the necessity of weights. Metabochip data imputed with the Haplotype Reference Consortium reference panel were used. Their final score comprised 50,633 weighted SNPs. The authors of the Gola et al. (2020) publication provided us with variant names, positions, chromosome numbers, weights, weighted alleles, and non-weighted alleles of these 50,633 SNPs, which we used to form GRSGola in the MONICA/KORA trainingset and the KORA testset.
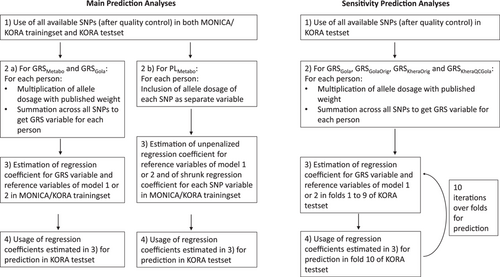
We added the genetic information of all three approaches to the two reference models. Regression coefficients of reference models alone and with the addition of PLMetabo, GRSMetabo, and GRSGola were calculated in the MONICA/KORA trainingset and the predictive value of each model was evaluated in the KORA testset. GRSMetabo and GRSGola were also evaluated without additional variables (crude models). Priority-Lasso regression is not possible without reference variables of first priority. Therefore no crude PLMetabo model was estimated.
Only SNPs available in both the MONICA/KORA trainingset and the KORA testset were used. We excluded SNPs with Hardy–Weinberg equilibrium (HWE) test p < 10−20 and minor allele frequency (MAF) < 1% as in Gola et al. (2020). We further excluded SNPs with imputation quality R² < 0.3, which was not as strict as in Gola et al. (2020), where a threshold of 0.8 was chosen, to keep as much genetic information as possible (Table S2).
2.5 Compared genetic models—Sensitivity analyses
Four sensitivity analyses were carried out via 10-fold cross-validation in the KORA testset only (Figure 2 and Table S2).
- (1)
To verify that the genetic data quality of the MONICA/KORA trainingset was sufficient, GRSGola was evaluated in the KORA testset, according to the same protocol as in the main analysis.
- (2)
Furthermore, to recreate the original risk score by Gola et al. (2020) as closely as possible, we used all available SNPs in the KORA testset, including also those which were unavailable in the MONICA/KORA trainingset, after exclusion of SNPs with HWE test p < 10−20, MAF < 1%, and imputation quality R² < 0.8 (termed GRSGolaOrig).
- (3)
To take full advantage of the more comprehensive genetic data in the KORA testset and to compare the performance of GRSGola to an established benchmark GRS, we additionally built the weighted GRS, developed by Khera et al. (2018). This score was derived from a GWAS and two genome-wide genotyped validation studies, imputed with 1000 g reference panel. It includes 6,630,150 genome-wide distributed SNPs. We built this GRS variable after applying two different QC schemes. On the one hand, we used only imputation quality R² < 0.3 as an exclusion criterion, as in the original publication by Khera et al. (2018) (termed GRSKheraOrig).
- (4)
On the other hand, we applied the same criteria as for GRSGolaOrig, namely HWE p < 10−20, MAF < 1%, and imputation quality R² < 0.8 (termed GRSKheraQCGola).
All GRS variables were added separately to the variables of the two reference models used for the main analyses. Crude models were also evaluated.
2.6 Statistical methods
Descriptive analyses of baseline characteristics were computed separately for cases and noncases. For the MONICA/KORA trainingset, weighting was incorporated using the survey- and sex-specific sampling weights to account for the case-cohort design. Multiple results of the 20 imputations were summed up as medians for percentiles and arithmetic means for proportions. Descriptive analyses for the MONICA/KORA trainingset were calculated using SAS (Version 9.4; SAS Institute Inc).
To assess the association of risk factor variables with incident CHD, Cox proportional hazard regression was applied. To take the case-cohort design into consideration, regression coefficients (ln-transformed hazard ratios) in the MONICA/KORA trainingset were calculated using Barlow's weighting method (Barlow et al., 1999). Standard error estimates for the regression coefficients in the MONICA/KORA trainingset were obtained from robust variance estimation (Barlow, 1994). Additional variation due to imputation was incorporated using Rubin's rules for Model 2 (Rubin, 1987). There were no missing values in the Model 1 variables, which had therefore not been imputed.
Priority-Lasso, which was used for the selection of predictive SNP variables, was extended by Barlow's weighting method to account for the case-cohort design of the MONICA/KORA trainingset. Details of priority-Lasso have previously been described in Klau et al. (2018). Briefly, priority-Lasso is an enhancement of the standard Lasso method with a fixed block structure of the variables. Each variable belongs to exactly one block and each of these blocks is assigned a priority level. Unpenalized Cox regression is applied to all variables of the first block. Subsequently, a Lasso regression is conducted among all variables of the second block, using the linear predictor of the first step as offset. Through this method, variables from blocks of lower priority are only included in the model, if they explain variability that was not explained by blocks of higher priority. We assigned the variables of our reference models the first priority (Block 1) and all SNP variables the second priority (Block 2). The R package priority-Lasso (Klau et al., 2017) was used after having been adapted to the case-cohort design, which includes also an adapted glmnet version. The glmnet version can be downloaded from https://github.com/mu4bu2/glmnet. The tuning parameter lambda was chosen via 10-fold cross-validation in the MONICA/KORA trainingset. We used the default cross-validation procedure as described in Simon et al. (2011). Briefly, we evaluated 100 different lambda values, with λmin and λmax defined as in Simon et al. (2011). Deviance, which uses the partial-likelihood for the Cox model, was used as the loss function. That lambda resulting in the minimal mean cross-validated prediction error was selected as the final tuning parameter and used in the complete MONICA/KORA trainingset for priority-Lasso estimations.
To take the uncertainty of genetic imputation into account, instead of best-guess genotypes (“hard calls”), the estimated alternate allele dosage for each SNP was used and treated as metric quantity. For priority-Lasso we included the (0,1)-standardized dosage of each SNP as a separate variable. For the construction of all GRS (GRSMetabo, GRSGola, GRSGolaOrig, GRSKheraOrig, GRSKheraQCGola), dosages were adapted to the published regression coefficients (two-dosage, if the regression coefficient referred to the other allele). The GRS variables were built by multiplying the adapted dosage with the published regression coefficient (ln-transformed odds ratio) and by summing across all SNPs, followed by (0,1)-standardization. Khera et al. (2018) published positive regression coefficients and therefore solely risk alleles were weighted. All other published regression coefficients used in the present analyses were a mixture of positive and negative values (Deloukas et al., 2013; Gola et al., 2020; Howson et al., 2017; van der Harst & Verweij, 2018).
The added predictive value of the genetic information was evaluated by three performance measures: (1) Harrel's concordance index (C-index) and the difference between the reference and the genetically extended C-indices (delta C-index) (Harrell et al., 1982); (2) the category-free net reclassification index (Pencina et al., 2011) calculated overall (cfNRI) and separately for cases (cfNRIcases) and controls (cfNRIcontr); (3) the absolute integrated discrimination improvement (IDI) and additionally the relative IDI (IDIrel), as the IDI depends on the incidence of the outcome (Pencina et al., 2010).
The performance measures in the KORA testset were calculated from the regression coefficients derived in the MONICA/KORA trainingset. As there were no missing values in the reference model 1 variables, the linear predictor was built as usual. The regression coefficients of Model 2 alone and with extension of GRSMetabo or GRSGola were averaged over the 20 imputations. Confidence intervals (CIs) were calculated with standard errors of an approximate normal-distribution for the C-index using the R package compareC (Kang et al., 2015) and via 1000-fold percentile bootstrapping for cfNRI and IDI using the R packages nricens and boot (Canty & Ripley, 2020; Davison & Hinkley, 1997; Inoue, 2018); for the PLMetabo—extended reference model 2, it was not possible to take the averaged regression coefficients due to different tuning parameters in the 20 imputations. Therefore, each performance measure was calculated separately in each imputation and the results were combined using Rubin's Rules (Rubin, 1987).
In the cross-validation of the sensitivity analyses, the arithmetic mean of all performance measures was calculated over the 10 folds. The CIs of the mean performance measures were calculated via 1000-fold percentile bootstrapping using the R package boot (Canty & Ripley, 2020; Davison & Hinkley, 1997).
Test results with two-sided p < 0.05 were considered statistically significant and all statistical analyses were performed with R version 4.0.0 unless specified otherwise.
3 RESULTS
3.1 Baseline characteristics
Table 1 shows baseline characteristics of the 451 incident CHD cases and 1488 noncases of the MONICA/KORA trainingset and the 160 incident CHD cases and 2749 noncases of the KORA testset. In both studies, the cases comprised more men than women. They were on average older and had a higher systolic blood pressure despite a more frequent use of antihypertensive medication. They were more likely to have diabetes at baseline and to be current or former smokers. Furthermore, the cases had higher levels of total cholesterol and lower levels of HDL cholesterol compared to noncases. Medians of GRSGola and GRSGolaOrig were substantially higher in cases than in noncases. There were only slight differences in GRSMetabo, GRSKheraOrig, and GRSKheraQCGola.
| MONICA/KORA trainingseta | KORA testset | |||
|---|---|---|---|---|
| Characteristics | CHD cases n = 451 | Noncases n = 1488 | CHD cases n = 160 | Noncases n = 2749 |
| Male (%) | 73.4 | 46.9 | 75.6 | 46.5 |
| Age (years) | 62 (55, 68) | 51 (43, 60) | 64 (54, 69) | 53 (44, 63) |
| Survey 1 (%) | 21.3 | 28.8 | – | – |
| Survey 2 (%) | 43.7 | 36.3 | – | – |
| Survey 3 (%) | 35.0 | 35.0 | – | – |
| Survey 4 (%) | – | – | 100.0 | 100.0 |
| Framingham risk score variables | ||||
| Prevalent diabetes mellitus at baseline (%) | 17.7 | 4.3 | 20.6 | 4.6 |
| Systolic blood pressure (mmHg) | 144.0 (130.0, 158.0) | 130.0 (120.0, 143.0) | 140.0 (126.4, 152.2) | 128.5 (117.0, 141.5) |
| Antihypertensive medication use (%) | 37.9 | 14.2 | 41.3 | 19.7 |
| Total cholesterol (mg/dl) | 253.1 (223.3, 286.0) | 233.0 (207.0, 265.9) | 238.7 (212.4, 265.7) | 229.7 (203.5, 260.2) |
| HDL cholesterol (mg/dl) | 48.0 (40.5, 58.4) | 55.3 (45.7, 67.3) | 50.0 (41.8, 61.7) | 56.2 (45.7, 68.7) |
| Smoking | ||||
| Former (%) | 36.8 | 27.3 | 36.3 | 33.5 |
| Current (%) | 30.8 | 23.1 | 28.8 | 22.7 |
| Never (%) | 32.4 | 49.6 | 35.0 | 43.8 |
| Genetic risk scores | ||||
| GRSMetabo | 0.26 (−0.13, 0.68) | 0.12 (−0.24, 0.54) | 0.22 (−0.21, 0.59) | 0.16 (−0.24, 0.54) |
| GRSGola | 4.45 (−0.10, 8.60) | 3.15 (−0.67, 6.97) | 2.85 (−2.95, 8.04) | 0.30 (−5.22, 5.86) |
| GRSGolaOrig | – | – | 1.42 (−5.41, 6.58) | −1.71 (−8.14, 4.60) |
| GRSKheraOrig | – | – | 17.60 (17.54, 17.67) | 17.58 (17.52, 17.64) |
| GRSKheraQCGola | – | – | 13.85 (13.79, 13.91) | 13.83 (13.77, 13.88) |
- Note: Categorical variables are presented as proportions and continuous variables as medians (25th; 75th percentile).
- Abbreviations: CHD, coronary heart disease; GRS, genetic risk score; HDL, high-density lipoprotein.
- a For the MONICA/KORA trainingset, weighting was incorporated using the survey- and sex-specific sampling weights to account for the case-cohort design.
3.2 Literature search for Metabochip SNPs associated with CHD
Our literature search yielded three studies reporting genome-wide significant association results between Metabochip SNPs and CHD. These were Deloukas et al. (2013), Howson et al. (2017), and van der Harst and Verweij (2018) who conducted large-scale, consecutive association studies based only or mainly on Metabochip data. All three studies were extensions of the largest GWAS meta-analysis at that time, performed by the Coronary ARtery DIsease Genomewide Replication and Meta-analysis (CARDIoGRAM) Consortium (Schunkert et al., 2011). We included all Metabochip SNPs that were reported to be genome-wide significantly associated with CHD either directly in one of the three studies, or in cited studies. The 138 identified SNPs were then used for selection in the priority-Lasso analysis as well as for building GRSMetabo, choosing the most recent published association estimate as weight, as the more recent studies had the larger sample sizes. The 138 extracted SNPs are listed with their weight (ln[odds ratio]) in Table S3.
3.3 Main analyses
The hazard ratios of the reference models, calculated in the MONICA/KORA trainingset and used for the prediction analyses in the KORA testset, are shown in Table S4. Except for survey and former smoking, all variables were significantly associated with incident CHD.
In the PLMetabo models, 10 SNPs were selected on top of Model 1; 7 of these also on top of Model 2 (Table S5).
The GRSMetabo was associated with incident CHD in the crude model (hazard ratio [95% CI] per SD: 1.2341 [1.1137, 1.3676]) and in Model 1 (1.2126 [1.0766, 1.3659]) but not in Model 2 (1.1300 [0.9989, 1.2784]); Table S6). The GRSGola was associated with CHD in all three models (crude: 1.2516 [1.1203, 1.3982]; Model 1: 1.2297 [1.0804, 1.3996]; Model 2: 1.1580 [1.0109, 1.3265]; Table S6).
The prediction performance measures in the KORA testset are presented in Tables 2 and S7. In the crude models, the GRSGola yielded a C-index of 0.5878 [95% CI: 0.5446, 0.6310] and thus performed better than the GRSMetabo (C-index: 0.5253 [0.4803, 0.5702]). Generally, the C-index is a measure to determine the power of a model to discriminate between cases and noncases based on predicted survival probabilities. A C-index of 0.5 indicates that the model is not able to distinguish between cases and noncases at all. A C-index significantly higher than 0.5 points at a better discrimination ability of the model than random guessing. On top of both reference models, there was no evidence that the SNPs that were selected by priority-Lasso, or the GRSMetabo improved the prediction. The extended models did not produce delta C-indices, cfNRIs nor IDIs significantly higher than 0. Whereas the delta C-index compares the discrimination ability of a reference and an extended model, cfNRI and IDI compare the reclassification ability of both. The cfNRI describes the proportion of people for whom the expanded model improved the prediction (predicted risk higher for cases, lower for noncases). The IDI quantifies the difference between a change on average sensitivity and a change on average 1–specificity. In IDIrel, this difference is considered in relation to the reference model to avoid dependence on incidence. In contrast to PLMetabo and GRSGola, the GRSGola in both models yielded improved C-indices (Model 2 delta C-index: 0.0087 [0.0044, 0.0130]) and IDI estimates (Model 2 IDIrel: 0.0509 [0.0131, 0.0894]). The overall cfNRI showed an improved reclassification in Model 1, although it was not statistically significant in Model 2.
| GRSMetaboa(138 SNPs) | |||||
|---|---|---|---|---|---|
| GRSMetabo | Model 1 | Model 1 + GRSMetabo | Model 2 | Model 2 + GRSMetabo | |
| C-index | 0.5253 [0.4803, 0.5702] | 0.7537 [0.7189, 0.7886]* | 0.7571 [0.7234, 0.7908]* | 0.7905 [0.7604, 0.8207]* | 0.7920 [0.7622, 0.8219]* |
| ∆C-index, cfNRIcases, cfNRIcontr, IDIabs, and IDIrel were not significant (Table S7). | |||||
| PLMetabob(Model 1: 10 SNPs, Model 2: 7 SNPs) | |||||
|---|---|---|---|---|---|
| PLMetabo | Model 1 | Model 1+ PLMetabo | Model 2 | Model 2+ PLMetabo | |
| C-index | –b | 0.7537 [0.7189, 0.7886]* | 0.7459 [0.7109, 0.7809]* | 0.7905 [0.7604, 0.8207]* | 0.7855 [0.7551, 0.8159]* |
| ∆C-index, cfNRIcases, cfNRIcontr, IDIabs, and IDIrel were not significant (Table S7). | |||||
| GRSGolac(39,570 SNPs) | |||||
|---|---|---|---|---|---|
| Performance measure | GRSGola | Model 1 | Model 1 + GRSGola | Model 2 | Model 2 + GRSGola |
| C-index | 0.5878 [0.5446, 0.6310]* | 0.7537 [0.7189, 0.7886]* | 0.7674 [0.7340, 0.8007]* | 0.7905 [0.7604, 0.8207]* | 0.7993 [0.7702, 0.8284]* |
| ∆C-index | – | 0.0136 [0.0067, 0.0205]* | 0.0087 [0.0044, 0.0130]* | ||
| cfNRI | – | 0.1817 [0.0280, 0.3341]* | 0.1158 [−0.0462, 0.2652] | ||
| cfNRIcases | – | 0.1667 [0.0166, 0.3172]* | 0.1761 [0.0253, 0.3219]* | ||
| cfNRIcontr | – | 0.0151 [−0.0225, 0.0516] | −0.0603 [−0.0991, −0.0250]* | ||
| IDI | – | 0.0071 [0.0028, 0.0119]* | 0.0048 [0.0011, 0.0091]* | ||
| IDIrel | – | 0.1055 [0.0445, 0.1741]* | 0.0509 [0.0131, 0.0894]* | ||
- Note: Model 1 includes age, sex, survey. Model 2 (Framingham risk score) additionally includes diabetes status, smoking status, systolic blood pressure, antihypertensive medication, HDL cholesterol, total cholesterol. Performance measures were derived from regression coefficients estimated in the MONICA/KORA trainingset (Tables S4–S6).
- Abbreviations: C-index, concordance Index; cfNRI, category-free net reclassification index; CI, confidence interval; GRS, genetic risk score; IDI, independent discrimination improvement; PL, priority-Lasso; SNP, single-nucleotide polymorphism.
- a GRSMetabo includes genome-wide associated Metabochip SNPs available in MONICA/KORA trainingset and KORA testset after exclusion of SNPs with Hardy–Weinberg equilibrium test p < 10−20, minor allele frequency < 1%, imputation quality R² < 0.3.
- b SNPs selected by priority-Lasso from Metabochip variants genome-wide significantly associated with CHD in the literature. At least two blocks of variables with different priorities are necessary to conduct a priority-Lasso regression.
- c GRS from Gola et al. (2020) with all SNPs available in MONICA/KORA trainingset and KORA testset after exclusion of SNPs with Hardy–Weinberg equilibrium test p < 10−20, minor allele frequency < 1%, imputation quality R² < 0.3.
- * Statistical significance at the 5% level.
3.4 Sensitivity analyses
In the sensitivity analyses that were solely based on data from the KORA testset (Table 3), the performance of the cross-validated GRSGola was slightly higher (C-index: 0.5881 [0.5435, 0.6299]) compared to the main analyses, where the regression coefficients were derived from the MONICA/KORA trainingset (C-index: 0.5878 [0.5446, 0.6310]; Table 2). Moreover, the cross-validated predictive performance of the GRSGola and the GRSGolaOrig, which differed in the application of QC criteria, were comparable. The crude GRSGolaOrig yielded a cross-validated C-index of 0.5910 [0.5488, 0.6356]. When added to the FRS variables, the C-index improved by 0.0106 [0.0019, 0.0237], the overall cfNRI was 0.2347 [0.0576, 0.4164], and the IDI and IDIrel estimates were 0.0116 [0.0021, 0.0295] and 0.1410 [0.0201, 0.3526], respectively.
The cross-validated performance measures of GRSKheraOrig and GRSKheraQCGola were comparable and yielded similar improved C-indices and cfNRI estimates as the GRSGola and GRSGolaOrig, but no significant improvement by IDI and IDIrel (Table 3).
| GRSGolaa (39,570 SNPs) | |||||
|---|---|---|---|---|---|
| GRSGola | Model 1 | Model 1 + GRSGola | Model 2 | Model 2 + GRSGola | |
| C-index | 0.5881 [0.5435, 0.6299]* | 0.7594 [0.7206, 0.7907]* | 0.7770 [0.7409, 0.8068]* | 0.7906 [0.7623, 0.8254]* | 0.8015 [0.7736, 0.8349]* |
| ∆C-index | – | 0.0176 [0.0051, 0.0340]* | 0.0109 [0.0019, 0.0236]* | ||
| cfNRI | – | 0.2616 [0.1020, 0.4202]* | 0.2107 [−0.0028, 0.3867] | ||
| cfNRIcases | – | 0.1634 [0.0010, 0.3113]* | 0.0214 [−0.1954, 0.2153] | ||
| cfNRIcontr | – | 0.0982 [0.0440, 0.1591]* | 0.1893 [0.0634, 0.2910]* | ||
| IDI | – | 0.0125 [0.0029, 0.0282]* | 0.0093 [0.0006, 0.0247]* | ||
| IDIrel | – | 0.2083 [0.0494, 0.5510]* | 0.1038 [0.0028, 0.2994]* | ||
| GRSGolaOrigb (49,918 SNPs) | |||||
|---|---|---|---|---|---|
| GRSGolaOrig | Model 1 | Model 1 + GRSGolaOrig | Model 2 | Model 2 + GRSGolaOrig | |
| C-index | 0.5910 [0.5488, 0.6356]* | 0.7594 [0.7206, 0.7907]* | 0.7761 [0.7397, 0.8065]* | 0.7906 [0.7623, 0.8254]* | 0.8012 [0.7741, 0.8349]* |
| ∆C-index | – | 0.0167 [0.0048, 0.0326]* | 0.0106 [0.0019, 0.0237]* | ||
| cfNRI | – | 0.2911 [0.0982, 0.4295]* | 0.2347 [0.0576, 0.4164]* | ||
| cfNRIcases | – | 0.1912 [−0.0084, 0.3299] | 0.0741 [−0.1221, 0.2652] | ||
| cfNRIcontr | – | 0.0999 [0.0437, 0.1532]* | 0.1605 [0.0520, 0.2770]* | ||
| IDI | – | 0.0157 [0.0049, 0.0329]* | 0.0116 [0.0021, 0.0295]* | ||
| IDIrel | – | 0.2729 [0.0851, 0.6257]* | 0.1410 [0.0201, 0.3526]* | ||
| GRSKheraOrigc (6,481,934 SNPs) | |||||
|---|---|---|---|---|---|
| GRSKheraOrig | Model 1 | Model 1 + GRSKheraOrig | Model 2 | Model 2 + GRSKheraOrig | |
| C-index | 0.5730 [0.5254, 0.6212]* | 0.7594 [0.7206, 0.7907]* | 0.7752 [0.7443, 0.8029]* | 0.7906 [0.7623, 0.8254]* | 0.8012 [0.7775, 0.8353]* |
| ∆C-index | – | 0.0158 [0.0038, 0.0373]* | 0.0105 [0.0018, 0.0259]* | ||
| cfNRI | – | 0.2258 [0.0650, 0.3867]* | 0.2036 [0.0523, 0.4222]* | ||
| cfNRIcases | – | 0.1769 [0.0248, 0.3399]* | 0.1010 [−0.0512, 0.3254] | ||
| cfNRIcontr | – | 0.0489 [−0.0078, 0.0996] | 0.1026 [0.0005, 0.2104]* | ||
| IDI | – | 0.0046 [−0.0011, 0.0171] | 0.0059 [−0.0011, 0.0213] | ||
| IDIrel | – | 0.0782 [−0.0212, 0.4034] | 0.0632 [−0.0138, 0.2844] | ||
| GRSKheraQCGolad(5,453,218 SNPs) | |||||
|---|---|---|---|---|---|
| GRSKheraQCGola | Model 1 | Model 1 + GRSKheraQCGola | Model 2 | Model 2 + GRSKheraQCGola | |
| C-index | 0.5712 [0.5233, 0.6179]* | 0.7594 [0.7206, 0.7907]* | 0.7737 [0.7426, 0.8029]* | 0.7906 [0.7623; 0.8254]* | 0.8004 [0.7766, 0.8352]* |
| ∆C-index | – | 0.0143 [0.0030, 0.0362]* | 0.0098 [0.0014, 0.0252]* | ||
| cfNRI | – | 0.2136 [0.0281, 0.3695]* | 0.2397 [0.0727, 0.4293]* | ||
| cfNRIcases | – | 0.1632 [−0.0214, 0.3071] | 0.1486 [−0.0389, 0.3410] | ||
| cfNRIcontr | – | 0.0503 [0.0025, 0.0995]* | 0.0911 [−0.0018, 0.2040] | ||
| IDI | – | 0.0038 [−0.0012, 0.0157] | 0.0056 [−0.0011, 0.0204] | ||
| IDIrel | – | 0.0739 [−0.0235, 0.3782] | 0.0692 [−0.0136, 0.2696] | ||
- Note: Model 1 includes age, sex, survey. Model 2 (Framingham risk score) additionally includes diabetes status, smoking status, systolic blood pressure, antihypertensive medication, HDL cholesterol, total cholesterol. Calculations are based only on data from KORA testset.
- Abbreviations: C-index, concordance Index; cfNRI, category-free net reclassification index; CI, confidence interval; GRS, genetic risk score; IDI, independent discrimination improvement; PL, priority-Lasso; SNP, single-nucleotide polymorphism.
- a GRS from Gola et al. (2020) with all SNPs available in MONICA/KORA trainingset and KORA testset after exclusion of SNPs with Hardy–Weinberg equilibrium test p < 10−20, minor allele frequency < 1%, imputation quality R² < 0.3.
- b GRS from Gola et al. (2020) with all SNPs available in KORA testset after exclusion of SNPs with Hardy–Weinberg equilibrium test p < 10−20, minor allele frequency < 1%, R² < 0.8.
- c GRS from Khera et al. (2018) in KORA testset after exclusion of SNPs with imputation quality R² < 0.3.
- d GRS from Khera et al. (2018) in KORA testset after exclusion of SNPs with Hardy–Weinberg equilibrium test p < 10−20, minor allele frequency < 1%, R² < 0.8.
- * Statistical significance at the 5% level.
4 DISCUSSION
The present study compared different approaches based on genetic data to improve CHD prediction beyond the established FRS variables.
One of our approaches was to select the most predictive variants among Metabochip SNPs genome-wide associated with CHD. We chose the novel priority-Lasso algorithm which allowed to select predictive SNP variables on top of our fixed variables from the FRS, aiming at highlighting strongly predictive SNPs (Klau et al., 2018). Their shrunk coefficients counteract overfitting and thus are expected to lead to better prediction results than unpenalized coefficients. However, although some Metabochip SNPs were selected because they explained more CHD outcome variability than the reference models alone, they were not able to increase the prediction accuracy in the KORA testset. The predictive value of the selected SNPs seemed to be too small.
We also built a weighted GRS including all genome-wide associated Metabochip SNPs (GRSMetabo). Here, the summing of SNP dosages weighted with the respective published ln(odds ratio) estimates takes differences in predictive importance of the variants into consideration. Usage of published weights, instead of estimates from the own data set, reduces overfitting. Our results indicated that the exclusive usage of genome-wide associated Metabochip variants, despite weighting, was not sufficient to improve the prediction. A similar approach with a prospective cohort and Metabochip data was conducted by Morris et al. (2016). They evaluated a 53-SNP GRS mainly based on variants from Deloukas et al. (2013) and found minimal incremental utility beyond clinical risk variables (age, sex, smoking, family history of cardiovascular disease, body mass index, blood pressure, treatment for hypertension, total and HDL cholesterol). We extended this approach by including further Metabochip variants identified by Howson et al. (2017) and van der Harst and Verweij (2018) as well as by estimating the predictive performance in an independent test data set. Further studies which included GRSs from established SNPs not limited to Metabochip data, reported inconclusive results regarding the added predictive value for incident cardiovascular diseases (Antiochos et al., 2016; Beaney et al., 2017; Paynter et al., 2010; Tada et al., 2016; de Vries et al., 2015). Variables preliminarily selected from univariable association tests, which is the approach of most GWAS, do not necessarily perform optimally in combination. Preselection based on machine learning algorithms can alternatively be performed (Boulesteix et al., 2020).
Our third approach was to evaluate a GRS which had been developed by Gola et al. (2020) in a case-control study on coronary artery disease (GRSGola). In this study, the variants had been preselected using random forest with variable importance. The score comprises variants that are not genome-wide significantly associated with the disease. We chose this score for comparison, as it had been derived from Metabochip data, showed very promising prediction results, and to our knowledge has not been validated yet. Indeed, the addition of GRSGola significantly improved the C-index, cfNRIcases, IDI, and IDIrel beyond both reference models.
Due to the limited imputation quality of the Metabochip data in our MONICA/KORA trainingset, we could not use about 20% of the SNPs included in the original GRS described by Gola et al. (2020) when we formed the GRSGola. We, therefore, repeated the analysis based only on data from our KORA testset where we could build the GRS nearly identical to the original description (GRSGolaOrig). The resulting predictive performance slightly improved but stayed far below the original published estimate. The C-index of GRSGolaOrig in our study was 0.5910 [95% CI: 0.5488, 0.6356] in the crude model, although the originally published value was 0.9222 [0.9045, 0.9400]. Our study design differed compared to Gola et al. (2020); we used cohort instead of case-control data. The transferability of results based on prevalent outcome data to incident outcome data might be limited due to survival bias occurring especially in studies on highly lethal diseases like CHD (Anderson et al., 2011). Moreover, we applied Cox proportional hazard regression instead of logistic regression. However, these reasons are not likely to explain all of the huge discrepancies. Further studies are needed to replicate our results.
Another GRS derived by Khera et al. (2018) in a study on prevalent coronary artery disease based on genetically imputed UK BiLEVE Axiom Array (807,411 variants) or UK Biobank Axiom Array (825,927 variants) data also yielded promising prediction results in the initial description. Preselection of variants for this comprehensive score was done using the LDPred algorithm (Vilhjálmsson et al., 2015). Like GRSGola it is not limited to genome-wide significantly associated variants. We included the score in our comparison to evaluate whether the prediction accuracy could be further improved without being limited to Metabochip data. The cross-validated C-index for the model including the GRSKheraOrig, age, and sex was 0.7752 [0.7443, 0.8029], which was below the originally published value of 0.81 [0.81, 0.81] (model: GRS, age, sex, ancestry, genotyping array). The discrepancy may be explained by our usage of prospective data. Weights and score derivation were both based on datasets with prevalent outcomes, leading to possible survival bias (Anderson et al., 2011). Furthermore, weights were obtained from the CARDIOGRAMplusC4D GWAS, which only included 77% of participants of European ancestry (Nikpay et al., 2015). Mosley et al. (2020) validated the GRS developed by Khera et al. (2018) in two studies on incident CHD using Cox regression. They achieved C-indices of 0.549 [0.521, 0.571] and 0.587 [0.532, 0.623] (model: GRS, ancestry), but did not yield additional predictive value beyond a clinical risk model. Hindy et al. (2020) found C-indices of 0.759 [0.724, 0.794] and 0.756 [0.750, 0.762] (model: age, sex, ancestry, GRS) in two prospective studies on coronary artery disease using Cox-regression. After addition to traditional risk factors, C-indices significantly increased from 0.776 to 0.802 and from 0.748 to and 0.768. Dikilitas et al. (2020) reported improved C-indices of the Khera GRS beyond a base model (age-as-time-scale Cox model with sex, site, ancestry) in three different racial and ethnic groups on incident CHD (European ancestry: improvement from 0.690 to 0.719). The Khera GRS was additionally evaluated by Wünnemann et al. (2019) in two studies on incident CHD. Using logistic regression, they retrieved C-indices of 0.57 [0.51, 0.62] and 0.60 [0.51, 0.62] in models which included age, sex, and ancestry in addition to the GRS. Finally, Aragam et al. (2020) computed the Khera GRS in three cohorts on prevalent coronary artery disease yielding crude C-indices ranging from 0.59 to 0.61.
To directly compare the scores derived by Gola et al. (2020) and Khera et al. (2018), we applied the same post-imputation exclusion criteria for both scores (GRSGolaOrig, GRSKheraQCGola). The GRSKheraQCGola and the GRSKheraOrig yielded similar results. However, both were slightly worse than the GRSGolaOrig, which overall produced the best improvement in the prediction accuracy of our comparison. This superiority might partly be due to the fact that Gola et al. (2020) derived their GRS from six studies of European descent from the German population, while Khera et al. (2018) used the more heterogenous UK Biobank to construct their score. Nevertheless, comprising more than six million SNPs, the GRSKheraOrig is considerably more extensive than GRSGolaOrig with about 50,000 SNPs, pointing out that the number of SNPs alone is not decisive.
We are aware of four further prospective cardiovascular disease prediction studies that have derived comprehensive GRSs including nonsignificant SNPs, all of them using Cox regression (Abraham et al., 2016; Elliott et al., 2020; Inouye et al., 2018; Mars et al., 2020). Elliott et al. (2020) and Inouye et al. (2018) achieved crude C-indices of 0.61 and 0.62. When added to traditional CHD risk factors, their C-indices increased from 0.76 to 0.78 and from 0.67 to 0.70. In Elliott et al. (2020) this increase was statistically significant, whereas significance remained unclear in Inouye et al. (2018). Abraham et al. (2016) also reported significantly improved C-indices by their derived GRS beyond the FRS variables in two prospective studies (C-indices improved from 0.731 to 0.742 and from 0.848 to 0.866). Elliott et al. (2020) and Abraham et al. (2016) additionally showed an improvement in overall cfNRI and IDI. Mars et al. (2020) reported an impairment in discrimination after the addition of their GRS to clinical risk factors (C-index decreased from 0.823 to 0.820), but showed an improved NRI in early-onset cases and late-onset controls.
Our study has several strengths. We evaluated two well-characterized longitudinal population-based studies with a long follow-up of up to 14 years and physician-validated CHD diagnoses. Our outcome was time-to-event, which enabled us to use survival analysis that is more informative and therefore more clinically relevant than the usage of logistic regression (George et al., 2014). We have developed the analysis plan a priori and followed it rigorously. To our knowledge, we are the first to externally investigate the score derived by Gola et al. (2020). We accounted for imputation uncertainty by using estimated posterior allele dosages and not hard calls. We have conducted an extensive literature search to find all genome-wide CHD-associated Metabochip variants and weighted them with the ln(odds ratio) derived from the study with the largest sample size.
Although there are various studies examining the predictive value of genetics (alone or in extension to clinical risk models), most of them lack truly independent validation in terms of model fit and predictive performance measures. As building GRSs usually involves two different data sets, it is a common misconception to believe that independent validation has already been performed in most genetic prediction studies (Choi et al., 2020). So called “discovery datasets” (usually GWAS) are commonly used to derive GRSs and to calculate weights. Estimation of regression coefficients (ln[hazard ratio] or ln[odds ratio]) and performance measures of the GRSs and possible clinical variables are then derived from the same population, often called “validation-” or “testset” (Abraham et al., 2016; Beaney et al., 2017; Elliott et al., 2020; Inouye et al., 2018; Iribarren et al., 2016; R. W. Morris et al., 2016; Mosley et al., 2020; Tada et al., 2016). This makes cross-validation or bootstrapping crucial to at least attenuate overfitting of the GRS (and possible other variables) to the testset (Choi et al., 2020). However, such approaches are rare (Antiochos et al., 2016; Müller et al., 2016). To our knowledge, we are the first to use two independent studies for model fit and evaluation to calculate the incremental predictive value of genetic information beyond a clinical risk model in the context of cardiovascular diseases.
Our study has several limitations. Compared to several other genetic prediction studies, we are limited in sample size. In addition, our population is restricted to European ancestry, and therefore the transferability of our findings to other ethnicities may be limited. Furthermore, the MONICA/KORA trainingset was not genome-wide genotyped. Therefore, our main analyses, which followed the advantageous independent validation approach, were limited to Metabochip SNPs, while the evaluation of GRSKheraOrig and GRSKheraQCGola was limited to a cross-validation approach in the KORA testset. However, as the Metabochip has been designed to cover DNA regions enriched with CHD-associated SNPs, we assume that we did not miss many strongly associated SNPs in our Metabochip-based approaches. We could show that GRSGola, which was derived from Metabochip data only, was sufficient to improve prediction beyond FRS variables. A further limitation is that our Metabochip imputation yielded a worse result compared to the Affymetrix imputation of the KORA testset. Generally, this can be explained by the smaller number of SNPs present on Metabochip compared to Affymetrix, the uneven scattering of variants across the genome and the larger proportion of lower frequency variants on Metabochip, which all makes it harder to impute (Liu et al., 2012). In addition, our Metabochip genotyped data set had a higher proportion of missing values compared to our Affymetrix genotyped data set. Altogether we had to exclude more than 10,000 SNPs due to bad imputation quality (R² < 0.3) for GRSGola in the MONICA/KORA trainingset. To make sure that the quality of the remaining SNPs was sufficient, we additionally evaluated GRSGola via cross-validation in the KORA testset as a sensitivity analysis. Cross-validated performance measures were then compared to performance measures produced in the KORA testset with the use of regression coefficients derived in the MONICA/KORA trainingset. A substantial improvement in cross-validated prediction would have indicated that genotyping quality of the discovery case-cohort data were not reliable. However, we could verify that cross-validation produced comparable results. All analyses were performed in a fairly homogeneous population of predominantly Caucasian individuals with German citizenship and residing in or near Augsburg. Thus, the participants from the MONICA/KORA trainingset and the KORA testset are very similar with regard to ethnic background. The KORA S4 cohort study has been used in various genome-wide studies, most recently in a study by Winkler et al. (2020), where no evidence of population stratification was found. The same applies for previous analyses of Metabochip data from MONICA/KORA including parts of the S1–S3 case-cohort (Gaulton et al., 2015; A. P. Morris et al., 2012). Because our final analyzed samples slightly differ from previous analyses, we have calculated genomic control inflation factors and conducted permutation tests in both studies, which also did not indicate population stratification. Nevertheless, it has to be mentioned that genome-wide coverage of SNPs without enrichment for disease loci is required to correctly define population outliers, which is not fully given in the MONICA/KORA trainingset. A small degree of population stratification in the MONICA/KORA trainingset, which we might have missed to detect, would lead to a slight underestimation of the predictive power of the models in the KORA testset.
In conclusion, the findings of our study confirm previously published results stating the predictive superiority of comprehensive polygenic risk scores that are not restricted to genome-wide associated variants. However, none of our examined strategies added a major predictive value beyond the FRS variables. We were not able to confirm the high predictive performance of the initial reports by Gola et al. (2020) and Khera et al. (2018). This confirms that weights and variant selection should be derived from prospective cohorts for incident disease prediction in future studies to avoid survival bias and underlines the importance of external validation.
ACKNOWLEDGMENTS
The authors thank Professor Anne-Laure Boulesteix, Institute for Medical Information Processing, Biometry and Epidemiology, Ludwig-Maximilians-University, Munich, Germany, for excellent statistical advice. This study was supported by a research grant from the Else Kröner-Fresenius-Stiftung (2015_A130), grants from the German Research Foundation (TH-784/2-1 and TH-784/2-2), from the Helmholtz Alliance “Aging and Metabolic Programming, AMPro,” from the German Federal Ministry of Education and Research to the German Center for Diabetes Research (DZD), and by the Bavarian State Ministry of Health and Care through the research project DigiMed Bayern (www.digimed-bayern.de). The KORA study was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. Metabochip genotyping was funded by the German Federal Ministry of Education and Research (BMBF), Germany, and the State of Bavaria through the research project Atherogenomics (01GS0834). Cornelia Huth was supported in part by intramural funding for Translational & Clinical Projects of the Helmholtz Zentrum München. Holger Prokisch is supported by the Free State of Bavaria, Ministry of Health (DigiMed Bayern). The German Diabetes Center (DDZ) is supported by the Ministry of Culture and Science of the State of North Rhine-Westphalia and the German Federal Ministry of Health. The sponsors had no further role in study design, data analysis, data interpretation, writing of the report and decision to publish the manuscript.
CONFLICT OF INTERESTS
Michael Roden reports personal fees from Eli Lilly, Poxel S. A. Société, Boehringer-Ingelheim Pharma, Terra Firma, Sanofi US, Servier Labatories, Novartis Pharma GmbH, Fishawack Group, PROSCIENTO, Inc., Target Pharmasolutions, Gilead Sciences, Kenes Group, Bristol Myers Squibb, Intercept Pharma, Inventiva, AstraZeneca, and Allergan GmbH outside the submitted work. Wolfgang Koenig reports personal fees from AstraZeneca, Novartis, Pfizer, The Medicines Company, DalCor, Kowa, Amgen, Corvidia, Daiichi-Sankyo, Berlin-Chemie, Sanofi, Bristol-Myers Squibb, and grants and nonfinancial support from Singulex, Abbott, Roche Diagnostics, and Beckmann outside the submitted work. Christian Herder reports personal fees from Sanofi and Lilly, and grants from Sanofi outside the submitted work. The other authors report no conflict of interests.
Open Research
DATA AVAILABILITY STATEMENT
The data are subject to national data protection laws and restrictions were imposed by the Ethics Committee of the Bavarian Chamber of Physicians to ensure the data privacy of the participants. The data can be requested through a project agreement via the online portal KORA. passt (https://epi.helmholtz-muenchen.de/). Please contact the corresponding author in case of questions.




