Differential micronucleus frequency in isogenic human cells deficient in DNA repair pathways is a valuable indicator for evaluating genotoxic agents and their genotoxic mechanisms
Abstract
The micronucleus (MN) test has become an attractive tool both for evaluating the genotoxicity of test chemicals because of its ability to detect clastogenic and aneugenic events and for its convenience. As the MN assay has been mostly performed using only DNA repair-proficient mammalian cells, we believed that the comparison of the MN frequency between DNA repair-proficient and -deficient human cells may be an excellent indicator for detecting the genotoxic potential of test chemicals and for understanding their mode of action. To address this issue, the following five genes encoding DNA-damage-response (DDR) factors were disrupted in the TK6 B cell line, a human cell line widely used for the MN test: FANCD2, DNA polymerase ζ (REV3), XRCC1, RAD54, and/or LIG4. Using these isogenic TK6 cell lines, the MN test was conducted for four widely-used DNA-damaging agents: methyl methanesulfonate (MMS), hydrogen peroxide (H2O2), γ-rays, and mitomycin C (MMC). The frequency of micronuclei in the double strand break repair-deficient RAD54−/−/LIG4−/− cells after exposure to γ-rays, H2O2, MMS and MMC was 6.2-7.5 times higher than that of parental wild-type TK6 cells. The percentages of cells exhibiting micronuclei in the base excision repair- and single strand break repair-deficient XRCC1−/− cells after exposure to H2O2, MMC and MMS were all ∼5 times higher than those of wild-type cells. In summary, a supplementary MN assay using the combination of RAD54−/−/LIG4−/−, XRCC1−/− and wild-type TK6 cells is a promising method for detecting the genotoxic potential of test chemicals and their mode of action. Environ. Mol. Mutagen., 2018. © 2018 Wiley Periodicals, Inc.
INTRODUCTION
A great number of new chemicals are being synthesized and registered every year; thus, a convenient in vitro assay is needed for assessing their genotoxicity and understanding the details of their action. Currently, the induction of DNA damage in mammalian cells by physical and chemical agents is evaluated by a number of short-term in vitro genotoxicity bioassays, including those that measure the frequency of gene mutations, chromosomal aberrations, and micronuclei [Yamamoto et al., 2011]. The in vitro micronucleus (MN) test has become an attractive tool for genotoxicity testing because of its capacity to detect not only clastogenic and aneugenic events but also because of its simplicity in scoring, accuracy, wide applicability in different cell types, and amenability to automation. The OECD guideline 487 for the MN test was recently made available, and it refers to the extensive data supporting the validity of the assay using various rodent cell lines (CHO, V79, CHL and L5178Y) and human cell lines such as TK6 [OECD, 2014]. The human lymphoblastoid TK6 cell line has been widely used for several in vitro genotoxicity tests including the MN test [Liber and Thilly, 1982; Olive et al., 1993; Greenwood et al., 1998; Amundson et al., 2005; Hastwell et al., 2009]. For the in vitro MN assay, DNA-repair-proficient cells have been routinely utilized; however, one major issue with the current method is that these cells can quickly and accurately repair DNA damage before the bioassay can detect any positive genotoxic outcomes. Therefore, we posited that comparing MN frequency between DNA-repair-proficient and -deficient cells could be a valuable indicator to detect the genotoxic potential of various chemicals and physical agents and their mode of action.
In the current study, we applied the usage of isogenic DNA-repair-deficient human cell lines to the in vitro MN test. To generate isogenic DNA-repair-deficient clones, we chose the TK6 cells as a parental cell line because these cells express functional p53 similarly to normal human tissues [Honma and Hayashi, 2011], rapidly proliferate (13 hrs per cycle), and are very sensitive to genotoxic chemicals. The rapid proliferation of these cells makes them susceptible to substantial fractions of lesions induced by genotoxic chemicals that interfere with DNA replication, leading to MN formation particularly in cells deficient in pathways that normally protect against responding to the interference the interference of DNA replication. We generated RAD54-/-/LIG4-/-, XRCC1−/−, FANCD2−/−, and REV3−/− TK6 cells by gene disruption (Tables 1 and 2) as these five DDR factors cover a wide range of proteins important in the event of DNA lesions. The disrupted genes include FANCD2 for interstrand crosslink repair, DNA polymerase zeta (REV3) for translesion DNA synthesis (TLS), and XRCC1 for base excision repair and single-strand break (SSB) repair, leading to the generation of FANCD2−/−, REV3−/−, and XRCC1−/− cells. We also simultaneously disrupted two genes involved in double-strand break (DSB) repair and generated RAD54−/−/LIG4−/− cells. Additionally, we included XPA-/- cells in this study to cover all possible DDR pathways in humans; XPA is an essential protein in the nucleotide excision repair pathway to repair bulky DNA adducts caused by UV and intrastrand crosslink agents. The frequency of MN in these mutant cells was then compared to that of the parental TK6 cells after exposure to DNA-damaging agents such as mitomycin C (MMC), an inter-strand crosslinking agent; methyl methanesulfonate (MMS), an alkylating agent; hydrogen peroxide (H2O2); and γ-rays. MMC and MMS act by crosslinking and methylating DNA, respectively. H2O2 causes oxidative base damage to DNA, and γ-rays directly generate DNA double strand breaks. Among the four TK6 mutants, RAD54−/−/LIG4−/− and XRCC1−/− cells showed markedly efficient induction of MNs following treatment of the four agents. In summary, we propose a supplementary MN test using the combination of RAD54−/−/LIG4−/−, XRCC1−/− and wild-type TK6 cells to measure genotoxic potential as well as to investigate the mode of action of chemicals, routinely used for screening.
| DNA-damage-response factor | Function | Reference |
|---|---|---|
| DNA polymerase ζ (REV3) | TLS | Hochegger et al. [ 2004] |
| FANCD2 | Fanconi-anemia interstrand crosslink repair | Kim and D'Andrea [ 2012] |
| Ligase 4 (LIG4) | DSB repair by NHEJ | Goodarzi and Jeggo [ 2013] |
| RAD54 | DSB repair by HR | Holthausen et al. [ 2010] |
| XRCC1 | Base excision repair and SSB repair | Horton et al. [ 2008] |
| XPA | Nucleotide excision repair | Musich et al. [ 2017] |
- TLS = Translesion DNA synthesis, FANCD2= Fanconi anemia group D2 protein, NHEJ = Non-homologous end joining, HR = Homologous recombination, DSB = Double strand break, SSB = Single strand break, RAD54 = Radiation damage protein 54, XRCC1= X-ray repair cross-complementing protein 1, XPA= Xeroderma Pigmentosum, Complementation Group A.
| Genotype | Marker genes | Type of genome editing techniques | References |
|---|---|---|---|
| REV3−/− | puroR, neoR | TALEN | This study |
| XRCC1−/− | bsrR, hisR | TALEN | This study |
| FANCD2−/− | puroR, neoR | CRISPR/Cas9 | This study |
| RAD54−/−/LIG4−/− | puroR, neoR, hygroR | CRISPR/Cas9 and TALEN | This study |
| RAD54−/− | puroR, neoR | TALEN | Keka et al. [2015] |
| LIG4−/− | puroR, neoR | CRISPR/Cas9 | Keka et al. [2015] |
| XPA−/− | puroR, hygroR | CRISPR/Cas9 | Mohiuddin et al. [2018] |
- *TSCER2 is derived from the TK6 cell line and carries a marker gene for measuring heteroallelic recombination in the thymidine kinase gene [Yatagai et al., 2008].
- REV3= Protein reversionless 3-like (REV3L) or DNA polymerase ζ, XRCC1= X-ray repair cross-complementing protein 1, FANCD2= Fanconi anemia group D2 protein, RAD54 = Radiation damage protein 54, LIG4 = Ligase 4, TALEN= Transcription Activator-Like Effector Nucleases, CRISPR/Cas9= Clustered Regulatory Interspaced Short Palindromic Repeats)/Cas9, XPA= Xeroderma Pigmentosum, Complementation Group A.
MATERIALS AND METHODS
Cell Line and Culture Conditions
DNA-repair-deficient TK6 cell lines (Table 2) and wild-type cells were cultured in RPMI 1640 medium (Life Technologies, Grand Island, NY, USA) supplemented with 10% (vol/vol) heat-inactivated horse serum (Cell Culture Bioscience, Nichirei Biosciences, Inc., Tokyo, Japan), 200 μg/ml sodium pyruvate, 100 U/ml penicillin and 100 μg/ml streptomycin, in a culture flask at 37°C, 5% CO2 and 100% humidity.
Generation of Human DNA-Repair-Deficient Human TK6 Cells
The TK6 mutants used in this study are shown in Table 2. To create the mutant cells, either guide RNAs targeting the exons using the Zhang CRISPR tool [Ran et al., 2013] or TALEN expression plasmids using the Golden Gate TALEN Kit and the TAL Effector Kit (Addgene) [Cermak et al., 2011; Sakuma et al., 2013] were generated together with gene-targeting constructs carrying selection markers. Either 6 μg of the CRISPR plasmid or a pair of TALEN expression plasmids and 2 μg of each of the two gene-targeting vectors carrying different selection marker genes were transfected into 4 × 106 TK6 cells using the Neon Transfection System (Life Technologies, US) with three 1350-volt pulses and 10 msec pulse widths. After electroporation, the cells were rested in 20 ml of drug-free medium. 48 hrs later, the cells were seeded into 96-well plates for selection for two weeks with two antibiotics against the selection marker genes in the transfected gene-targeting constructs indicated in Table 2. Gene-targeting events were confirmed by western blot analysis and/or RT-PCR.
RT-PCR
The loss of RAD54 transcript was confirmed by RT-PCR using the primers 5′-ACTGCAGCAAGTTCAGTGCC-3′ and 5′-CTCTACTATGCTATTAGGAG-3′. The loss of REV3 transcript was also confirmed by RT-PCR using the primers 5′- GGCGACATGTTTTCAGTAAGGATAG-3′and 5′-GTGTCTTTCCTTCTCATGATAACCA-3′. GAPDH transcripts were analyzed as a positive control for the RT-PCR analysis using the primers 5′-CATTGCTGACAGGATGCAGAAGG-3′ and 5′-TGCTTGCTGATCCACATCTGCTGG-3′.
Western Blot
Human XRCC1 and FANCD2 were detected using the following primary antibodies: anti-XRCC1 (1/1000, mouse monoclonal, Abcam, 33-2-5:ab1838) and anti-FANCD2 (1/1000, mouse monoclonal, Santa Cruz Biotechnology Inc., F-117: sc-20022). Anti-mouse IgG conjugated with HRP (NA931, GE Healthcare) was used as a secondary antibody.
Test Chemicals and Agent
MMS, (CAS 22801-14) and H2O2 (CAS 081–04215) were purchased from Nacalai Tesque Inc. (Kyoto, Japan) and Wako Pure Chemical Industries, Ltd (Osaka, Japan), respectively. MMC (CAS M4287) was purchased from Sigma-Aldrich Inc. (CA, USA). All test chemicals were dissolved in phosphate-buffered saline (Takara Bio Inc., Shiga, Japan). All test chemicals were prepared immediately prior to treatment. Caesium 137 installed in a Gamma cell R40 Exactor (Best Theratronics Ltd, Ontario, Canada) was the source for gamma-ray radiation.
Colony Formation Assay
To measure the sensitivity of TK6 cell lines to genotoxic agents, a colony formation assay was conducted. The sensitivity was evaluated by counting colony formation in methylcellulose plates as described previously [Zhao et al., 2007; Qing et al., 2011]. The cells were treated with various concentrations of MMS, MMC and H2O2 for 4 hrs. During the treatment with MMC and H2O2, the cells were exposed in complete media whereas in case of MMS, the cells were exposed in serum-free media. Irradiation of the cells using a 137Cs γ-ray source was done after putting the cells into 6-well cluster plates. Serially-diluted TK6 cells were then plated onto triplicated wells of 6-well cluster plates in 5 ml/well of D-MEM/F-12 (Life Technologies) supplemented with 10% horse serum, 2 mM L-Glutamine, 200 μg/ml sodium pyruvate and 1.5% (weight/vol) methylcellulose (Wako, Osaka, Japan). Colonies were counted 10–14 days after the irradiation or chemical treatment. The percentage of surviving colonies after the irradiation or chemical treatment was determined relative to the percentage of surviving untreated colonies.
Treatment with Genotoxic Agents for the MN Test
Cell density was maintained under 1.5 × 106 cells/ml. The population doubling time is ∼13 hrs for TK6 cells. Cell suspensions were prepared at 2 × 105 cells/ml and treated with serial dilutions of one of the chemicals described above for 4 hrs at 37°C or irradiated with gamma-rays at a dosage rate of 1 Gy/min. Vehicle or serial dilutions of the chemicals (500 μl) were added to 4.95 ml of cell suspension in a 6 well plate (single cell culture). Cell suspensions (5 ml) in a 6 well plate were irradiated for 0 (control) to 1 min on ice (single cell culture).
Relative Survival Measurements for the MN Test
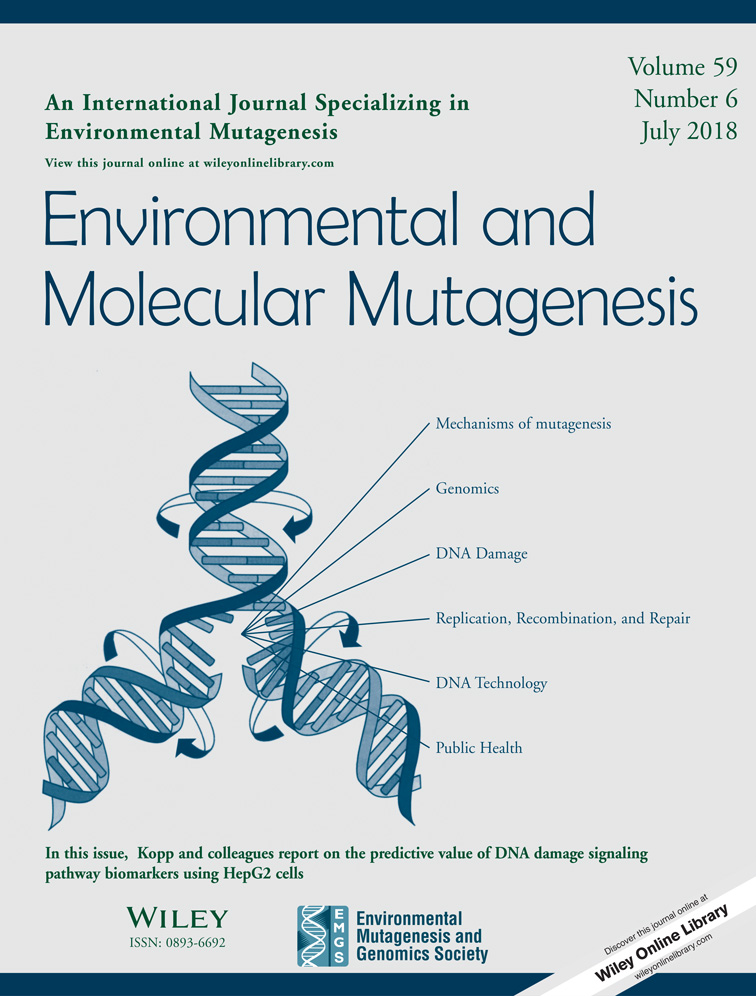
Cytotoxicity was evaluated by cell viability measured via trypan blue dye exclusion (TBDE) and cytostatic activity on the basis of cell growth. The TBDE assay was conducted just after the treatment (Day 0) and 2 days later (Days 0 and 2). The cells in an aliquot of the cell suspension were counted just after the treatment and on Day 2. The relative increase in cell count (RICC) for Day 2 was calculated using the following formula [Greenwood et al., 2004; Fellows et al., 2008]:
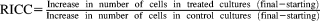
Dose Selection for the MN Test
The dose that reduced the viability of individual gene-disrupted cells to 50 to 60% was selected following OECD guideline 487. TK6 mutants (RAD54-/-/LIG4-/-, XRCC1-/-, and REV3-/-) and wild-type cells were exposed to all four DNA-damaging agents. The viability of FANCD2−/− cells was tested only with MMC, since the FANC-deficient cells are well known to be highly sensitive to crosslinking agents [Kim and D'Andrea, 2012]. We then used the doses of the DNA-damaging agents indicated in Supporting Information, Figure 1, which caused 50% to 71% survival relative to that of non-treated cells.
MN Scoring via Microscopy
In accordance with a previously reported method [Honma and Hayashi, 2011], the micronuclei were counted at 24 and 48 hrs after treatment. A small portion of the cell culture (∼1 × 106 cells) from the flask was transferred to a centrifuge tube and then centrifuged. The pellet was suspended in hypotonic 75 mM KCl solution for 10 min at room temperature, rinsed twice with ice-cold fixative (glacial acetic acid: methanol of 1:3) and then resuspended in methanol containing 1% (vol/vol) acetic acid. A drop of the suspension was placed on a clean glass slide, which was air-dried, and the cells were stained with 40 μg/ml acridine orange solution. The slides were cover-slipped, and the cells (only those showing well-outlined interphase and mononucleated cells) were immediately examined at 400x magnification under an Olympus BX50 Fluorescence Microscope (excitation: 460–490 nm, absorbing: 520 nm; Olympus Corporation, Tokyo, Japan). In total, at least three thousand cells (those exhibiting well-outlined interphase and mononucleated cells) per dose were examined (one thousand cells per replicate), and the frequency (%) of micronucleated cells was determined. The induced MN frequency was calculated by subtracting the percentage of micronucleated cells without exposure to DNA-damaging agents from that after exposure to DNA-damaging agents.
Statistical Analyses
Analysis of covariance (ANCOVA) was used to test for mean intercept differences and differences in the slopes of the linear dose-response curves in cell survival analyses between wild-type and a series of mutant cells using Graph Pad Prism 7. P-values were two-sided and adjusted for multiple comparisons, as noted. Statistical significance of the induced micronucleus frequency in the mutants at a defined dose, in comparison with that in the wild-type, was determined using a two-tailed t-test. The induced micronucleus frequency was calculated considering propagation of errors following the formula B – A ± (SD(a) + SD(b)), where B= MN frequency after treatment and A = MN frequency without treatment of DNA damaging agent.
RESULTS
Generation of DNA-Repair-Deficient TK6 Cell Lines
We previously generated RAD54−/− and LIG4−/− TK6 cells (Table 2) [Keka et al., 2015], and here, we disrupted the RAD54 gene in the LIG4−/− TK6 cells. The genotype of the resulting RAD54−/−/LIG4−/− TK6 cell lines was verified via RT-PCR and western blot (Fig. 1A). XRCC1 (Supporting Information, Fig. 2), FANCD2 (Supporting Information, Fig. 3), and REV3 (Supporting Information, Fig. 4) genes were also disrupted in TK6 cells. Inactivation of the XRCC1 and FANCD2 genes was evaluated via western blot analysis (Figs. 1B and 1C). Disruption of the REV3 gene was confirmed via RT-PCR (Fig. 1D).
Gene disruption of TK6 cells. (A-C) Western blot analysis of indicated gene-disrupted TK6 clones using the antibody against the disrupted gene and (D) RT-PCR analysis to determine transcript levels of the gene-of-interest in the mutant and wild-type TK6 cells.
Analysis of Cellular Toxicity of DNA-Damaging Agents in the Mutant Clones
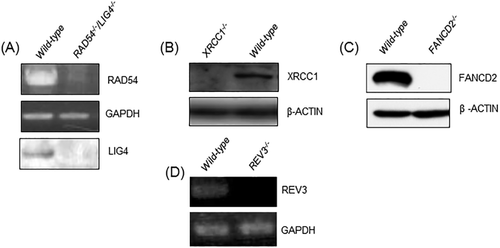
We evaluated the cell toxicity profiles of the TK6 mutant clones to γ-rays, H2O2, MMC and MMS using the colony formation assay. The sensitivity profiles of all of the tested mutants are shown in Figure 2. RAD54-/-/LIG4-/- cells displayed the highest sensitivity to γ-rays followed by RAD54-/- and LIG4-/- cells (Fig. 2A). With regards to cellular sensitivity to MMS, XRCC1-/- cells showed the greatest sensitivity. All of the mutant lines, including RAD54-/-/LIG4-/- cells, showed higher sensitivity to MMS than did the parental TK6 cells (Fig. 2B). RAD54-/-/LIG4-/- cells showed the most elevated sensitivity to H2O2, followed by XRCC1-/- cells, and RAD54-/- and FANCD2-/- cells showed mild sensitivity to H2O2 (Fig. 2C). Following exposure to MMC, FANCD2−/− cells showed the most sensitivity. The RAD54-/-/LIG4-/-, RAD54-/- and LIG4-/- cells showed mild sensitivity to MMC (Fig. 2D).
Sensitivity of DNA-repair-deficient TK6 mutant cells to γ-rays (A), MMS (B), H2O2 (C), and MMC (D) as measured via colony formation assay. The error bars represent SD from three independent experiments. (E) Sensitivity profiles of the indicated DNA-damaging agents in the selected DNA repair-deficient TK6 panel. Negative (the left side) and positive (the right side) scores indicate that the indicated cells are sensitive and resistant to the DNA-damaging agents, respectively. Wild-type cells are shown at the bottom with a reference value of 0. The average IC50 and standard deviations (n =3) of each drug in wild-type cells were calculated. P ≤ 0.01, (black-colored bar); 0.01 < P < 0.05, (gray-colored bar); and not significant, (white-colored bar); in comparison with wild-type cells as analyzed by ANCOVA.
MN Induction by DNA-Damaging Agents
While RAD54-/-/LIG4-/-, XRCC1-/-, REV3-/- and wild-type cells were exposed to γ-rays, MMS, MMC and H2O2, FANCD2-/- cells were treated with MMC only. The frequency of cells displaying MNs (the MN frequency) at 48 hrs (Fig. 3) was counted, following a protocol described in previous reports [Honma and Hayashi, 2011; Kimura et al., 2013]. Some of the MN assays were also repeated at 24 hrs (Supporting Information, Fig. 5) following OECD guideline 487, which requires the measurement of MNs after 1.5 to 2 divisions following exposure of cells to DNA-damaging agents.

The MN test in wild-type and indicated DNA- repair-deficient TK6 clones at 48 hrs after treatment with indicated DNA-damaging agents. (A, C, E, and G). B, D, F, and H indicate the frequency of micronucleated cells (%) induced by the indicated DNA-damaging agents. Error bars represent SD from at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
γ-rays: The cells were treated with γ-rays at the following levels: 0.06 Gy for RAD54-/-/LIG4-/- (RICC: 69%), 0.13 Gy for REV3-/- (RICC: 55%) and 0.25 Gy for XRCC1-/- (RICC: 69%) (Supporting Information, Fig. 1). The wild-type cells were exposed to γ-rays at all three different doses to compare the MN frequency in each mutant (Figs. 3A, 3C, and 3E). To calculate the MN frequency induced by γ-rays, we subtracted the MN frequency in irradiated cells by that in non-irradiated cells (Subtracted values shown in Figs. 3B, 3D, and 3F). The MN frequency induced by the irradiation was 6.2, 5.3 and 4 times higher in RAD54-/-/LIG4-/-, XRCC1-/- and REV3-/- cells, respectively, when compared to wild-type cells. Thus, the loss of XRCC1 and REV3 increases the MN frequency to comparable extents as the simultaneous loss of the two major DSB factors, LIG4 and RAD54. The MN frequency was also measured at 24 hrs after 0.06 Gy γ-irradiation (Supporting Information, Fig. 5A). The MN frequency induced by the irradiation was 4.1 times higher in RAD54-/-/LIG4-/- cells in comparison with wild-type cells (Supporting Information, Fig. 5A). We also treated the cells with a lower dose of γ-rays at the following levels: 0.03 Gy for RAD54-/-/LIG4-/- cells (RICC: 83%) and 0.13 Gy for XRCC1-/- cells (RICC: 116%). The MN frequency induced by the irradiation was 5.9 and 3.9 times higher in RAD54-/-/LIG4-/- and XRCC1-/- cells, respectively, when compared to wild-type cells (Supporting Information, Fig. 7).
H2O2: We exposed RAD54-/-/LIG4-/- cells (RICC: 58%) to 5 μM H2O2 and also exposed XRCC1-/- (RICC: 80%) and REV3-/- (RICC: 65%) cells to 40 μM H2O2 (Supporting Information, Fig. 1). The wild-type cells were exposed to both 5 μM and 40 μM H2O2. We then measured the MN frequency at 48 hrs (Figs. 3A, 3C, and 3E). The subtracted MN frequency values were 6.4, 5.2, and 6.3 times higher for RAD54-/-/LIG4-/-, XRCC1-/- cells and REV3-/- cells, respectively (Figs. 3B, 3D, and 3F). The MN frequency was also evaluated at 24 hrs after treatment of XRCC1-/- and wild-type cells with 40 μM H2O2 (Supporting Information, Fig. 5B). The MN frequency induced by H2O2 was 4.9 times higher in XRCC1-/- cells, when compared with wild-type cells (Supporting Information, Fig. 5B). We also exposed RAD54-/-/LIG4-/- cells (RICC: 65%) to 2.5 μM H2O2 and XRCC1-/- cells (RICC: 63%) to 20 μM H2O2. The wild-type cells were exposed to both 2.5 μM and 20 μM H2O2. The subtracted MN frequency values were 4.5 and 3.7 times higher for RAD54-/-/LIG4-/- and XRCC1-/- cells respectively, in comparison with wild-type cells. (Supporting Information, Fig. 7)
MMS: We exposed XRCC1-/- (RICC: 50%) and wild-type cells to 250 ng/ml MMS and also exposed RAD54-/-/LIG4-/- (RICC: 57%) and REV3-/- (RICC: 50%) cells to 500 ng/ml MMS (Supporting Information, Fig. 1) for measuring the MN frequency at 48 hrs. The MMS-induced MN frequency was 7.1 times higher in RAD54-/-/LIG4-/-, 5 times higher in XRCC1-/-, and 3.6 times higher in REV3-/- cells, in comparison with wild-type cells (Figs. 3B, 3D, and 3F). The measurement of the MN frequency at 24 hrs indicates that the MMS-induced MN frequency was 5.8 times higher in XRCC1-/- cells when compared to wild-type cells (Supporting Information, Fig. 5B). We also exposed XRCC1-/- (RICC: 67%) and wild-type cells to 125 ng/ml MMS and RAD54-/-/LIG4-/- (RICC: 57%) cells to 250 ng/ml MMS (Supporting Information, Fig. 1) to measure the MN frequency at 48 hrs. The MMS-induced MN frequency was 4.4 times higher in RAD54-/-/LIG4-/- and 4.8 times higher in XRCC1-/-cells, in comparison with wild-type cells (Supporting Information, Fig. 7).
MMC: The MN frequency was quantitated at 48 hrs after treatment of cells with 0.06 μg/ml MMC. The RICC values were 61% for RAD54-/-/LIG4-/-, 63% for XRCC1-/-, 51% for REV3-/-, and 71% for FANCD2-/- cells (Supporting Information, Fig. 1). The MMC-induced MN frequency was 7.5 times higher in RAD54-/-/LIG4-/-, 5.5 times higher in XRCC1-/-, 5.7 times higher in REV3-/- cells, and 5.3 times higher in FANCD2-/- cells, in comparison with wild-type cells (Figs. 3B, 3D, 3F, and 3H). We then measured the MN frequency at 24 hrs after treatment of XRCC1-/- cells with 0.06 μg/ml MMC (Supporting Information, Fig. 5A). The MMC-induced MN frequency was 6 times higher in XRCC1-/- cells in comparison with wild-type cells (Supporting Information, Fig. 5B). The MN frequency was also quantitated at 48 hrs after treatment of cells with 0.03 μg/ml MMC. The RICC values were 69% for RAD54-/-/LIG4-/- and 81% for XRCC1-/- cells. The MMC-induced MN frequency was 6.5 times higher in RAD54-/-/LIG4-/- and 4 times higher in XRCC1-/- cells, in comparison with wild-type cells (Supporting Information, Fig. 7).
DISCUSSION
We prepared a panel of isogenic DNA-repair-deficient TK6 cell lines and measured the MN frequency following exposure of the cells to four widely used DNA-damaging agents: γ-rays, H2O2, MMC, and MMS. All TK6 mutants tested in the present study showed markedly higher MN frequency over parental TK6 cells. The number of MNs induced by γ-rays, H2O2, MMS and MMC increased 6.2, 6.4, 7.1 and 7.5 times, respectively, in RAD54-/-/LIG4-/- cells in comparison with wild-type cells. The number of MNs induced by γ-rays, H2O2, MMS and MMC all increased over 5 times in XRCC1-/- cells in comparison with wild-type cells. These results strongly suggest that examining the differential MN frequency between isogenic TK6 cells proficient and deficient in DNA repair pathways could be a valuable supplementary analysis to clarify the genotoxicity of presumed non-genotoxic chemicals and physical agents as well as to investigate their mode of action.
We next investigated the dose response of DNA damaging agent-treated RAD54-/-/LIG4-/- and XRCC1-/- cells by performing the MN test at a dose/concentration two times lower than that at the highest RICC. We found a dose-dependent increase in the MN formation in RAD54-/-/LIG4-/- and XRCC1-/- cells. RAD54-/-/LIG4-/- cells showed a 1.6 times higher level of MN induction than XRCC1-/- cells at a two times lower dose of gamma-rays (RAD54-/-/LIG4-/- cells, 6.2 times higher MN induction at 0.06 Gy vs XRCC1-/- cells, 3.9 times higher at 0.13 Gy, all compared to wild-type). Moreover, even at very low dose of gamma rays (0.03 Gy), RAD54-/-/LIG4-/- cells displayed a 5.9 times higher induction of MN than wild-type cells. These MN data suggest that in order to examine and understand gamma-ray-induced genotoxicity, RAD54-/-/LIG4-/- cells are more useful than XRCC1-/- cells. This observation is plausible, as gamma rays directly cause double strand break (DSB) formation to DNA, and RAD54 and LIG4 are involved in the DSB repair by HR and NHEJ pathways, respectively.
In terms of the MN induction by H2O2, we found that at 20 µM of H2O2, XRCC1-/- cells show an induction of MN frequency at 3.7 times higher in comparison with wild-type cells whereas even at a low dose of H2O2 (5 µM), induction of MN frequency in RAD54-/-/LIG4-/- cells is 6.4 times higher in comparison with wild-type cells. Therefore, a 4-fold lower concentration of H2O2 can induce a 1.7 times higher level of MN in RAD54-/-/LIG4-/- cells over XRCC1-/- cells. These MN data also suggest that in order to examine and understand H2O2-induced genotoxicity, RAD54-/-/LIG4-/- cells may be more useful than XRCC1-/- cells because H2O2 efficiently generates single-strand breaks (SSBs) and SSBs are often converted to double-strand breaks (DSBs).
The current study demonstrates the utility of RAD54-/-/LIG4-/- TK6 cells for detecting various DNA-damaging agents and their mode of action. The data are in agreement with the reported efficient detection of the genotoxic potential of chemicals from the National Toxicology Program chemical library using RAD54-/-/KU70-/- chicken DT40 cells [Nishihara et al., 2016]. The clear advantage of the RAD54-/-/LIG4-/- TK6 mutant is that they are capable of proliferating with nearly normal kinetics, show a plating efficiency (60%) comparable to wild-type cells (70%), and exhibit relatively low spontaneously arising MNs (Fig. 3A). As expected from previous studies analyzing cells deficient in both HR and NHEJ [Fukushima et al., 2001; Couëdel et al., 2004; Mills et al., 2004], the RAD54-/-/LIG4-/- TK6 cell line is valuable for detecting MNs caused by agents that directly generate DSBs as well as for determining their mode of action. Moreover, the increased detection of MN in RAD54-/-/LIG4-/- cells after H2O2 and MMS treatment is interesting since these two agents generate DSBs with low efficiencies; instead, they have been found to induce mutagenesis and apoptosis mainly though interference of DNA replication by base damage [Hochegger et al., 2004; Branzei and Foiani, 2010]. The LIG4-dependent DSB repair pathway (NHEJ) is incapable of repairing DSBs that occur during DNA replication, such as DSBs induced by the topoisomerase I poison, camptothecin and the poison against poly[ADP ribose]polymerase, olaparib [Hochegger et al., 2006; Murai et al., 2012; Maede et al., 2014; Kobayashi et al., 2015]. In addition, RAD54 plays only a minor role in repairing DSBs that occur during DNA replication in comparison with RAD51, a factor essential for HR; as such, RAD54-deficient mice are viable while the loss of RAD51 is lethal to cells, causing numerous DSBs during DNA replication [Sonoda et al., 1998; Eppink et al., 2011]. Considering the fact that ionizing-radiation induces far more base damage and SSBs compared to DSBs [Bradley and Kohn, 1979], efficient induction of MNs in XRCC1-/- and REV3-/- cells likely reflects the important roles for REV3 and XRCC1 in preventing conversion of base damage and SSBs to DSBs and MNs through DNA replication. Further studies are required for elucidating the molecular mechanism underlying MN induction associated with H2O2 and MMS exposure. Based on these results, we propose that the application of RAD54-/-/LIG4-/- TK6 cells in the conventional MN test would be significant for understanding the mode of action of genotoxic agents.
In the present study, XRCC1-/- and REV3-/- TK6 cells were created, as we expected a marked increase in the MN frequency caused by genotoxic agents that introduce base damage. In fact, REV3-/- chicken DT40 cells are very sensitive to a wide variety of DNA-damaging agents including tamoxifen, 4-hydroxyestradiol and nitric oxide [Mizutani et al., 2004; Wu et al., 2006]. The current study indicates that XRCC1-/- TK6 cells are more useful for the supplemental MN test than REV3-/- TK6 cells for the following reasons. 1) The plating efficiency of XRCC1-/- cells is better than that of REV3-/- cells (data not shown). 2) XRCC1-/- cells show a more prominent phenotype in terms of MMS-induced toxicity (Fig. 2) and MN frequency (Fig. 3) in comparison with REV3-/- cells. A disadvantage of XRCC1-/- cells is that bulky adducts, including UV-lesions, sensitize REV3-/- cells but not XRCC1-/- cells [Horton et al., 2008; Takezawa et al., 2011]. Thus, including translesion DNA synthesis- (TLS-) deficient TK6 cells other than REV3-/- cells in the MN assay may be recommended for determining the mode of action of a wide variety of mutagenic chemical compounds. This is because a number of TLS polymerases collaboratively undergo DNA synthesis past various types of DNA lesions [Sale et al., 2012]. Moreover, TLS dominantly contributes to mutagenesis during the physiological cell cycle [Gibbs et al., 2005]. To create a TLS-deficient TK6 mutant to detect a wide variety of mutagenic potential, we need to create TK6 cells deficient in multiple TLS polymerases, as we have done for the DT40 cell line [Yoshimura et al., 2006; Kohzaki et al., 2010]. Another possible strategy is disruption of a gene responsible for excision-repair of UV-lesions in the XRCC1-/- background.
In this study, we also conducted a MN test using a XPA-/- cell line after exposure to four DNA damaging agents. As expected from the role of XPA in nucleotide excision repair but not in BER or SSB repair, XPA-/- cells showed no significant increases in MN induction in comparison with wild-type cells upon exposure to H2O2 or MMS (Supporting Information, Fig. 6), which is in contrast with the prominent phenotypes in RAD54-/-/LIG4-/- and XRCC1-/- cells (Fig. 3). Neither gamma rays nor MMC increased MN induction in XPA-/- cells in comparison with wild-type cells.
After the identification of the genotoxicity of chemical compounds via in vitro assay screening, molecular mechanisms underlying the genotoxicity may need to be clarified. The genotoxicity of chemicals is often caused by a few independent mechanisms. In fact, we previously revealed that sodium meta arsenite causes DNA damage through at least two different mechanisms, one depending on the formation of reactive-oxygen-species and the other being independent [Ji et al., 2009]. Thus, to understand mechanisms underlying the mutagenic potential of individual chemical compounds, we need to accurately quantitate the relative contribution of multiple DDR pathways to the repair of different types of lesions induced by each test chemical. Accurate quantitation may be a formidable challenge since the shape of dose response curves of cellular survival may distinctly differ depending on the DDR genes disrupted in mutant cells as well as on chemical compounds. In the current study, we simply compared the induced MN frequency between one of the mutant clones and wild-type cells after exposure of these two clones to the same dose of DNA-damaging agents. While this protocol shows higher MN frequency of the isogenic mutants to typical DNA-damaging agents in the MN test, more time-consuming experimentation is required to accurately quantitate the relative contribution of multiple DDR pathways as we have done for DT40 isogenic mutants [Maede et al., 2014; Nishihara et al., 2016]. Thus, in the future, a practical MN protocol needs to be established to comprehensively investigate the molecular mechanisms underlying the genotoxicity, in addition to a MN protocol for simply identifying the genotoxic potential of test chemical compounds.
ACKNOWLEDGMENTS
We are grateful to M. Kato and A. Kobayashi for technical assistance.
STATEMENT OF AUTHOR CONTRIBUTIONS
S.T. and L.K.S. designed the basic framework of this study. L.K.S performed all experiments with the help of S.K., S.A. and H.K. L.K.S. prepared the manuscript draft with intellectual input from S.T. J.N. and S.T. finalized the manuscript. T.S. and T.Y. provided information for designing some of the experiments. M.H. provided isogenic TK6 wild-type cells. H.S and K.H advised L.K.S about technical issues during the experimentation. All authors have approved the final manuscript.