Identification of a new epitope for HIV-neutralizing antibodies in the gp41 membrane proximal external region by an Env-tailored phage display library
Abstract
HIV controllers are a valuable source for the identification of HIV-neutralizing antibodies, as chronic infection over decades allows extensive affinity maturation of antibodies for improved Ag recognition. We analyzed a small cohort of elite controllers (ECs) for HIV-neutralizing antibodies using a panel of standardized HIV-1 pseudovirions on TZM-bl cells. An HIV-1 Env-tailored phage display library was generated to select epitopes targeted by neutralizing antibodies in the EC26 plasma sample showing the broadest neutralizing activity. Selected Env fragments were mostly allocated to the membrane proximal external region of gp41. After preabsorbing the EC26 plasma with the selected phage EC26-2A4, we achieved 50% depletion of its neutralizing activity. Furthermore, antibodies affinity-purified with the EC26-2A4 epitope from EC26 plasma showed neutralizing activity, proving that the selected phage indeed contains an epitope targeted by neutralizing plasma antibodies. Epitope fine mapping of the purified plasma antibodies on peptide arrays identified a new epitope overlapping, but clearly distinct, from the prominent 2F5 epitope. Of note, the purified antibodies did not show autoreactivity with cardiolipin, whereas low reactivity with phosphatidylserine comparable to mAb 2F5 was observed. Thus, this new epitope represents a promising candidate for further analysis in view of HIV vaccine development.
Introduction
Eliciting broadly neutralizing antibodies (bnAbs) against HIV-1 is a major goal in HIV vaccine development 1. However, despite the identification of a number of epitopes for bnAbs present in patient sera 2, these epitopes failed so far to induce such antibodies after vaccination in vivo 3-8. The mechanism behind the elicitation of bnAbs during natural infection remains unclear. Emerging evidence indicates that most HIV-1-specific antibodies elicited during early infection fail to neutralize a wide-spectrum of HIV-1 strains and emphasizes the importance of analyzing the Ab profile of patients showing delayed disease progression to allow for sufficient affinity maturation 9. A subgroup of HIV-1-infected individuals controlling the infection over years in the absence of antiretroviral therapy and maintaining serum viral loads (VL) below the detection limit (<50 copies/mL) is termed “elite controllers” (ECs) 10. Although virus control is certainly due to a potent HIV-specific immune response 10, few qualitative differences in immune responses have been identified between controllers and viremic individuals. The differences encountered so far mainly concern the cellular immune response and revealed an overrepresentation of certain HLA alleles in controllers, in particular B*57:01 and B*27:05, as well as an enrichment of the CCR5Δ32bp haplotype 11. Regarding humoral immunity in controllers and progressors, no major qualitative differences have been reported so far 12. In particular concerning ECs, mapping of epitopes targeted by cross-clade bnAbs is still rudimentary. Deciphering these immunogenic epitopes is of considerable interest since they may represent potential vulnerabilities of the HIV-1 envelope and may help to design more effective immunogens able to elicit neutralizing antibodies upon vaccination 13.
We previously identified epitopes for bnAbs in sera from long-term nonprogressors (LTNPs) by dissecting the humoral immune response based on phage libraries displaying random peptides from 7 to 12 aas in length 14. In the present study, we extended these analyses to ECs and identified a subgroup with bnAbs in plasma by standardized pseudovirus-based neutralization assays 15. In order to dissect the HIV-specific Ab profile in these ECs, we generated an Env-tailored phage library displaying Env fragments from 30 to 100 aas from the HIV-1ADA strain. A similar approach has been previously used to identify immunogenic polypeptides from Mycoplasma hypopneumoniae and M. Mycoides 16, 17. The Env-tailored phage library was used for affinity selection (biopanning) of Env-epitopes targeted by bnAbs from EC26 plasma, which ranked top in our neutralization assays.
Analysis of the selected clones revealed that the majority could be allocated to the viral transmembrane protein gp41 including the membrane proximal external region (MPER). This region is known to contain the epitopes for the potent bnmAbs 2F5, 4E10, and Z13, which neutralize 80–100% of the tested primary HIV-1 across the clades 18, 19. Usually, neutralizing antibodies targeting the MPER are not frequently detected in patient plasma 9, 20-24, which was mainly attributed to their polyreactivity with autoantigens 25-27. So far, only one study described neutralizing antibodies directed against an epitope overlapping with that of mAb 2F5 28, 29. By screening the Env library with IgG from plasma EC26, we identified a novel epitope in the MPER region of gp41, which overlaps, but is clearly distinct from the 2F5 epitope. This epitope, EC26-2A4, may represent an interesting vaccine candidate, as (i) antibodies affinity purified with the epitope from the parental EC plasma show neutralizing activity, and (ii) the purified antibodies, despite reactivity with phosphatidylserine comparable to mAb 2F5, do not exhibit significant autoreactivity with cardiolipin.
Results
Identification of EC plasma with broad neutralizing capacity
We recruited a cohort of 12 ECs and 2 viremic controllers (VCs) for this study, the characteristics of which are summarized in Supporting Information Table 1. To characterize the nAb profile in plasma from these ECs/VCs, we first used tier 1 Env-pseudotyped viruses HIV-1SF162.LS, HIV-1Bal.26, and HIV-1ss1196.1 in a standardized TZM-bl assay 15. Supporting Information Table 2 summarizes the neutralizing Ab titers of the plasma samples expressed as the reciprocal plasma dilution resulting in 50% inhibition of the infection (IC50). Strain SF162.LS was neutralized by all EC/VC plasma samples except for EC06 and VC12, with IC50 values ranging from 178 to >12,150. Plasma samples from EC07, EC08, EC09, EC10, EC26, and VC11 neutralized all three subtype B tier 1 strains with titers comparable or better than the broadly neutralizing control sera VI-2992 and VI-3196, which are, however, derived from a subtype A infected individual 30. Plasma from EC26 ranked top against HIV-1SF162.LS and HIV-1ss1196.1 and ranked second against HIV-1Bal.26. The breadth of nAbs of EC26 plasma was therefore further determined against an Env pseudotyped virus panel of 11 tier 2 isolates from subtypes A, B, C, and CRF BC. EC26 plasma neutralized 10/11 viruses out of the tier 2 panel with titers comparable with broadly neutralizing plasma samples from the control patient ITM1 (IC50 values between 21 and 76, data not shown). Due to the broad neutralizing activity of the EC26 plasma sample, we continued using this sample to identify the respective epitopes targeted by neutralizing antibodies.
Selection of epitopes for nAbs in EC26 plasma based on an HIV-1 Env-tailored phage display library
Neutralizing antibodies target various regions of the HIV-1 envelope involved in virus entry. In order to select such Env epitopes, we generated a phage display library displaying Env peptides from shotgun cloned PCR fragments derived from the entire gp160 sequence of the HIV-1ADA strain at the N-terminus of the pIII phage protein. Randomly analyzed library clones showed that about 15% of the clones contained HIV-1ADA env DNA fragments positioned in frame with the pelB leader and the phage gIII gene and encoding Env open reading frames of 30–100 aas.
The biopanning procedure was initiated by a negative selection with immobilized IgGs from pooled HIV negative plasma. Subsequently, positive selections were performed with the depleted library and IgG from the EC26 plasma sample. After two rounds of negative and positive selections, 150 phage clones were analyzed by ELISA to determine their binding specificity for EC26 IgG. From these, the 22 most reactive clones were subjected to sequence analysis. By comparing the peptide insert sequences to HIV-1ADA Env sequences, the 22 sequenced clones could be grouped into five distinct clusters. The ELISA reactivity of one representative phage of each group is shown in Figure 1A. Peptide inserts showed linear homology to Env or to the second exon of Rev. Two sequences could be allocated completely to gp41, one mapped to the C-terminus of gp120, but extended into the N-terminal part of gp41 (Supporting Information Fig. 1). Clone EC26-2H10 covered the well-known KLIC motif, a linear immunodominant region corresponding to gp41 cluster II 21, 31. An interesting clone, EC26-2A4, mapped to the MPER region of gp41 known to be targeted by broadly neutralizing mAbs 2F5, 4E10, and Z13. The peptide insert of the clone EC26-2A4 (IEESQNQQEKNEQELLELDKWASLWNWFD) contains the entire 2F5 epitope (ELDKWA), whereas the 4E10 epitope (NWFDIT) is only partially contained at the C-terminus. To characterize further the binding activity of the epitope EC26-2A4 with mAbs 2F5 and 4E10, we performed Ab phage ELISA. As expected from the sequence data, EC26-2A4 strongly bound mAb 2F5, whereas mAb 4E10 was not bound (Fig. 1B).
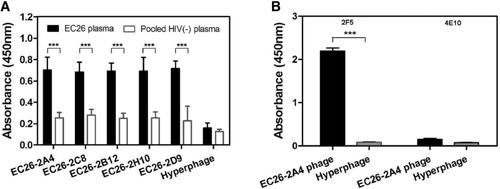
Epitope EC26-2A4 is targeted by HIV-1 neutralizing antibodies present in plasma EC26
To clarify whether the selected EC26-2A4 peptide is targeted by neutralizing antibodies contributing to the neutralizing activity of plasma from EC26, we first preabsorbed EC26 plasma with the purified phage EC26-2A4 or Hyperphage (control without peptide insert), followed by neutralization studies of the depleted plasma. Preincubation with phage EC26-2A4 resulted in a significant reduction in the neutralizing activity of EC26 plasma against HIV-1SF162.LS compared with that of plasma preincubated with the control Hyperphage (Fig. 2B). The IC50 values of EC26 plasma samples preincubated with EC26-2A4 phage and Hyperphage, respectively, differed significantly (11,547 versus 20,535). Likewise, preincubation of mAb 2F5 with the EC26-2A4 phage resulted in reduced neutralization compared with that of preincubation with Hyperphage (Fig. 2A). The fact, that preincubation with Hyperphage also reduced the mAb 2F5 neutralizing activity, is probably due to unspecific blocking due to the large size of the phage coat (>2500 copies for the pVIII protein) as opposed to the three to five copies of the specific epitope displayed on the phage linked to the pIII protein.
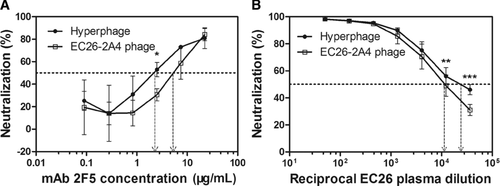
We then affinity purified IgG specific for the EC26-2A4 peptide from EC26 plasma with a fusion protein (EC26-2A4-MBP), where EC26-2A4 has been fused N-terminally to the maltose-binding protein (MBP). This fusion protein showed similar binding for EC26 IgG as the original EC26-2A4 phage (data not shown). The purified IgG fraction (referred to as EC26-2A4-IgG) was then analyzed for neutralization of HIV-1 on TZM-bl cells. A total of 50 μg/mL EC26-2A4-IgG neutralized 91.8 % of SF162.LS, 53.1% of ss1196.1, and 48.3% of Bal.26, respectively (Fig. 3). Thus, EC26 plasma indeed contains neutralizing antibodies targeting the MPER peptide EC26-2A4, which contribute significantly to the neutralizing activity in the plasma.
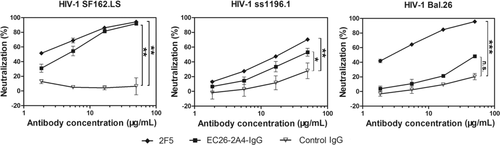
As neutralizing mAbs against MPER, in particular mAb 4E10 and to a lesser extent 2F5, are associated with autoreactivity against phospholipids, we analyzed the reactivity of plasma EC26 and the antibodies affinity purified with EC26-2A4-MBP for reactivity with phospholipids. The plasma as well as the purified EC26-2A4-IgG did not show significant reactivity with cardiolipin by ELISA at 100 μg/mL, which is a clinically relevant concentration 32, 33 (Fig. 4A); however, the purified antibodies showed low reactivity with phosphatidylserine, which was comparable with that of mAb 2F5 (Fig. 4B). The positive control, mAb 4E10, showed strong reactivity with cardiolipin as well as with phosphatidylserine at the same concentration, whereas the negative control mAb 447-52D did not react with either of the Ags.
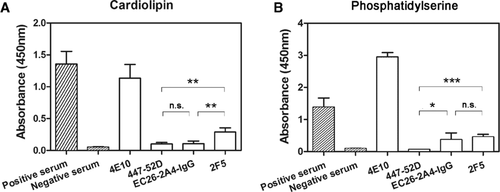
The core epitope of EC26-2A4-IgG is distinct but overlaps with the epitope of mAb 2F5
The core epitope for purified EC26-2A4-IgG was further fine mapped on peptide arrays (Fig. 5). By comparing the reactivity of sets of overlapping peptides shifted by one aa with mAb 2F5 and purified EC26-2A4-IgG we could define more precisely the core epitope recognized by EC26-2A4-IgG. As expected from published studies, mAb 2F5 recognized the core epitope LDKW on these arrays, whereas purified EC26-2A4 IgG targeted an epitope partially overlapping the 2F5 epitope, but slightly shifted toward the N-terminus. This new core epitope with the sequence (659ELLELDKW666) is termed EC26-2A4MZ. W666 was absolutely essential for the reactivity with mAb2F5, whereas the absence of this aa weakened the reactivity with EC26-2A4-IgG, but did not abolish it completely. Furthermore, the EC26-2A4MZ core epitope includes E659, which is neither present in the 2F5 epitope nor in the recently described m66.6 epitope (see below).

Discussion
bnAbs are a major component of protective immunity against human/simian immunodeficiency viruses (SHIV), as shown in numerous animal studies 34-41. Therefore, the identification of such antibodies as well as their epitopes in HIV-infected persons is a high priority in HIV vaccine development 1, 42-46. In humans, bnAbs have mostly been identified in persons chronically infected with HIV-1, as broad neutralizing activity usually is associated with extensive Ab affinity maturation 30, 47. A particular rich source for bnAbs are LTNPs or viremic controllers, who are infected by HIV-1 over decades and show low but detectable virus replication that stimulates an efficient immune response. For ECs, generally, the autologous neutralizing Ab titers have been reported to be lower than in viremic patients 48. Here, we identified ECs with bnAbs from a small cohort and profiled the immune response of the most active plasma (EC26) using an Env-tailored phage displayed library.
Besides the few well-known bnmAbs b12, 2G12, 2F5, 4E10, and Z13 1, recently exceptionally broad neutralizing antibodies have been identified using new methodological approaches based on sorting and cloning of single Env-specific B cells from patients with broadly neutralizing activity in plasma and the use of engineered Env Ags for screenings 2, 7, 47, 49-53. Other HIV-neutralizing antibodies as well as the corresponding epitopes were identified by the phage display technology 14, 54-58. Interestingly, profiling the neutralizing specificities in the plasma samples with broad neutralizing activities showed that these were generally directed against the gp120 part of the envelope spike, especially the CD4-binding site, and very rarely targeted the gp41 component 20, 22, 23, 30, 51, 59-62. Consequently, only a few neutralizing mAbs targeting gp41 are known. Most prominent are the broadly neutralizing patient-derived mAbs 2F5, 4E10, and Z13, targeting the MPER region of gp41 in the context of the viral membrane 19, 63-69. Recently, a new mAb, CAP206-CH12, targeting an MPER epitope overlapping with the 4E10 epitope was identified by Ag-specific B-cell sorting 7. During submission of this manuscript, another broadly neutralizing mAb (10E8) was published online, that was isolated from an individual with broadly neutralizing mAbs and recognizes an epitope overlapping with mAb 4E10 70. Additional Ab fragments have been derived from phage libraries and include D5 71, m44 58, and Fab 3674 72, as well as mAb HK20 73, all interfering with the formation of the six helix bundle structure, an essential fusion intermediate.
Although the MPER region is immunogenic, and occasionally neutralizing activities of broadly neutralizing patient sera can be mapped to this region 20, 22, 61, 74, neutralizing antibodies overlapping with the 2F5 epitope are extremely rare. This may be due to the lipophilic character and limited accessibility of the MPER epitopes as well as the different conformations the MPER region adopts during virus maturation and entry 75. Nevertheless, such antibodies are functionally important and have recently been associated with protection in mother-to-child transmission 76. Besides that, only one study by Shen et al. 28 identified neutralizing antibodies overlapping with the 2F5 epitope in 1 out of 311 plasma samples (<0.3%) from subtype B infected subjects. Recently, an optimized mAb (m66.6) from this patient recognizing the core sequence of the 2F5 epitope in conjunction with two more upstream leucines was described, which had a more narrow neutralizing profile compared with mAb 2F5 and showed strong polyreactivity with a number of autoantigens like mAb 2F5 29, 77.
In the present study, we identified neutralizing antibodies from a broadly neutralizing EC plasma sample (EC26) that also target an MPER epitope partially overlapping with the 2F5 epitope. This epitope, EC26-2A4, was selected from our Env-tailored phage display library with IgG from patient EC26. Interestingly, most epitopes selected with EC26-IgG clustered in gp41 or the second exon of Rev. This was not due to a bias in our Env-tailored phage library, since (i) epitopes in gp120 are also present in our library (Supporting Information Fig. 1) and (ii) screening of the same library with a different EC sample resulted preferentially in V3 epitopes (Zhou et al., unpublished data). This rather points at a patient-specific dominance of certain epitopes recognized by the respective antibodies in the plasma. Although the epitopes selected from the Env-tailored phage library, which were analyzed in this study, correspond to linear epitope sequences, potentially the library could also contain conformational epitopes due to the large insert sizes or to random junction of Env fragments during shotgun cloning.
The EC26-2A4 peptide was used to affinity-purify binding IgG from EC26 plasma. Purified EC26-2A4-IgG neutralized HIV-1SF162.LS with an IC50 < 6 μg/mL validating the EC26-2A4 epitope as target for neutralizing antibodies present in plasma EC26. Although the corresponding phage peptide contains the entire 2F5 epitope at its C-terminus, fine mapping of the epitope with the purified neutralizing EC26-2A4-IgG on peptide arrays (Fig. 5) clearly showed that the core epitope (EC26-2A4MZ: 659ELLELDKW666) is shifted toward the N-terminus with respect to the 2F5 epitope (LDKW). Of note, whereas the presence of W666 is absolutely essential for the reactivity with mAb 2F5 as shown in Figure 5 and published studies 19, 78, the absence of W666 only reduces slightly the reactivity with EC26-2A4-IgG, further underlining the differences between the two epitopes. Interestingly, L660 and L663 also contribute to the epitope of mAb 66.6 mentioned above 29. However, E659 is part of the core epitope EC26-2A4MZ, but not of the m66.6 epitope. Thus, the EC26-2A4MZ core epitope overlaps, but is different from the epitopes for mAbs 2F5 and m66.6.
A characteristic of broadly neutralizing MPER mAbs 4E10, 2F5, and m66.6, but not the recently described mAb 10E8 70, is their polyreactivity with autoantigens 28, 29, 77. Therefore, we analyzed the reactivity of the purified EC26-2A4-IgG with cardiolipin and phosphatidylserine. Whereas the low reactivity of EC26-2A4-IgG with phosphatidylserine was comparable to mAb 2F5, interestingly, there was no significant reactivity with cardiolipin at a clinically relevant Ab concentration 77. Thus, the EC26-2A4 epitope may represent an interesting vaccine candidate. In this regard, it is remarkable that we could induce moderate epitope-specific neutralizing activity against HIV-1SF162 in mouse sera upon vaccination with a DNA prime-phage boost protocol 56, 79 with EC26-2A4 epitope coupled to different carriers 80. However, further studies have to show the specificity and breadth of neutralization for the new MPER epitope. Concerning the EC26-2A4-IgG, once the Ab genes have been isolated, Ab affinity and breadth of neutralization potentially could be further optimized.
Materials and methods
Subjects studied
Plasma samples from ECs and viremic controllers were obtained from the Hospital Carlos III in Madrid (Spain) and the HIV Center from the Goethe Medical School in Frankfurt (Germany) after receiving informed consent. Twelve ECs enrolled had (i) documented HIV-1 infection for more than 10 years and (ii) persistently undetectable VL < 50 copies per mL plasma in the absence of any antiretroviral therapy. Two viremic controllers had VL from 100 to 150 copies/mL plasma. Control sera VI2992 and VI3196, kindly provided by Guido Vanham (Antwerpen), were from a therapy naïve HIV-1 subtype A infected patient ITM-1, and previously reported to broadly neutralize primary isolates on PBMC and TZM-bl cells 30. Further, control sera for the neutralization studies were previously published 14 and include LTNPs MH01 and MH04 with high titers of neutralizing antibodies against a panel of recombinant isolates from clade B and C and sera from four progressors with comparable CD4 counts (>400 per mL) and low VL (<11,000 copies per mL) (Supporting Information Table 1). The study was approved by the Ethics Commission of the Goethe University of Frankfurt am Main, Germany.
Neutralization assays on TZM-bl cells
Neutralization studies were performed using a panel of standardized HIV-1 pseudoviruses on TZM-bl cells 15. All plasma samples were heat-inactivated at 56°C for 1 h prior to use. Neutralizing Ab titers of all samples were first analyzed against three standard HIV-1 reference subtype B tier 1 strains (SF162.LS, SS1196.1, and Bal.26). The breadth of nAbs in the best neutralizing samples was further determined against HIV-1 tier 2 pseudoviruses derived from standard primary isolates of subtype B (THRO4156.18, WITO4160.33, RHPA4359.7, TRJO4551.58, and REJO4541.67), subtype A (Q769.d22 and Q23.17 ), subtype C (DU156.12 and DU422.1), and CRF BC (CH110.2 and CH117.4) strains as previously described 15. Briefly, pseudoviruses were produced by cotransfection of 293T/17 cells with an HIV-1 env-expressing vector and an env-deficient backbone vector (pSG3Δenv) and titrated on TZM-bl cells. After preincubation of the pseudovirus stock with serial dilutions of plasma or Abs for 1 h at 37°C, TZM-bl cells were infected in a 96-well plate for 48 h. Relative luminescence units were determined and the 50% inhibitory dose (IC50) was determined compared with virus only controls.
Construction of an HIV-1 Env-tailored phage display library
The env gene of HIV-1ADA was amplified from the pSV3-ADA env plasmid (kindly provided by Dr. Rolf Kaiser, Cologne) and the PCR product was sonicated to generate random DNA fragments with a size of 100–1200 bp. After purification and polishing, the fragments were cloned into the pHORF3 vector 16 and electroporated into electro-competent TOP10F′ (Invitrogen). The grown colonies were harvested by suspending in 40 mL 2× tryptone yeast medium. The library was either directly used for phage packaging using Hyperphage 81, 82, or after addition of glycerin (20%), the library was stored at −80°C in 1 mL aliquots.
Colony PCR and sequence analysis of inserts
The size distribution of the inserts from single clones bearing pHORF3 was analyzed by colony PCR using the MHLacZ-Pro-f primer (5′-GGCTCGTATGTTGTGTGG-3′) and MHgIII-r primer (5′-GGAAAGACGACAAAACTTTAG-3′) 83 following the protocol: 94°C, 1 min; 56°C, 1 min; 72°C, 2 min; 30 cycles. Plasmids from 136 clones bearing pHORF3 were purified and sequenced using the same primer pair on an ABI PRISM 310 Genetic Analyzer to determine the representation of different env regions in the library.
Selection of phage reactive with HIV antibodies from EC
Plasma samples from ECs and pooled HIV negative plasma were diluted 1:100 in 2% MPBST and added to microtiter plates coated with a goat antihuman Fc Ab for 2 h at RT. After washing, 100 μL (∼107 phage particles) of the HIV-1 Env-tailored phage display library were first incubated with the wells containing pooled HIV negative IgGs for 2 h at RT (negative selection). Then supernatants were transferred into wells coated with the IgGs from EC for 2 h at RT. The nonbinding polypeptide phage particles were removed by ten stringent washing steps with PBST. After washing, the bound phage were eluted with 200 μL/well Trypsin (10 μg/mL) for 30 min at 37°C. Ten microliters of the eluted phage were used for titration as reported 84. A total of two rounds consisting of negative and positive selections were performed. Single phage clones were produced as previously described 16.
Phage capture ELISA
Twenty-five nanograms of mouse anti-M13 (B62-FE2, Progen) in PBS were coated at 4°C overnight followed by blocking with 2% MPBST. After washing three times with PBST, 100 μL of the monoclonal phage were added for 2 h. Plasma from ECs or pooled HIV negative plasma samples were diluted 1:100 in 2% MPBST supplemented with 10% v/v TOP10F' cell lysate and 1 × 108 CFU/mL Hyperphage, added to the captured phage particles and incubated for 2 h at room temperature. Bound IgGs were detected with HRP-conjugated goat antihuman IgG for 1 h and visualized with TMB microwell peroxidase substrate (KPL, USA). The staining reaction was stopped by adding 100 μL 1 N HCl. The absorbance at 450 nm and scattered light at 620 nm were measured as reference using an ELISA reader (MWG-BIOTECH).
Identification of selected epitopes
Plasmids from clones with strong ELISA reactivity were purified and the inserts sequenced with the above-mentioned MHLacZ-Pro-f and MHgIII-r primers. Deduced aa sequences were compared with published HIV-1 protein sequences (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) and similarities with known HIV-1 neutralizing Ab epitopes were studied using the online HIV database (http://www.hiv.lanl.gov/content/index).
Depletion of epitope-specific neutralizing antibodies from plasma
Plasma or antibodies were preincubated with 106 CFU of twice PEG-purified and filtered EC26-2A4 phage/Hyperphage or MBP-EC26-2A4 fusion protein (see below, 10 μg/mL) for 1 h at 37°C followed by neutralization assays on TZM-bl cells as described above.
Cloning of selected phage epitopes in fusion with MBP
The peptide insert EC26-2A4 was amplified from the phage by PCR and cloned N-terminally to the MBP into the expression vector pMAL-pIII (NEB). Briefly, EC26-2A4 was amplified using the primers EC26-2A4-fw (5′- CGG GGT ACC TAT TGA AGA ATC GCA GAA CCA GCA A-3′) and EC26-2A4-rev (5′-TCC CCG GCC GAG TCA AAC CAA TTC CAC AAA CTT GC-3′) to introduce restriction sites KpnI and EagI, respectively, for cloning. After sequence analysis, the EC26-2A4 peptide fused to MBP was expressed following the NEB manual, purified by amylose column (NEB) and quantified using the BCA assay kit (Thermo).
ELISA
Plates were coated with 200 ng/well proteins in PBS at 4°C overnight. After washing three times, plates were blocked with 5% MPBST for 2 h at room temperature and incubated overnight with plasma diluted 1:100. Plates were washed three times and incubated with HRP-conjugated antihuman IgG antibodies for 1 h at room temperature. After washing five times, plates were developed using 100 μL/well Sureblue TMB substrate (KPL), stopped with 100 μL 1 N HCl and read at 450 nm and 620 nm as a reference. Reactivity of selected phage clones with mAbs 2F5 and 4E10 was tested by coating the plates with 200 ng/well antibodies, washing and blocking as above, followed by incubation with selected monoclonal phage, which were then detected by anti-M13-HRP Ab. ELISAs to determine the reactivity of plasma and purified IgG against cardiolipin and antiphosphatidylserine IgG were performed using the Cardiolipin IgG ELISA kit (Abnova) and the antiphosphatidylserine ELISA kit (Biorad) using the included controls and according to the manufacturer's instructions.
Affinity purification of plasma antibodies with the selected epitope
Purification of EC26-2A4-binding antibodies from EC26 plasma was performed using the AminoLink Plus kit (Thermo) according to the manufacturer's instructions. Briefly, 1.5 mg EC26-2A4-MBP proteins were coupled to the resin. Plasma samples were injected into the fusion protein conjugated column and incubated overnight at 4°C. After washing, the binding fraction was eluted by IgG elution buffer (Thermo). The eluted fractions were combined and concentrated to 100 μg/mL using Amicon 30 centrifugal filter units.
Mapping the core epitope of EC26-2A4 purified IgG by spot synthesis peptide arrays
Peptide arrays were synthesized by Fmoc chemistry at activated PEG spacers on cellulose membranes by automated parallel peptide synthesis on a MultiPep RS instrument (Intavis) 85. Overlapping peptides of the EC26-2A4-epitope were synthesized consisting of 18mers, 15mers, 12mers, 10mers, 8mers, and 6mers, each shifted by one aa. After rehydration, washing, and blocking with 5% (w/v) skimmed milk powder in PBS supplemented with 0.5% Tween-20 (5% MPBST) for 3 h at RT, membranes were incubated with 500 ng/mL 2% MPBST diluted affinity purified plasma antibodies or 2F5 mAb as positive control at 4°C overnight. After washing three times with PBS, bound antibodies were detected using the ECL kit (Thermo).
Statistical analysis
The graphs including mean and SD were performed using Graph Pad Prism5 (Graph-Pad Inc., San Diego, CA, USA). To strengthen our results, we used two-way or one-way ANOVA followed by t-test for all statistical analyses with the help of Graph Pad Prism5, if not indicated otherwise. Statistically significant p values are marked by asterisks in the graphs (***p < 0.001; **p < 0.01; *p < 0.05).
Acknowledgments
We thank Margot Landersz and Peter Prochir for sequencing. Special thanks to Guido Vanham and Leo Heyndrickx (Institute of Tropical Medicine, Antwerp) for providing broadly neutralizing control sera. The pseudovirus stocks were generated in close collaboration with the Comprehensive Antibody Vaccine Immune Monitoring Center CA-VIMC at Duke University. Special thanks to Michael Humbert for critical reading of the manuscript. This project was supported by the Deutsche Forschungsgemeinschaft (GRK1172; UD/MZ). The Georg-Speyer-Haus is supported by the Federal Ministry of Health and the Ministry of Higher Education, Research and the Arts from the state of Hessen. The HIV Specimen Cryorepository HSC greatfully acknowledges the support by the Bill & Melinda Gates Foundation (Collaboration for AIDS Vaccine Discovery (CAVD), Grant no. OPP38580_01; HvB).
Conflict of interests
The authors declare no financial or commercial conflict of interest.
References
Abbreviations
-
- bnAb
-
- broadly neutralizing antibodie
-
- EC
-
- elite controllers
-
- env
-
- envelope
-
- LTNP
-
- long-term nonprogressor
-
- MPER
-
- membrane proximal external region




