Deep-learning approaches to identify critically Ill patients at emergency department triage using limited information
Supervising Editor: Austin Johnson, MD, PhD
Funding and support: This work was conducted with support from CRICO-Risk Management Foundation of the Harvard Medical Institutions Incorporated (Improving Patient Safety Grant).
Abstract
Study objective
Triage quickly identifies critically ill patients, facilitating timely interventions. Many emergency departments (EDs) use emergency severity index (ESI) or abnormal vital sign triggers to guide triage. However, both use fixed thresholds, and false activations are costly. Prior approaches using machinelearning have relied on information that is often unavailable during the triage process. We examined whether deep-learning approaches could identify critically ill patients only using data immediately available at triage.
Methods
We conducted a retrospective, cross-sectional study at an urban tertiary care center, from January 1, 2012–January 1, 2020. De-identified triage information included structured (age, sex, initial vital signs) and textual (chief complaint) data, with critical illness (mortality or ICU admission within 24 hours) as the outcome. Four progressively complex deep-learning models were trained and applied to triage information from all patients. We compared the accuracy of the models against ESI as the standard diagnostic test, using area under the receiver-operator curve (AUC).
Results
A total of 445,925 patients were included, with 60,901 (13.7%) critically ill. Vital sign thresholds identified critically ill patients with AUC 0.521 (95% confidence interval [CI] = 0.519–0.522), and ESI <3 demonstrated AUC 0.672 (95% CI = 0.671–0.674), logistic regression classified patients with AUC 0.803 (95% CI = 0.802–0.804), 2-layer neural network with structured data with AUC 0.811 (95% CI = 0.807–0.815), gradient tree boosting with AUC 0.820 (95% CI = 0.818–0.821), and the neural network model with textual data with AUC 0.851 (95% CI = 0.849–0.852). All successive increases in AUC were statistically significant.
Conclusion
Deep-learning techniques represent a promising method of augmenting triage, even with limited information. Further research is needed to determine if improved predictions yield clinical and operational benefits.
1 INTRODUCTION
1.1 Background
Triage quickly identifies critically ill patients, helping to facilitate rapid interventions with the goal of altering the course of disease. Many emergency departments (EDs) use the emergency severity index (ESI) or other standardized scores to facilitate triage and prioritize patients.1 Concurrently, many EDs combine this with a clinical trigger system, which mobilizes available physicians and nurses to see patients with acute ESI scores or abnormal vital signs immediately after initial triage. The use of clinical triggers to mobilize clinicians has been demonstrated to improve patients’ time to physician evaluation and time to antibiotics.2
1.2 Importance
Both ESI and vital sign triggers rely on specific vital sign thresholds. Although ESI is among the most validated algorithms for triage, numerous studies have shown that both under-triage and over-triage remain persistent issues.3, 4 When patients are under-triaged, opportunities to change the course of disease are missed, whereas over-triage has the potential to disrupt physicians’ and nurses’ workflows, detracting from safe and efficient care for other patients in the ED. In particular, better understanding of the effects of advanced age, the influence of specific chief complaints, and more robust criteria for vital sign abnormalities have been highlighted as areas for improvement to the current ESI.4, 5 Studies examining machine-learning approaches have shown promise in supplementing the ESI score at triage, including random forest models to help differentiate outcomes for patients within ESI categories,6 gradient boosting algorithms to predict admission,7 and outcomes in specific conditions, such as mortality in sepsis.8
However, many of these studies have leveraged information, such as structured diagnosis lists and past medical histories, which is unavailable for many patients at the time of triage. In particular, patients who are making their first contact with an ED rarely bring medical history information in a readily accessible electronic format. Patients also may be unable to meaningfully provide a clear past medical history, whether because of dementia, language barriers, or limited health literacy. Depending on their complexity, some machine-learning approaches are not readily integrated with commercial electronic health record (EHR) systems, and may require considerable effort to tune and adapt to a health system's specific population.
Deep neural networks are a family of machine-learning algorithms that have led to rapid improvements across a variety of domains, including computer vision and natural language processing and have made progress toward automated diagnosis in subfields of radiology and pathology. Compared to traditional methods of regression analysis, neural networks are intended to model multiple levels of complex, high dimensional interaction terms between independent variables without loss of specificity, and can be rapidly retrained to account for subtle differences between populations. In the last few years, several open source frameworks have made it simple to develop, deploy, and share these algorithms without the need for specialized equipment.
1.3 Goals of this investigation
We examined whether a set of progressively complex deep-learning algorithms could identify critically ill patients with greater discriminative power than ESI or vital sign triggers alone using information immediately available at triage, as measured by the area under the receiver-operator curve (AUC).
2 METHODS
2.1 Study design and selection of participants
This was an observational study examining a retrospective cohort of adult patients who visited an academic, urban ED at a tertiary care center in the Northeastern United States with an average volume of 55,000 visits annually. All patients between January 1, 2012–January 1, 2020 were screened for the study. Patients were included if their data included triage vital signs (patients with one or more vital signs were included, patients with none were not). ESI score, whether a vital sign trigger had been activated, and ultimate disposition (including whether they expired within the ED) were recorded for all patients. Clinical data was obtained from an automated quality assurance database for the ED information system. Data were de-identified during extraction using the HIPAA SAFE HARBOR method, and the study authors were blinded to identifying information. The study was structured with respect to the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) statement.9 The host institutional review board granted an exemption for de-identified data used.
2.2 Measurements
Vital signs were defined as a patient's initial triage measurement of temperature, heart rate, systolic and diastolic blood pressure, respiratory rate, and percent oxygenation. To exclude potentially erroneous data entries, vital signs were included in the analysis provided they fell within broad physiologically feasible ranges, including temperature below 110°, heart rate below 300 beats/min, systolic, and diastolic measurements below 300 mm Hg, respiratory rate below 80 respirations/min, and oxygenation at or below 100%. Missing or spurious data were considered as null values for the analysis.
Triage chief complaints were included as free text entered immediately at a patient's arrival at triage. Chief complaints were not standardized during the study period, and typographic errors and blank entries were included in the analysis for fidelity. Nursing documentation after the patient's initial registration was not included. As a result of de-identification, patients older than 89 years of age were included within a single 90+ age category.
Vital sign trigger criteria were defined as heart rate <40 or >130 beats/min, respiratory rate below 8 or above 30 respirations/min, systolic blood pressure below 90 mm Hg, or an oxygen saturation below 90% on room air, which is standard practice for trigger activations at triage for the institution.2 As a result of the clinical system at our institution, all patients who meet clinical trigger criteria are classified as having an ESI score of 1 or 2.
2.3 Outcomes
The primary outcome was whether a patient was critically ill, defined as whether they expired within 24 hours of arrival, required ICU admission from the ED, or were transferred from an inpatient ward to the ICU (or for an emergent procedure) within 24 hours of admission. The data abstraction process included the possibility of a patient being discharged and returning to the ED as critically ill within 24 hours. There have been many different measures of resource use and illness severity used across studies evaluating the efficacy of triage and machine-learning predictions, including specific diagnoses such as sepsis,8 admission,7 and overall mortality.10 The composite of mortality and ICU admission is an appealing compromise metric, as it identifies a population that is more likely to require rapid intervention than the larger population of patients needing admission.
The Bottom Line
Initial patient triage is designed to rapidly identify which patients will require the greatest resources in the emergency department and the hospital. In this manuscript, the authors demonstrate that deep learning techniques can improve the accuracy of model to predict the need for ICU admission and or death when compared to traditional triage methods. Future prospective studies are needed to determine the benefit of these new models.
2.4 Neural network model creation and derivation
Neural networks consist of a series of nodes known as neurons, which take input variables, apply an affine transformation to the inputs based on a set of weights, and yield an output based on whether a discrete threshold, known as the activation function, has been met. A loss function measuring the neuron's output relative to the correct diagnostic labels is used to adjust the neuron's weights repeatedly until loss has been minimized. Accordingly, logistic regression can be thought of as a single neuron with a sigmoid activation function.
Neural networks leverage multiple layers of interconnected neurons to model high-degree interaction effects. For instance, a neuron within a deep neural network layer might recognize specific combinations of elevated heart rate and temperature, whereas a separate neuron within the same layer might recognize separate thresholds for the same variables when associated with a different age range or set of chief complaints.
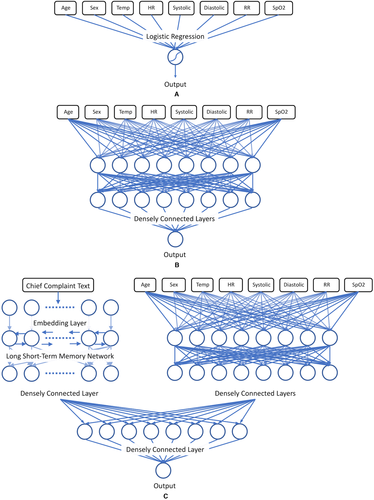
Our models were created in TensorFlow, an open-source framework for deep-learning.11 The triage data was split between the structured vital sign data and the chief complaint text data. The vital sign data was normalized and used as the input for a logistic regression (with L2 normalization), and for a 2-layer deep neural network. A third, deep neural network combining both structured and text data likewise used the vital sign data as the input to a smaller 2-layer deep neural network. The chief complaint text data was first embedded into a text vector and input into a long short-term memory network, a standard architecture for text processing and in clinical natural language processing.12, 13 As tree-based approaches have been used in a number of other recent studies of machine learning in the ED,6 we also provided a tree-based model on the structured data for comparison, using the XGBoost framework.14
The outputs of these 2 subnetworks were then concatenated and used as the cumulative input to a densely connected layer, with a final logistic prediction layer. Hyperparameters of the model, such as the number of neurons per layer and the total number of layers in the overall model, were adjusted using random search and subsequently manually adjusted, with adaptive moment estimation for optimization.15 Data was divided into a randomized 80:10:10 training:validation:testing split to avoid overfitting and run in a 10-fold cross-validation to ensure that all data were tested.
2.5 Analysis
We report the diagnostic accuracy of the methods evaluated (both current triage methods and machine-learning methods) in this study in terms of their sensitivity, specificity, accuracy, and AUC with 95% CI, relative to the reference standard of critical illness abstracted from the medical record. Statistical analysis was carried out in Python 3.8 using the SciPy and SciKit-Learn scientific and machine-learning libraries.16, 17 Differences between group means were tested at an alpha level of 0.05, with strict Bonferroni correction for multiple comparisons (0.0065 for 8 comparisons).
Prior studies of machine-learning at triage have suggested that critically ill patients represent a fraction of the total patients seen within the ED, estimated as 2% of patients by Raita et al.18 and Levin et al.6, who examined a large random sample of patients from the National Hospital Ambulatory Medical Care Survey (NHAMCS) and the yearly volume of an urban ED, respectively. As a result of the high proportion of lower-acuity patients in the underlying population, we used the AUC as the primary measure of test accuracy for comparison between the existing triage models and the deep-learning models. AUC is reported with a 95% CI and compared using DeLong's test.19, 20 Comparisons were made between tests based on ascending AUC values, using an alpha level of 0.05, with Bonferroni correction (0.01 for 5 comparisons).
3 RESULTS
3.1 Characteristics of the study subjects
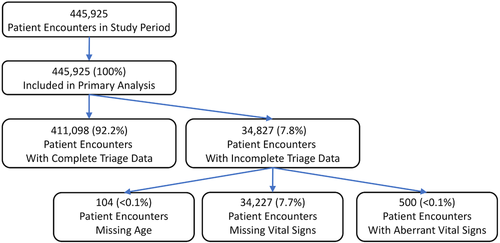
From January 1, 2012–January 1, 2020, 445,925 adult patients met inclusion criteria, detailed in Figure 1A. Patient demographic and vital sign characteristics are detailed in Table 1. A total of 60,901 (13.7%) patients met criteria for critical illness. Vital sign information contained a missing or spurious data point for 34,827 (7.6%) of patients, most commonly in the form of a missing temperature measurement at triage (24,872; 5.6%). Missing or spurious vital signs (500, <0.1%) or ages (104, <0.1%) were entered into the neural network as null values and were included in the analysis. The full enrollment process is described in Figure 1.
| All patients | Not critically Ill | Critically ill | |
|---|---|---|---|
| Characteristic (IQR) | (n = 445,925) | (n = 384,917, 86.3%) | (n = 60,901, 13.7%) |
| Age | 53 (34 –68) | 50 (32–65) | 69 (57–81) |
| Sex (female, n, %) | 241,412 (54.1) | 212,657 (55.2) | 28,755 (47.2) |
| Triage temperature | 98.0 (97.5–98.6) | 98.0 (97.0–98.5) | 98.0 (97.6- 98.6) |
| Triage heart rate | 84 (72–96) | 83 (72–95) | 86 (73–100) |
| Triage systolic BP | 133 (120–148) | 133 (120–148) | 130 (113–149) |
| Triage diastolic BP | 77 (68–87) | 78 (69–77) | 72 (61–83) |
| Triage respiratory rate | 18 (16–18) | 18 (16–18) | 18 (16–20) |
| Triage SpO2% | 99 (97–100) | 99 (98–100) | 98 (96–100) |
There were significant differences between the groups of patients assessed at triage who were critically ill and those who were not. Critically ill patients typically were older, more likely to be male, had faster heart rates and respiratory rates, higher temperatures, lower blood pressures, and lower oxygen saturations, all of which were significant (P < 0.00625). Admitted patients who were transferred to the ICU within 24 hours constituted a small portion of the overall population of the critically ill (n = 4,623; 7.6%). The full details of the population cohorts are detailed in Table 1.
3.2 Main results
The existing standards of triage evaluation to identify critical patients at our institution, abnormal vital sign triggers and ESI scores ≤2, demonstrated limited overall accuracy, and divergent sensitivity and specificity. Strict abnormal vital sign triggers demonstrated low discrimination (AUC, 0.521; 95% CI = 0.519–0.522), very low sensitivity (0.050; 95% CI = 0.050–0.051) but very strong specificity (0.991; 95% CI = 0.991–0.991). Conversely, the ESI score demonstrated greater discrimination (AUC, 0.697; 95% CI = 0.696–0.699; difference in AUC, P < 0.01), representing the product of significantly increased sensitivity but more modest specificity. The full details of the models’ diagnostic scores are presented in Table 2 and ROC curves in Figure 2.
| Method | Sensitivity (95% CI) | Specificity (95% CI) | AUC (95% CI) |
|---|---|---|---|
| Abnormal vital sign trigger | 0.050 (0.050–0.051) | 0.991 (0.991–0.991) | 0.521 (0.519–0.522) |
| Triage ESI ≤2 | 0.697 (0.696–0.699) | 0.647 (0.646–0.649) | 0.672 (0.671–0.674) |
| Logistic regression | 0.778 (0.775–0.782) | 0.673 (0.669–0.678) | 0.805 (0.801–0.808) |
| Neural network–structured data | 0.813 (0.811–0.814) | 0.653 (0.652–0.655) | 0.812 (0.811–0.814) |
| XGBoost structured data | 0.814 (0.813–0.815) | 0.666 (0.665–0.667) | 0.820 (0.818–0.821) |
| Neural network combined data | 0.845 (0.844–0.846) | 0.704 (0.702–0.705) | 0.857 (0.856–0.858) |
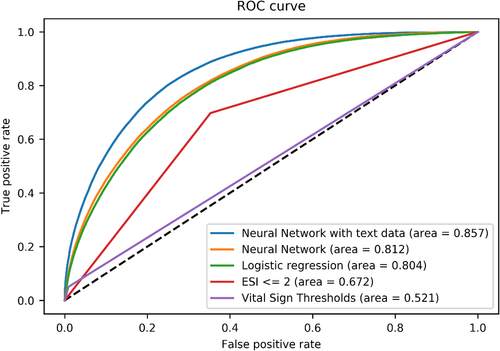
The deep-learning approaches demonstrated progressive increases in sensitivity and AUC as the models became more complex. The design of the deep-learning models is illustrated in Figures 3A–3C. The initial logistic regression on structured data yielded an AUC of 0.805 (95% CI = 0.801–0.808), with a sensitivity of 0.778 (95% CI = 0.775–0.782). The 2-layer neural network on this same structured data demonstrated modest increases in AUC (0.812; 95% CI = 0.811–0.814) and sensitivity (0.813; 95% CI = 0.811–0.814), but was slightly below that of the tree-based model, which had an AUC of 0.820 (0.818–0.821), representing a slightly higher specificity (0.666; 95% CI = 0.665–0.667). All pairwise comparisons of increasing AUC were significant (P < 0.01).
The addition of the unstructured chief complaint data provided further discriminatory power. After training and hyperparameter optimization, the final neural network model classified critically ill patients with AUC 0.851 (95% CI = 0.849–0.852), reflecting a total sensitivity of 0.845 (95% CI = 0.844–0.846). Compared with the tree-based model, this increase in AUC was likewise significant (P < 0.01).
4 DISCUSSION
Our study examines several models of triage that demonstrate increasing accuracy in tandem with increasing complexity. For those models relying on vital sign data alone, this progression is logical, as more complex models can draw more granular borders between data. The discrete vital sign cutoffs used for trigger criteria are simple to remember and demonstrate considerable specificity but miss a substantial number of critically ill patients. The enhanced accuracy of the more complex models likely reflects 2 factors—interaction effects between different vital signs and the potential effects of age.
Early in their disease course, a critically ill patient may demonstrate subtle changes in multiple vital signs, which may be difficult to recognize individually, but meaningful in the aggregate. We are primed to recognize that a heart rate above 100 or a respiratory rate above 20 is abnormal because these numbers are salient.21, 22 Comparatively, a heart rate of 95 combined with a respiratory rate of 18 might be equally predictive of illness, which may be easy for a clinician to miss, but will not elude a regression. Similarly, there exist meaningful age-related variations within vital signs, particularly for the elderly, which the regression and neural network models can recognize, but might be lost on all but the most meticulous clinicians.23, 24 Cognitive aids exist for recognizing abnormal vital signs over broad age ranges, such as the Broselow tape in pediatric resuscitation,25 but no cognate tool exists for the adult and elderly populations. For the deep-learning models examined in our study, however, abnormal vital signs can be redefined to a patient's age on a year-by-year basis.
The modest improvement in accuracy between the logistic regression model and the neural network model and tree-based models examining vital signs and demographic variables alone may reflect an underlying information-theoretic limit to a single set of measurements. Although these models are more accurate than rigid vital sign cutoffs and use of the ESI score, it is notable that optimal neural network model examining vital signs in our study was only 2 layers deep and was not improved by adding additional layers representing higher-dimension interaction effects. This suggests that although vital signs are essential to the triage process, and their interpretation can be improved substantially, alone they are not sufficient to identify all critical patients.
Although the addition of textual chief complaint data entails only a small amount of additional data per patient, it was associated with small but significant improvements in both the sensitivity and specificity of the neural network model. This likely reflects the critical contextual information that a patient's chief complaint provides. A young patient presenting with tachycardia and tachypnea may not be critically ill if their complaint is anxiety, and the additional attention of being taken to a critical care bay might exacerbate their symptoms. However, the same vital signs in a patient with a chief complaint of abdominal pain could be essential to identifying a ruptured ectopic pregnancy. Integrating free-text data directly into the analysis is a particular strength of the neural networks, as many other approaches (eg, regression) may require either entering completely standardized chief complaints (for use as categorical variables), or extensive pre-processing. For example, a system that requires a nurse to choose between “chest pain” and “back pain” will not capture the information that “sudden chest pain radiating to back” can signify as a chief complaint.
Our models demonstrate similar levels of accuracy to prior machine-learning approaches to predict admission from the ED. The neural network in our study achieved similar accuracy to that examined by Hong et al7 that predicted the larger category of all patients requiring admission, and Levin et al,6 which additionally predicted a patient's specific ESI score. However, a significant distinction between our model and similar approaches is that our model depends only on information immediately available at triage. The models examined by Hong et al7 had the benefit of using the triage ESI score as an input variable. Although predicting the larger population of patients requiring admission is important for operations management, and the triage ESI score represents a rich source of data, it represents a prediction that is informed by the triage process, rather than informing it. Conversely, the e-triage system outlined by Levin et al leverages pre-existing data within the medical record, which may disadvantage patients without prior access to care, or patients who cannot provide a history.26 As a result of using an open source framework, the neural networks examined in this study can be adapted for use in an EHR or web-browser, and the model parameters are available to interested researchers by request.
It is important to note that, as with any decision support tool, it would be a mistake to use our model as a substitute for the judgement of emergency physicians and nurses. The triage process is multifaceted, and the definition of critical illness used in this study excludes many patients who require immediate attention and would be appropriately identified at an ESI level 1 or 2. For instance, patients presenting with testicular or ovarian torsion will rarely require ICU admission but clearly need immediate attention. Similarly, many triage protocols, such as those for patients being ruled out for stroke, require patients to be appropriately triaged to levels 1 and 2, even if many ultimately do not require critical interventions. However, even accounting for the effect of these patients on the specificity of ESI, within our study, the current ESI algorithm fails to identify nearly a third of critically ill patients as meeting high acuity level criteria.
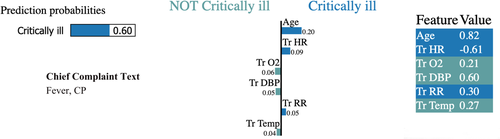
Accordingly, our results suggest that a neural network model can be a powerful supplement to clinicians’ immediate evaluations during the triage process. Although the population of patients within this study represented a higher rate of critically ill patients than found in similar datasets, a distinct advantage of neural network models is that they can be rapidly retrained to reflect the characteristics of different population with similar performance. Combined with the use of frameworks such as Local Interpretable Model-Agnostic Explanations,27 these models can also provide clinicians with real-time insight about features that are particularly suggestive of critical illness (Figure 4) (ie, subtle but important changes in vital signs) relative to patient age and chief complaint. Thus, clinicians can think of a neural network model at triage as akin to having an automated, finely grained Broselow tape for adults.
5 LIMITATIONS
This was a retrospective study conducted at a single urban tertiary care center with a significantly higher proportion of critically ill patients than has been reported in similar studies. This may be partly explained in terms of the relatively high average age of the patient population but could also reflect institutional bias. The use of ICU admission as a proxy for critical illness also introduces several significant limitations. Depending on a hospital's ICU and floor capabilities, as well as its institutional culture, a patient requiring ICU admission at one institution (for frequent vital sign checks, vasoactive drips, VIP status, or as part of certain post-operative protocols) might qualify for a stepdown unit at another. Similarly, because abnormal vital signs may serve as an independent criteria for admitting patients to the ICU, the presence of abnormal vital signs may create a self-fulfilling prophecy—artificially enhancing the accuracy of any predictive test of ICU admission, independently of the severity of underlying illness. Finally, because of the fact that our composite metric examines outcomes after care in the ED, a portion of patients who are triaged appropriately as critically ill but who respond rapidly to treatment and ultimately do not need admission to an ICU, may be inappropriately mislabeled as not critically ill, artificially decreasing the measured sensitivity of triage.
An important technical and clinical consideration is that although our study examined tests in terms of AUC and optimal test characteristics, many clinicians will prefer to use diagnostic thresholds that maximize test sensitivity at triage at the expense of reduced specificity. These preferences, and their operational consequences, should be examined carefully before adapting any clinical decision support system.
6 CONCLUSIONS
In this single-center, retrospective study of deep-learning approaches to identifying critically ill patients at ED triage, neural network and gradient-boosting models demonstrated significantly higher accuracy than traditional methods of triage, suggesting that these models have the potential to significantly enhance the triage process. Although diagnosing patients who are critically ill can more accurately help to more appropriately mobilize resources within the ED to treat them, future studies are needed to assess the clinical and operational impact of using neural networks to enhance the triage process and to identify which critically ill patients can benefit most from rapid intervention.
AUTHORS CONTRIBUTIONS
JWJ, ELL, LDS, and NE conceived of the overall study design. Initial data gathering was performed by ELL and LAN, with model development and initial statistical analysis performed by JWJ. NE provided critical review of model design. AVG provided additional statistical review. JWJ drafted the initial manuscript. LJJ provided initial manuscript review. All authors contributed significantly to its revision. LDS and MWD provided supervision of the study. JWJ takes responsibility for the integrity of the data and the accuracy of the data analysis.
CONFLICTS OF INTEREST
Risk Management Foundation of the Harvard Medical Institutions Incorporated, was not involved in the design and conduct of the study; collection, management, analysis, or interpretation of the data; or preparation and review of the manuscript. CRICO did not approve the manuscript and had no input in the decision to submit the manuscript for publication. No publication restrictions apply. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard University and its affiliated academic healthcare centers.
Biography
Open Research
DATA AVAILABILITY STATEMENT
Source code for the triage neural network models are available at: https://github.com/jwjoseph/NN_triage_predict. Code is free for use and adaptation under the MIT License.





