Vβ cluster sequences reduce the frequency of primary Vβ2 and Vβ14 rearrangements†
Abbreviations
ESembryonic stem
RSSrecombination signal sequences
Abstract
T-cell receptor (TCR) β variable region exons are assembled from numerous gene segments in a highly ordered and regulated manner. To elucidate mechanisms and identify cis-acting elements that control Vβ rearrangement, we generated an endogenous TCR-β allele with only the Vβ2, Vβ4, and Vβ14 segments. We found that αβ T lineage cells containing this Vβ2–4–14 allele and a wild-type TCR-β allele developed normally, but exhibited a significant increase in Vβ2+ and Vβ14+ cells. To quantify Vβ rearrangements on the Vβ2–4–14 allele, we generated αβ T-cell hybridomas and analyzed TCR-β rearrangements. Despite the deletion of almost all Vβ segments and 234 kb of Vβ cluster sequences, the Vβ2–4–14 allele exhibited only a slight decrease in Vβ rearrangement as compared with the wild-type TCR-β allele. Thus, cis-acting control elements essential for directing Vβ rearrangement across large chromosomal distances are not located within the Vβ cluster. We also found a significant increase in the frequency of Vβ rearrangements involving Vβ2 and Vβ14, but not Vβ4, on the Vβ2–4–14 allele. Collectively, our data suggest that Vβ cluster sequences reduce the frequency of Vβ2 and Vβ14 rearrangements by competing with the productive coupling of accessible Vβ2 and Vβ14 segments with DJβ1 complexes.
Introduction
T-cell receptor (TCR) and immunoglobulin (Ig) genes are each composed of variable region exons and constant (C) region exons. In developing T and B lymphocytes, TCR and Ig variable region exons are assembled from germline variable (V), diversity (D), and joining (J) segments 1. The initiation of chromosomal V(D)J recombination is regulated in a lineage-specific and developmental stage-specific manner through modulation of recombination signal sequence (RSS) accessibility to the RAG1/RAG2 (RAG) endonuclease 2. Despite intense efforts, little is understood about the precise molecular mechanisms that determine recombinational accessibility. However, experimental data indicate that multiple factors likely contribute to render antigen receptor loci RAG accessible, including cis-acting transcriptional elements, transcription factors, nucleosome positioning, epigenetic chromatin modifications, nuclear localization, and higher-order locus topology 3, 4.
TCR-β variable region exons are assembled from multiple Vβ, Dβ, and Jβ segments in a step-wise and regulated manner 5. The mouse TCR-β locus consists of approximately 35 Vβ segments, 2 Dβ–Jβ clusters (Dβ1–Jβ1 and Dβ2–Jβ2), and 2 Cβ (Cβ1 and Cβ2) that span 685 kb on chromosome 6 (Fig. 1) 6. The Dβ1–Jβ1 cluster, Cβ1, the Dβ2–Jβ2 cluster, and Cβ2 span only 13 kb. All Vβ, except Vβ2 and Vβ14, reside within a 234-kb cluster that lies between Vβ4 and Vβ18, with Vβ18 being located 250 kb upstream of Dβ1. The Vβ2 segment is located 156 kb upstream of Vβ4, while the Vβ14 segment resides 10 kb downstream of Cβ2. Arrays of trypsinogen genes are situated within the TCR-β locus both between Vβ2 and Vβ4 and between Vβ18 and Dβ1. In CD4−/CD8− (double-negative or DN) thymocytes, Dβ to Jβ recombination occurs across short distances on both alleles, followed by Vβ recombination to an assembled DJβ complex across large chromosomal distances, over the trypsinogen genes, and on one allele at a time 5. The assembly and expression of an in-frame (productive) VβDJβ rearrangement on the first allele drives further thymocyte development to the CD4+/CD8+ (double-positive or DP) stage and prevents Vβ to DJβ recombination on the second allele to enforce TCR-β locus allelic exclusion 7, 8. Thymocytes that assemble an out-of-frame (non-productive) VβDJβ rearrangement on the first allele can initiate Vβ to DJβ recombination on the second allele in an attempt to assemble a productive VβDJβ rearrangement and signal further thymocyte differentiation 7, 8.
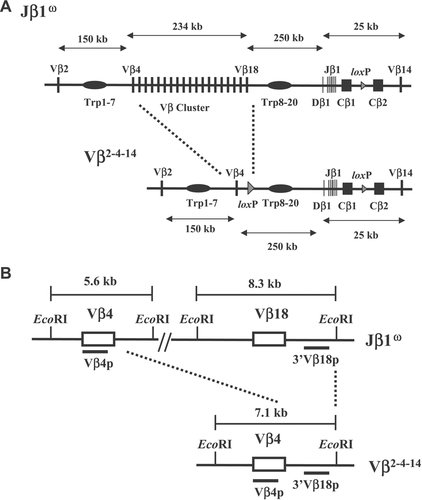
Schematic representation of the Vβω and Vβ2–4–14 alleles. (A) Schematic diagrams of the entire TCR-β loci on the Vβω and Vβ2–4–14 alleles. The loxP sites inserted in place of the DJβ2 clusters and during gene-targeting simplification of the Vβ cluster are indicated by triangles. The relative size of the Vβ cluster and the Dβ–Jβ–Cβ regions are not drawn to scale. (B) Schematic diagrams of the EcoRI restriction fragment length polymorphisms created on the Vβω and Vβ2–4–14 alleles. Black bars indicate the locations of the Vβ4 and 3′Vβ18 probes.
The assembly of TCR-β variable region exons is regulated by cis-acting transcriptional elements that promote recombinational accessibility of particular Vβ, Dβ, and Jβ segments 5. The TCR-β enhancer (Eβ), which resides between Cβ2 and Vβ14, is required for Dβ to Jβ and Vβ to DJβ rearrangement 9, 10. The germline Dβ1 promoter (pDβ1), which resides immediately upstream of Dβ1, is required only for TCRβ rearrangements involving Dβ1 and Jβ1 segments 11, 12. Eβ directs general chromatin opening across both Dβ–Jβ–Cβ clusters and also forms a holoenzyme complex with pDβ1 to remodel nucleosome positioning over the Dβ1 RSS and to allow RAG access 13, 14. Upstream of each Vβ segment resides a promoter that drives Vβ transcription and, at least for Vβ13, mediates RAG access to the downstream Vβ RSS and directs Vβ to DJβ rearrangement of that Vβ 15. Notably, Eβ neither activates germline Vβ transcription nor directs general chromatin opening over Vβ segments 13, suggesting Eβ may direct Vβ to DJβ rearrangement only by promoting recombinational accessibility of Dβ and Jβ segments. Thus, perhaps other cis-acting control transcriptional elements activate germline Vβ promoters, mediate Vβ recombinational accessibility, and/or direct Vβ to DJβ rearrangement.
The complex organization and overall size of the TCR-β locus present significant obstacles to the elucidation of mechanisms and identification of potential cis-acting elements that regulate Vβ rearrangement. Thus, we have taken a gene-targeted mutation approach to simplify the endogenous TCR-β locus and, thereby, to potentially facilitate control features that cannot be readily analyzed in the large, complicated wild-type locus. Here, we describe the generation of an endogenous TCR-β locus with only three Vβ segments. Our analysis of Vβ rearrangements on this allele provides insights into potential mechanisms through which these particular Vβ segments may rearrange to DJβ complexes.
Results
Generation of an endogenous TCR-β locus with only the Vβ2, Vβ4, and Vβ14 segments
To elucidate mechanisms and identify potential cis-acting elements that regulate Vβ rearrangement, we used sequential gene-targeted mutation to delete 234 kb of Vβ cluster sequence and generate an endogenous TCR-β locus with only three Vβ segments. The initial targeting events were performed in Jβ1ω/ω embryonic stem (ES) cells that lack the Dβ2–Jβ2 cluster on both alleles 16. This gene-targeted modification resulted in the generation of Vβ2–4–14/ω ES cells in which one chromosome 6 allele contains the Vβ2–4–14 TCR-β locus consisting of Vβ2, Vβ4, the Dβ1–Jβ1–Cβ1 cluster, Cβ2, and Vβ14 and the other chromosome 6 allele contains a “wild-type” TCR-β (Vβω) locus consisting of Vβ2, the Vβ cluster, the Dβ1–Jβ1–Cβ1 cluster, Cβ2, and Vβ14 (Fig. 1). The Vβ2–4–14 and Vβω allele both contain the same length of intervening DNA sequence and number of trypsinogen genes between their Dβ1–Jβ1 proximal Vβ segment and the Dβ1–Jβ1 region. We chose to simplify the TCR-β locus in this manner to both evaluate whether any putative cis-acting elements that regulate Vβ rearrangement reside within the deleted Vβ cluster sequences and to generate a simplified TCR-β locus in which the rearrangement of upstream Vβ segments could still occur across large chromosomal distances and over the trypsinogen genes. The sequential targeting strategy also created EcoRI fragment length polymorphisms spanning Vβ4 and Vβ18 on the Vβω allele and Vβ4 on the Vβ2–4–14 allele that can be used to distinguish between Vβ rearrangements on each allele (Fig. 1).
Normal development and altered Vβ repertoire of Vβ2–4–14/ω αβ T lineage cells
To characterize αβ T-cell development in Vβ2–4–14/ω mice, we used Vβ2–4–14/ω ES cells and RAG-2-deficient blastocyst complementation 17 to generate chimeric Vβ2–4–14/ω mice in which all lymphocytes are derived from Vβ2–4–14/ω ES cells. The numbers of thymocytes and peripheral αβ T cells in multiple Vβ2–4–14/ω mice analyzed was comparable to those in Jβ1ω/ωcontrol mice (data not shown). Flow cytometry analysis conducted with anti-CD4 and anti-CD8 antibodies on thymocytes isolated from Vβ2–4–14/ω and Jβ1ω/ωcontrol mice revealed a normal distribution of DN, DP, and SP populations (Fig. 2). The same flow cytometry analysis of spleen and lymph node cells from Vβ2–4–14/ω and Jβ1ω/ωcontrol mice showed a normal distribution of CD4+ and CD8+ peripheral αβ T cells (Fig. 2; spleen data not shown). Thus, αβ T-cell development appears grossly normal in Vβ2–4–14/ω chimeric mice.
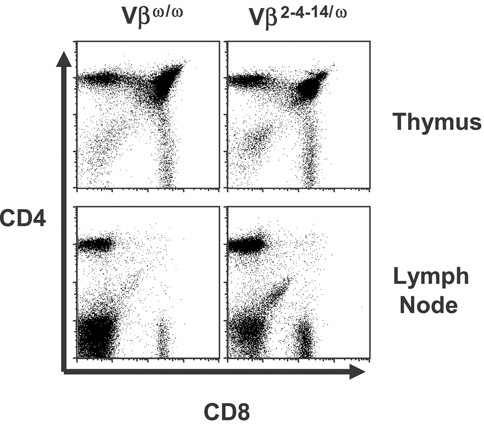
Normal αβ T-cell development in Vβ2–4–14/ω chimeric mice. Representative CD4-PE and CD8-FITC flow cytometry analyses of cells isolated from the thymuses or lymph nodes of Vβω/ω and Vβ2–4–14/ω mice are shown.
To evaluate Vβ repertoire in Vβ2–4–14/ω thymocytes, we conducted flow cytometry analysis with antibodies specific for the TCR-β chain and either Vβ2, Vβ4, Vβ5, Vβ8, or Vβ14 on thymocytes isolated from three Vβ2–4–14/ω mice and three Jβ1ω/ω control mice. In contrast to Jβ1ω/ω thymocytes, which can express Vβ from either TCR-β allele, Vβ2–4–14/ω thymocytes can express Vβ2, Vβ4, or Vβ14 from both alleles, but Vβ5 from only one allele. Thus, we expected to observe a modest increase in the percentage of Vβ2–4–14/ω thymocytes expressing cell surface Vβ2, Vβ4, and Vβ14 as compared with Jβ1ω/ω thymocytes, with a corresponding decrease in the percentage of Vβ2–4–14/ω thymocytes expressing cell surface Vβ5 as compared with Jβ1ω/ω thymocytes cells. As expected, we found a small, but significant, decrease in the percentage of Vβ5+ thymocytes in Vβ2–4–14/ω mice (4.4±0.2%) as compared with Jβ1ω/ω mice (7.7±1.8%) (Fig. 3). However, we found a substantial increase in the percentage of Vβ2+ thymocytes in Vβ2–4–14/ω mice (29.7±2.0%) as compared with Jβ1ω/ω mice (6.1±1.2%), a smaller but significant increase in the percentage of Vβ14+ thymocytes in Vβ2–4–14/ω mice (11.7±1.0%) as compared with Jβ1ω/ω mice (5.7±1.0%), and no change in the percentage of Vβ4+ thymocytes in Vβ2–4–14/ω mice (5.9±0.7%) as compared with Jβ1ω/ω mice (6.3±1.2%) (Fig. 3).
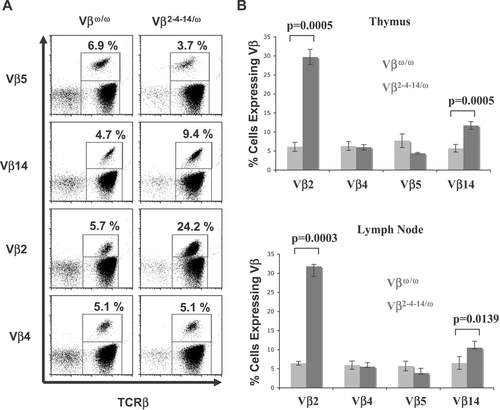
Altered Vβ repertoire in Vβ2–4–14/ω thymocytes and peripheral αβ T cells. (A) Representative TCR-β-PE and Vβ5-FITC, Cβ-PE and Vβ14-FITC, Cβ-PE and Vβ2-FITC, Cβ-PE and Vβ4-FITC flow cytometry analyses of cells isolated from the lymph nodes of Vβω/ω and Vβ2–4–14/ω mice are shown. The percentage of TCR-β αβ T cells that express each particular Vβ is indicated. (B) Bar graphs showing the average percentage of TCR-β+ thymocytes and lymph node cells that express Vβ2, Vβ4, Vβ5, or Vβ14.
To evaluate Vβ repertoire in Vβ2–4–14/ω αβ T cells, we performed the same flow cytometry analysis on peripheral lymphocytes isolated from the lymph nodes of three Vβ2–4–14/ω mice and three Jβ1ω/ω control mice. We found a small, but significant, decrease in the percentage of Vβ5+ αβ T cells in Vβ2–4–14/ω mice (3.9±0.3%) as compared with Jβ1ω/ω mice (5.7±1.3%) (Fig. 3). However, we found a substantial increase in the percentage of Vβ2+ αβ T cells in Vβ2–4–14/ω mice (31.8±2.6%) as compared with Jβ1ω/ω mice (6.4±0.5%), a smaller but significant increase in the percentage of Vβ14+ αβ T cells in Vβ2–4–14/ω mice (10.5±0.3%) as compared with Jβ1ω/ω mice (6.5±1.7%), and no change in the percentage of Vβ4+αβ T cells in Vβ2–4–14/ω mice (5.5±0.1%) as compared with Jβ1ω/ω mice (5.9±1.8%) (Fig. 3).
These data indicate that deletion of 234 kb of Vβ cluster sequence and almost all Vβ segments on a single TCR-β allele significantly alters the Vβ repertoire of developing thymocytes and peripheral αβ T cells. Because the number of thymocytes and peripheral αβ T cells were comparable among Vβ2–4–14/ω mice and Jβ1ω/ωmice, Vβ2–4–14/ω mice develop five times more αβ T lineage cells that express Vβ2 and about two times more αβ T lineage cells that express Vβ14, with a corresponding decrease in the percentage of αβ T lineage cells that express Vβ5. The significant increased expression of Vβ2 and Vβ14 in Vβ2–4–14/ω mice as compared with Jβ1ω/ω mice suggests that the rearrangement of the Vβ2–4–14 allele can effectively compete with rearrangement of the Jβ1ω allele and/or cells with in-frame Vβ2 and Vβ14 rearrangements are preferentially selected during thymocyte development.
Efficient Vβ rearrangement on the Vβ2–4–14 allele
Since the assembly of an in-frame VDJβ rearrangement is required for αβ T-cell development and only one-third of VDJβ rearrangements occur in-frame, approximately 40% of αβ T cells contain VDJβ rearrangements on both alleles 5, 7, 8. The other 60% contain an in-frame VDJβ rearrangement on one allele and a DJβ complex on the other allele due to TCR-β-mediated inhibition of Vβ rearrangement 5, 7, 8. Thus, to determine whether overall Vβ rearrangements occurred at the normal level in Vβ2–4–14/ω mice, we generated a panel of 180 Vβ2–4–14/ω αβ T-cell hybridomas and analyzed TCR-β rearrangements in these cells by Southern blot analysis of EcoRI-digested genomic DNA using 3′Jβ1 and 5′Dβ1 probes. In this manner, we found that 37 of 180 (21%) Vβ2–4–14/ω αβ T-cell hybridomas contained VDJβ rearrangements on both alleles, while 143 of 180 (79%) contained VDJβ rearrangements on one allele and DJβ rearrangements on the other allele (Table 1). Given that this ratio of αβ T cells with the VDJβ/DJβ versus VDJβ/VDJβ configuration (79/21) does not correspond to the normal 60/40 ratio, these data suggest that overall level of Vβ to DJβ rearrangements may be reduced in Vβ2–4–14/ω αβ T cells.
| Number (% total) | |||
| Total number | VDJ/DJ | VDJ/DJ | |
| 180 | 143 (79%) | 37 (21%) | |
| Number VDJ/DJ | VDJ on allele | ||
| Vβω | Vβ2–4–14 | Unknown | |
| 143 | 89 | 42 | 12 |
| Number VDJ/DJ with upstream Vβ rearrangement on the Vβ2–4–14 allele | Upstream Vβ rearranged | ||
| Vβ2 | Vβ4 | ||
| 42 | 38 | 4 | |
To distinguish between Vβ rearrangements on the Vβ2–4–14 and Vβω alleles, we made use of a restriction fragment length polymorphism that was created upon deletion of Vβ cluster sequences. Southern blot analysis with the 3′Vβ18 probe on EcoRI-digested Vβ2–4–14/ω αβ T-cell hybridoma DNA detects a 7.1-kb germline fragment from the Vβ2–4–14 allele and a 8.3-kb germline fragment from the Vβω allele (Fig. 1). Upon the rearrangement of upstream Vβ segments, the genomic sequence to which the 3′Vβ18 probe hybridizes is excised from the chromosome and lost during DN to DP expansion. We found that 89 (62%) of the 143 Vβ2–4–14/ω αβ T-cell hybridomas containing VDJβ rearrangements on a single allele lost the 8.3-kb (Vβω) fragment and retained the 7.1-kb (Vβ2–4–14) fragment, while 42 (29%) of these lost the 7.1-kb (Vβ2–4–14) fragment and retained the 8.3-kb (Vβω) fragment. If the upstream Vβ on the Vβ2–4–14 allele rearranged with an equal probability as those on the Vβω allele, we would have expected to observe 50% of these hybridomas with upstream Vβ rearrangements on the Vβ2–4–14 allele. Consequently, these data indicate that the overall rearrangement efficiency of Vβ2 and Vβ4 rearrangements on the Vβ2–4–14 allele is only slightly lower than the overall rearrangement efficiency of ∼35 Vβ segments on the Vβω allele. The remaining 12 (9%) of these hybridomas retained both the 8.3- and 7.1-kb fragments, indicating that they contained Vβ14 to DJβ1 rearrangements since Vβ14 rearranges by inversion without deletion of TCR-β sequences. Although we confirmed by Southern blot analysis with the Vβ14 probe that these hybridomas contained Vβ14 to DJβ1 rearrangements (Table 1), we cannot distinguish whether these Vβ14 rearrangements occurred on the Vβ2–4–14 or Vβω allele. However, based upon our previous observation that Vβ14 to DJβ1 rearrangements occur on only 7% of Vβω alleles 18, we assume that the increased utilization of Vβ14 is due to an approximately twofold increase in the frequency of primary Vβ14 rearrangements on the Vβ2–4–14 alleles.
Increased frequency of primary Vβ2 rearrangements on the Vβ2–4–14 allele
The frequency at which particular Vβ segments are expressed in the Vβ repertoire is not detectably selected during DN to DP thymocyte development 19, 20. In addition, the frequency at which Vβ2 and Vβ4 are expressed in peripheral αβ T cells is the same frequency at which they are expressed in DN thymocytes (19, Fig. 3). Therefore, to determine the relative frequency of primary Vβ2 versus Vβ4 rearrangements on the Vβ2–4–14 allele, we further analyzed TCR-β rearrangements in the 42 Vβ2–4–14/ω αβ T-cell hybridomas with upstream Vβ rearrangements on only the Vβ2–4–14 allele. Southern blot analysis with the Vβ4 probe on EcoRI-digested Vβ2–4–14/ω αβ T-cell hybridoma DNA detected a 7.1-kb germline fragment from the Vβ2–4–14 allele and a 5.6-kb germline fragment from the Vβω allele (Fig. 1). Upon Vβ4 to DJβ rearrangement, Vβ4 was located within a novel-sized (non-germline) EcoR1 fragment that also hybridized with the 3′Jβ1 probe, while upon Vβ2 to DJβ rearrangement, Vβ4 was excised from the chromosome and lost during DN to DP expansion. We found that 38 of these hybridomas lost the 5.6-kb band, while only 4 lost the 5.6-kb band and gained a non-germline band. Thus, of the 42 Vβ2–4–14/ω αβ T-cell hybridomas with upstream Vβ rearrangements on the Vβ2–4–14 allele and DJβ rearrangements on the Vβω allele, 90% (38 of 42) contained Vβ2 rearrangements and only 10% (4 of 42) contained Vβ4 rearrangements (Table 1). The identity of these rearrangements was confirmed by PCR. Consequently, deletion of most Vβ cluster Vβ segments and 234 kb of Vβ locus sequence resulted in a substantial increase in the frequency of primary Vβ rearrangements, but not in the frequency of Vβ4 rearrangements, on the Vβ2–4–14 allele.
Discussion
We have shown here that the overall level of Vβ rearrangements on the Vβ2–4–14 allele containing just Vβ2, Vβ4, and Vβ14 was less than twofold lower than the overall level of Vβ rearrangements on the VβWT allele containing ∼35 Vβ segments. The overall level of Vβ rearrangement must be determined by the rate of Vβ to DJβ recombination and the time window in DN thymocytes during which Vβ to DJβ recombination can occur. Though our findings indicate that the Vβ2–4–14 allele exhibits a twofold decrease in the rate of Vβ rearrangement as compared with the Vβω allele, it is remarkable that Vβ rearrangements in a TCR-β locus with only three Vβ segments compete so effectively with Vβ rearrangements in a locus with 35 Vβ segments. Consequently, our observation suggests that the number of endogenous Vβ segments available for recombination with DJβ1 complexes alone is not a major determinant of the overall rate of Vβ to DJβ1 rearrangements in DN thymocytes. In this regard, we cannot exclude the possibility that a cis-element that inhibits Vβ rearrangements is located within the 234 kb of deleted Vβ cluster sequences. Our findings also indicate that, in each developing thymocyte, the two TCR-β alleles are almost equally chosen to rearrange, regardless of the large Vβ cluster deletion that generates a large discrepancy in the number of Vβ between each allele. Finally, our data demonstrate that cis-elements essential for promoting Vβ2, Vβ4, and Vβ14 accessibility and directing Vβ2 and Vβ4 rearrangements across large chromosomal distances over the trypsinogen genes are not located within the 234 kb of deleted Vβ cluster sequences. However, our observation that the Vβ2–4–14 allele exhibits a twofold decrease in the rate of Vβ rearrangement as compared with the Vβω allele might indicate that cis-elements that contribute to promote Vβ rearrangement reside within the deleted Vβ cluster sequences and/or that a full complement of Vβ within the Vβ cluster enhances recombinational accessibility of all the Vβ cluster segments.
We also found a dramatically altered Vβ repertoire in the peripheral αβ T cells of Vβ2–4–14/ω mice as compared with Jβ1ωω control mice, with a significant increase in the percentage of cells expressing Vβ2 and Vβ14, but not Vβ4. We demonstrated that the increased utilization of Vβ2 is due to a substantial increase in the frequency of primary Vβ2 rearrangements on the Vβ2–4–14 allele as compared with the Vβω allele. Although we also found that the increased utilization of Vβ14 is due to a corresponding increase in the frequency of primary Vβ14 rearrangements, we could not determine whether these Vβ14 rearrangements occurred on the Vβ2–4–14 or Vβω allele. However, based upon our previous observations that Vβ14 to DJβ1 rearrangements normally occur on approximately 7% of Vβω alleles, and the frequency of primary Vβ14 rearrangements determines the representation of Vβ14 in the peripheral Vβ repertoire 18, we conclude that the increased utilization of Vβ14 is due to an approximately twofold increase in the frequency of primary Vβ14 rearrangements on the Vβ2–4–14 allele as compared with the Vβω allele. Thus, our data demonstrate that deletion of 234 kb of Vβ cluster sequences containing 32 Vβ segments leads to an increase in the frequency of primary Vβ to DJβ1 rearrangements involving Vβ2 and Vβ14, but not Vβ4.
In DN thymocytes, Vβ to DJβ recombination must proceed through the physical juxtaposition (looping) of RAG-accessible Vβ segments and DJβ complexes, formation of RAG/Vβ/DJβ synaptic complexes, and RAG-mediated cleavage and joining 1, 20. Based upon our previous observation that replacement of the endogenous Vβ14 RSS with a tenfold more efficient RSS led to a corresponding increase in the frequency of primary Vβ14 to DJβ1 rearrangements, we suggested that Vβ14 to DJβ1 recombination likely proceeds through cycles of juxtaposition, synaptic complex formation, and release prior to RAG-mediated cleavage and joining 20. We also recently demonstrated that rate of Vβ14 to DJβ1 recombination is determined by the productive coupling of RAG-accessible Vβ14 segments and DJβ complexes 18. In this context, our current finding that deletion of 234 kb of Vβ cluster sequences containing 32 Vβ segments leads to an increase in the level of primary Vβ14 to DJβ1 rearrangements suggests that the productive coupling of RAG-accessible Vβ14 segments and DJβ complexes occurs at higher frequency on the Vβ2–4–14 allele as compared with Vβω allele. In this context, our data also suggest that the looping of Vβ cluster Vβ segments with accessible DJβ complexes and/or formation of RAG/Vβ/DJβ synaptic complexes involving Vβ cluster Vβ segments reduces, most likely via competition, the frequency of productive coupling of RAG-accessible Vβ14 segments and DJβ complexes.
We found a dramatic increase in upstream Vβ to DJβ1 rearrangements on the Vβ2–4–14 allele involving Vβ2, but not Vβ4. There could be several explanations as to why deletion of 234 kb of Vβ cluster sequences containing 32 Vβ segments results in a substantial increase in the frequency of primary Vβ2 to DJβ1 rearrangements, but not in the frequency of primary Vβ4 to DJβ1 rearrangements. Because deletion of 220 kb of Vβ cluster sequence and the trypsinogen genes between Vβ10 and the Dβ2–Jβ2 cluster increases accessibility and rearrangement of Vβ10, without increasing accessibility and rearrangement of either Vβ4 and Vβ16, which reside only 10 kb upstream of Vβ10, or Vβ2, which lies 164 kb upstream of Vβ10 21, it seems highly unlikely that our Vβ cluster deletion specifically increased Vβ2 RAG accessibility. Thus, we suspected that the insertion of a loxP site just downstream of Vβ4 inhibited RAG access to the Vβ4 RSS since this loxP site disrupts the Vβ16 promoter. Consistent with this notion, we found that insertion of a loxP site just downstream of Vβ4 on the Jβ1ω allele inhibited expression, and by extension the rearrangement, of Vβ4 (Supporting Information Fig. 1). Based upon these data, we assume that, if the loxP site on the Vβ2–4–14 allele were located elsewhere, Vβ4 rearrangements would also have increased. Consequently, we conclude that Vβ2 is normally RAG accessible in a much higher percentage of DN thymocytes than the percentage in which Vβ2 to DJβ1 rearrangements occur; however, the large number of Vβ segments within the Vβ cluster ordinarily compete with accessible Vβ2 segments for synaptic complex formation with DJβ1 complexes and, thereby, reduce the frequency of Vβ2 rearrangements.
Since Vβ2 and Vβ14 each normally rearrange to DJβ complexes at a normal frequency, each should rearrange at the same elevated frequency on the Vβ2–4–14 allele; yet, we observed an unequal increase in rearrangements involving Vβ2 as compared with Vβ14. Again, since it seems highly unlikely that our Vβ cluster deletion specifically increased Vβ2 RAG accessibility, we favor the explanation that the productive coupling between Vβ2 and DJβ complexes is enhanced on the Vβ2–4–14 allele. In this regard, perhaps deletion of Vβ cluster sequences on the Vβ2–4–14 allele alters the higher-order structure of the TCR-β locus such that RAG-accessible Vβ2 segments and DJβ1 complexes are more frequently able to loop together. The Vβ2 and Vβ14 segments are distinct from the other 33 Vβ segments due to their location outside of the Vβ cluster 6, their transcriptional regulation 22, 23, and the great efficiency of their flanking RSS for recombination with 5′Dβ RSS 24. Therefore, the generation and analysis of mice containing additional specific TCR-β locus deletions 21, Vβ RSS replacements 20, and Vβ recombination reporters 18 will be required to determine the mechanisms by which the rearrangement of Vβ cluster Vβ segments is directed.
Materials and methods
Generation of targeting construct and probes
The Vβ4loxP and Vβ18loxP targeting vectors were constructed in pLNTK 25. For Vβ4loxP, the 5′ homology arm is a 2.4-kb genomic fragment spanning the SacI site upstream of Vβ4 and an XhoI site inserted by PCR 110 bp 3′ of the Vβ4 RSS subcloned into the SalI site of pLNTK. The 3′ homology arm is a 3.2-kb genomic fragment containing sequences located 3′ of Vβ16 subcloned into the XhoI site of pLNTK. For Vβ18loxP, the 5′ homology arm is a 2.1-kb DrdI/SalI genomic fragment containing Vβ8 subcloned into the SalI site of pLNTK. The 3′ homology arm is a 2.7-kb SalI/BamHI genomic fragment containing sequences located 3′ of Vβ18 subcloned into the XhoI site of pLNTK. The 5′ homology arm also contained germline Dβ1–Jβ1 sequences inserted at the 3′ end. The 5′Vβ4 probe is a 234-bp HindIII/StuI fragment. The 3′Vβ4 probe is a 500-bp PCR product amplified with 5′-AGAATTTCTATTAGATCA-3′ and 5′-GGCACAGCTGTATGGACTTG-3′. The 5′IntVβ4 probe is a 1.7-kb PstI genomic fragment.
The 5′Vβ18 probe is a 650-bp PCR product amplified with 5′-CATCCATTTGCCTAAGAATTTCATG-3′ and 5′-GACAAATTGGCAACCAATAGAATGG-3′.
The 3′Vβ18 probe is 470-bp PCR product amplified with 5′-TTAGGCAGGCATAGGAACATAACTG-3′ and 5′-CTACTCACCTTCTGTATTTATTGG-3′. The 3′IntVβ18 probe is a 1-kb EcoRV/BamHI genomic fragment.
Gene-targeting and generation of ES cells
The Vβ4loxP targeting vector was electroporated into Jβ1ω/ω ES cells 16 as previously described 26 to generate Vβ4loxPNeo/ω ES cells. Targeted clones were identified by Southern blot analysis with the 5′Vβ4 probe on HindIII-digested genomic DNA (4.2 kb Vβ4loxPNeo, 6.6 kb Vβ4ω) and confirmed with the 3′Vβ4 probe on StuI-digested DNA (6.2 kb Vβ4loxPNeo, 9.2 kb Vβ4ω). Targeted ES cells were infected with recombinant AdenoCre and subcloned to identify Vβ4loxP/ω ES cells. The correct Cre deleted subclones were identified by Southern blot analysis with the 5′Vβ4 probe on HindIII-digested genomic DNA (5.7 kb Vβ4loxP, 6.6 kb Vβ4ω) and confirmed with the 3′Vβ4 probe on StuI-digested DNA (8.4 kb Vβ18loxP, 9.2 kb Vβ18ω). Next, the Vβ18loxP targeting vector was electroporated into Vβ4loxP/ω ES cells. Targeted clones were identified by Southern blot analysis with the 5′Vβ18 probe on EcoRI-digested genomic DNA (4.2 kb Vβ18loxPNeo, 8.3 kb Vβ18ω) and confirmed with the 3′Vβ18 probe on EcoRI-digested DNA (6.0 kb Vβ18loxPNeo, 8.3 kb Vβ18ω). Targeted ES cells were infected with recombinant AdenoCre and subcloned to identify Vβ182–4–14/ ES cells that contain deletion between the loxP site inserted 3′ of Vβ4 and the 3′ loxP site of Vβ18loxPNeo, which should only happen at an appreciable frequency if the two targeting events were on the same TCR-β allele. The correct Cre deleted subclones were identified by Southern blot analysis with the 5′IntVβ4 probe on EcoRI-digested genomic DNA (7.1 kb Vβ2–4–14/ω, 5.6 kb Vβ18ω) and confirmed with the 3′IntVβ18 probe on EcoRI-digested DNA (7.1 kb Vβ18loxP, 8.3 kb Vβ18ω).
Generation and analysis of chimeric mice
Vβ2–4–14/WT chimeric mice were generated through Rag2-deficient blastocyst complementation 17. Cells from the thymuses, spleens, and lymph nodes of 4–6-wk-old Vβ2–4–14/ω and wild-type 129SvEv (Taconic) control mice were isolated, counted, and then stained with FITC-conjugated anti-CD8 and PE-conjugated anti-CD4 antibodies or FITC-conjugated anti-Vβ and PE-conjugated anti-TCR-β chain antibodies (Pharmingen). For analysis of DN subsets, thymocytes were stained with a cocktail of PE-conjugated antibodies for TCR-β, TCR-δ, CD4, CD8, B220, and NK.1, as well as FITC-conjugated anti-CD25 and CYC-conjugated anti-CD44 antibodies (Pharmingen). Data acquisition was conducted on a FACSCalibur equipped with CellQuest and data analysis performed with FlowJo software. More than five mice of each genotype were analyzed.
Analysis of TCR-β rearrangements
Hybridomas were generated as previously described 26. The Southern blot analysis of TCR-β Dβ to Jβ and Vβ to DJβ rearrangements was conducted on EcoRI-digested genomic DNA with the 5′Dβ1 and 3′Jβ1 probes 16. The analysis of Vβ2–4–14 versus VβWT Vβ to DJβ rearrangements was performed on EcoRI-digested genomic DNA with the 3′Vβ18, 5′IntVβ4, Vβ2, and Vβ14 probes 20.
Acknowledgements
We thank Andrea Carpenter for helpful discussions of the manuscript and assistance for statistical analysis. C. H. B. is a Pew Scholar in the Biomedical Sciences. R. M. is a V Scholar. F. W. A. is an Investigator of the Howard Hughes Medical Institute. This work was supported by the National Institutes of Health Grant AI20047 (F. W. A.) and the Department of Pathology of the Children's Hospital of Philadelphia (C. H. B.).
Conflict of interest: The authors disclose no financial or commercial conflict of interest.




