Interferon regulatory factor 7-mediated responses are defective in cord blood plasmacytoid dendritic cells
Abstract
Plasmacytoid dendritic cells (pDC) are specialized in massive production of type I interferons (IFN) upon viral infections. Activation of IFN regulatory factor (IRF)-7 is critically required for the synthesis of type I IFN in pDC. IRF-7 is highly expressed by resting pDC and translocates into the nucleus to initiate type I IFN transcription. In a previous work, we observed an impaired IFN-α production in enriched cord blood pDC following a TLR9 stimulation using CpG oligonucleotides. Herein, we show that highly purified pDC from cord blood exhibit a profound defect in their capacity to produce IFN-α/β in response to TLR9 as well as to TLR7 ligation or human CMV or HSV-1 exposure. Microarray experiments indicate that expression of the majority of type I IFN subtypes induced by a TLR7 agonist is reduced in cord blood pDC. We next demonstrated a reduced nuclear translocation of IRF-7 in cord blood pDC following CpG and HSV stimulation as compared to adult pDC. We conclude that impaired IRF-7 translocation in cord blood pDC is associated with defective expression of type I IFN genes. Our data provide a molecular understanding for the decreased ability of cord blood pDC to produce type I IFN upon viral stimulation.
Abbreviations:
-
- CBMC:
-
cord blood mononuclear cells ; HCMV: human cytomegalovirus ; IRF: interferon regulatory factor ; pDC: plasmacytoid dendritic cell
-
- TRAF:
-
TNF receptor-associated factor
Introduction
Production of type I interferons (IFN) is a key feature of the host defense against invading pathogens. In addition to their anti-viral activities, type I IFN strongly activate both innate and adaptative immunity 1. Although most cell types can produce type I IFN, plasmacytoid dendritic cells (pDC) represent a major source of these cytokines, especially upon viral infections or exposure to ligands of Toll-like receptor 9 (TLR9) (e.g. unmethylated CpG-rich deoxynucleotides) or TLR7 (e.g. resiquimod (R-848)) 2, 3. Activation of IFN regulatory factor (IRF)-7 is critically required for the induction of type I IFN synthesis in pDC 4. Indeed, IRF-7 is highly expressed in the cytoplasm of resting pDC and translocates in the nucleus upon activation of pDC 5. Whereas type I IFN production is a key feature of pDC, they also mature and produce several inflammatory cytokines and chemokines upon TLR7/9 ligation. This pathway has been shown to be dependent on NF-κB activation 6, 7. Although pDC are present in significant numbers in human cord blood, there is evidence that they do not function as their adult counterparts since we found that they secrete less IFN-α upon exposure to CpG 8.
Herein, we first specified this defect by comparing the maturation and the production of type I IFN and inflammatory cytokines under different conditions of stimulation including exposure to viruses. The observation that IRF-7-dependent responses were profoundly defective whereas NF-κB-dependent responses were partially affected led us to compare the nuclear translocation of IRF-7 and NF-κB in activated pDC isolated from adult and cord blood.
Results
IFN-α/β induction through TLR7 is impaired in cord blood pDC
Using immunomagnetic selection procedure, pDC were purified from fresh blood obtained from human placental cords and adult donors. This method enabled us to obtain >95% pure pDC in both adult and cord blood cell populations. In order to study the ability of cord blood pDC to produce IFN-α/β in response to a TLR7 agonist, we used resiquimod (R-848), a small synthetic compound belonging to the family of imidazoquinolines. Dose-response experiments indicate that R-848 elicited IFN-α secretion in a dose-dependent manner with a maximal effect at 10 µg/mL of R-848, whereas IFN-α secretion by cord blood pDC was severely impaired whatever the dose of the stimulus used (Fig. 1A). We confirmed these results by stimulating pDC from a larger number of adult donors and cord blood samples with R-848 (10 µg/mL), showing that cord blood pDC produced significantly less IFN-α as compared to their adult counterparts (Fig. 1B). Moreover, we quantified IFN-β production using the same experimental conditions, showing that IFN-β levels are significantly lower when produced by cord blood pDC (Fig. 1C).
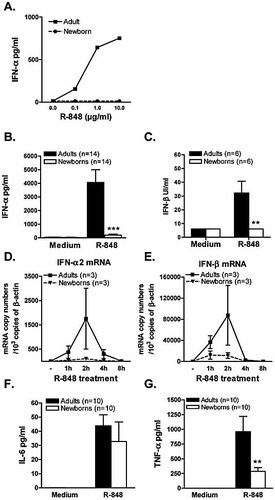
Defective IFN-α/β synthesis in R-848-stimulated cord blood pDC. (A) Purified pDC (2×105 cells/mL) were cultured with graded doses of R-848, at the concentration of 2×105 cells/mL for 48 h. Supernatants were collected and assayed by ELISA for detection of IFN-α levels. One representative adult and cord blood sample out of three is shown. (B, C) Adult and cord blood pDC were stimulated with R-848 (10 µg/mL), and IFN-α (B) and IFN-β (C) levels were quantified by ELISA. Data represent the means ± SEM of 14 adult and 14 cord blood pDC samples (B) (***p<0.001 cord blood versus adult pDC) or six adult and cord blood pDC samples (C) (**p<0.01 cord blood versus adult pDC). (D, E) Adult and cord blood pDC were incubated with R-848 (10 µg/mL) for the indicated times. Total RNA was extracted and real-time PCR was performed for IFN-α2 and IFN-β mRNA quantification. Results are expressed in mRNA copy numbers per 103 β-actin mRNA copies. The means ± SEM obtained from three different adult and cord blood samples are shown. (F, G) Adult and cord blood pDC were stimulated as described above with R-848 (10 µg/mL), and IL-6 (F) and TNF-α (G) levels were quantified by ELISA. Data represent the means ± SEM of ten adult and ten cord blood pDC samples; **p<0.01 cord blood versus adult pDC.
To determine whether the neonatal defect in type I IFN production was already apparent at the mRNA level, we used real-time RT-PCR to measure IFN-α2 and IFN-β gene expression in R-848-stimulated cord blood and adult pDC. As shown in Fig. 1D, IFN-α2 mRNA accumulation peaked in adult pDC at 2 h following R-848 incubation. IFN-β mRNA accumulation followed the same kinetics as shown in Fig. 1E. In contrast, R-848-induced IFN-α2 and IFN-β expression was severely decreased in cord blood pDC at any time point.
As R-848 also induces inflammatory cytokine production in pDC, we thus evaluated TNF-α and IL-6 secretion by adult and cord blood pDC using the same experimental conditions. As shown in Fig. 1F, IL-6 synthesis was comparable between adult and cord blood pDC following R-848 stimulation. On the other hand, cord blood pDC produced significantly lower levels of TNF-α than adult pDC in response to R-848 (Fig. 1G).
Cord blood pDC exhibit a reduced maturation in response to TLR7 ligation
In parallel to cytokine production analysis, we compared the phenotypical changes induced by R-848 on adult versus cord blood pDC. For this purpose, we stimulated adult and cord blood purified pDC with R-848 (10 µg/mL) for 24 h. Cells were collected and stained for cell surface molecules and their expression was analyzed by flow cytometry. As shown in Fig. 2, apart from CD86 marker, R-848 indeed induced a clear up-regulation of CD80, CD40, CCR7, CD54 and HLA-DR molecules on adult pDC. Cord blood pDC also responded to R-848, as all markers of maturation tested were up-regulated compared to resting pDC. However, the magnitude of CD80, CD40, CCR7 and HLA-DR up-regulation was significantly lower in cord blood pDC. In contrast, CD54 and CD86 expression in R-848-stimulated pDC did not differ between neonatal and adult cells. Taken together, these data reveal that the defect of cord blood pDC in response to R-848 is not limited to type I IFN production but also involves, albeit to a lower degree, the acquisition of a mature phenotype and the production of TNF-α.
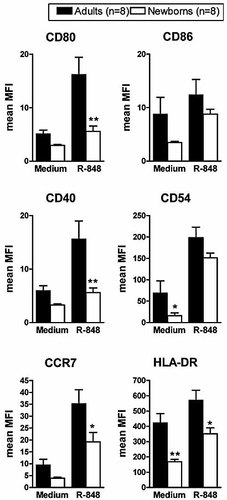
Incomplete maturation of cord blood pDC in response to R-848. Adult and cord blood pDC were incubated with R-848 (10 µg/mL) for 24 h. Cells were harvested and analyzed by flow cytometry after staining with the indicated mAb. Data are expressed as the means of MFI ± SEM obtained from eight different adult and cord blood samples; *p<0.05, **p<0.01 cord blood versus adult pDC.
Expression of type I and type III IFN subtypes is decreased in R-848-stimulated cord blood pDC
To get a broader overview of the TLR7-mediated responses in cord blood pDC, we compared the gene expression profile induced by R-848 in adult and cord blood pDC using oligonucleotide microarray technique. Adult and cord blood pDC were stimulated for 2 h with R-848 (10 µg/mL), a time point which corresponds to the peak of IFN-α/β mRNA induction, as previously shown. Thereafter, RNA samples were hybridized to microarrays designed to detect 54676 transcripts.
Our analysis indicates first that 1055 genes were significantly up-regulated in adult pDC in response to R-848 and among them, 256 were found to be significantly differentially regulated between adult and cord blood pDC (Supporting Information Table 1). We next compared in adult pDC versus cord blood pDC stimulated with R-848, expression of IRF-7-dependent genes, such as type I and type III IFN subtypes, and of NF-κB-dependent genes, like IL-6 and TNF-α genes.
|
Gene |
Fold change adultsa) |
Fold change newborns |
Ratio fold change adults/newborns |
|---|---|---|---|
|
IFN-α8 |
125.73 |
2.80 |
45.10 |
|
IFN-α1 |
666.01 |
20.86 |
31.93 |
|
IFN-ω1 |
213.76 |
9.10 |
23.60 |
|
IFN-α7 |
264.49 |
14.46 |
18.30 |
|
IFN-α10 |
489.92 |
27.07 |
18.10 |
|
IFN-α21 |
150.59 |
8.33 |
18.07 |
|
IFN-α4 |
369.82 |
20.64 |
17.92 |
|
IFN-α17 |
116.72 |
6.60 |
17.80 |
|
IFN-α5 |
656.74 |
41.79 |
15.71 |
|
IFN-α13 |
132.69 |
9.59 |
13.83 |
|
IFN-λ2 |
174.44 |
13.02 |
13.40 |
|
IFN-α16 |
842.47 |
66.26 |
12.71 |
|
IFN-λ3 |
60.77 |
5.90 |
10.30 |
|
IFN-α14 |
621.03 |
79.46 |
7.82 |
|
IFN-α6 |
8.61 |
1.30 |
6.70 |
|
IFN-α2 |
1203.34 |
185.98 |
6.47 |
|
IFN-β1, fibroblast |
1136.57 |
263.03 |
4.32 |
|
IFN-λ1 |
10.86 |
2.50 |
4.30 |
- a) Gene expression is described as the mean of the fold change (ratio between treated and untreated pDC) of three donors in each group.
Table 1 clearly shows that type I and type III IFN family subtypes were all induced upon TLR7 triggering by adult pDC. All of these genes were significantly less up-regulated in cord blood pDC. Indeed, the ratio of the fold change (fold change adult pDC/fold change cord blood pDC) ranged from 4.3 to 45.1. The decreased expression of IFN-α2 and IFN-β genes in R-848-stimulated cord blood pDC was thus also apparent in the microarray analysis.
In Table 2 we compared the expression of 18 NF-κB-inducible genes in adult and cord blood stimulated pDC. Among them, only five genes were found to be significantly differentially expressed between adult and cord blood pDC whereas the ratio of the fold change from 13 of those genes ranged from 0.87 to 1.92 (Table 2). Of note, IL-6 and TNF-α gene induction in cord blood pDC was in agreement with our observations presented in Fig. 1F, G. Indeed, IL-6 production was found to be comparable between adult and neonatal pDC whereas TNF-α production was found to be significantly lower in cord blood pDC. Overall, these data revealed a profound defect in the expression of type I and type III IFN family subtypes in R-848-stimulated cord blood pDC whereas NF-κB-dependent responses were less affected.
|
Gene |
Fold change adultsa) |
Fold change newborns |
Ratio fold change adults/newborns |
|---|---|---|---|
|
TNF |
58.85 |
25.93 |
2.27b) |
|
NF-κB2 (p49/p100) |
6.93 |
3.13 |
2.21b) |
|
Jun-B proto-oncogene |
4.43 |
2.05 |
2.16b) |
|
TRAF1 |
28.42 |
13.40 |
2.12b) |
|
CCL5 |
11.72 |
5.68 |
2.06b) |
|
Bcl2-related protein A1 |
559.73 |
291.18 |
1.92 |
|
NF-κBIA |
5.95 |
4.41 |
1.35 |
|
IL-6 |
39.38 |
29.90 |
1.32 |
|
NGFI-A-binding protein 1 |
7.01 |
5.39 |
1.30 |
|
IL-1α |
7.62 |
6.03 |
1.26 |
|
IL-8 |
209.75 |
176.61 |
1.19 |
|
CCL3 |
382.36 |
345.41 |
1.11 |
|
CCL4 |
186.06 |
178.45 |
1.04 |
|
Bcl2-like 1 |
15.05 |
15.36 |
0.98 |
|
pentraxin-related gene |
32.47 |
34.09 |
0.95 |
|
TNF-α-induced protein 2 |
9.80 |
10.85 |
0.90 |
|
CXCL 2 |
13.07 |
15.04 |
0.87 |
|
B-cell CLL/lymphoma 2 |
14.61 |
16.82 |
0.87 |
- a) Gene expression is described as the mean of the fold change (ratio between treated and untreated pDC) of three donors in each group.
- b) Gene significantly differentially regulated.
CpG-induced IFN-α/β production is impaired in cord blood pDC
In a previous work, we observed an impaired IFN-α production in enriched cord blood pDC following a TLR9 stimulation using CpG oligonucleotides 8. Herein, we first confirmed that highly purified cord blood pDC were impaired in their capacity to mount significant IFN-α response whatever the dose of CpG A used for their stimulation (Supporting Information Fig. 1). We next extended our findings to IFN-β production upon stimulation with CpG A (10 µg/mL) and we observed that IFN-β levels were also significantly lower in supernatants of activated cord blood pDC as compared to adult pDC (Fig. 3A). In the same time, we measured IFN-α2 and IFN-β mRNA accumulation in cord blood and adult pDC upon CpG stimulation (10 µg/mL). We could observe that IFN-α2 and IFN-β mRNA levels peaked 8 h following TLR9 ligation in adult pDC. In contrast, expression of IFN-α2 and IFN-β genes was profoundly decreased in cord blood pDC at any time point of the kinetic (Fig. 3B, C). Altogether, these data establish that type I IFN induction through TLR7 and TLR9 is impaired in cord blood pDC.
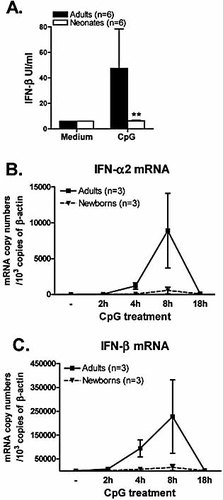
Impaired type I IFN synthesis in CpG-stimulated cord blood pDC. (A) Purified pDC (2×105 cells/mL) were stimulated with CpG A (10 µg/mL), and IFN-β levels were quantified by ELISA. Data represent the means ± SEM of six adult and six cord blood pDC samples; **p<0.01 cord blood versus adult pDC. (B, C) Adult and cord blood pDC were incubated with CpG A (10 µg/mL) for the indicated times. Total RNA was extracted and real-time PCR was performed for IFN-α2 and IFN-β mRNA quantification. Results are expressed in mRNA copy numbers per 103 β-actin mRNA copies. The means ± SEM obtained from three different adult and three cord blood samples are shown
Nuclear translocation of IRF-7 is defective in response to CpG stimulation in cord blood pDC
The findings that TLR7- and TLR9-dependent responses developed by cord blood pDC were defective led us to analyze TLR7/9 expression in adult and neonatal pDC at the basal level. As assessed by real-time RT-PCR shown in Fig. 4A, TLR7 and TLR9 mRNA levels were found to be comparable between resting adult and cord blood pDC. Therefore, the defective IFN-α/β production by cord blood pDC could not be attributed to a lower expression of TLR7 and TLR9.
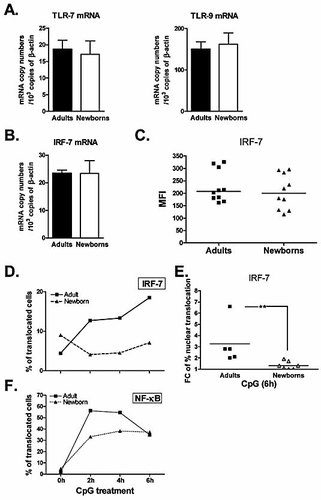
IRF-7 translocation is defective in cord blood pDC. (A) mRNA was extracted from resting purified adult and cord blood pDC, reverse-transcribed and amplified by real-time PCR using primers specific for TLR7 or TLR9. mRNA levels were quantified and normalized against β-actin. Data represent the means ± SEM of nine adult and nine newborn donors. (B) IRF-7 mRNA levels from resting adult and cord blood pDC were quantified by real-time PCR as described above and normalized against β-actin. The means ± SEM obtained from eight adult donors and eight newborn samples are shown. (C) Untreated PBMC or CBMC were stained for intracytoplasmic IRF-7 detection and analyzed by flow cytometry. pDC were identified as BDCA-2/4+ cells and analyzed for their IRF-7 expression. Scatter plot represents the mean fluorescence intensities for each individual donor (ten adult and ten cord blood samples). (D–F) Adult and cord blood enriched pDC were stimulated for the indicated times with CpG A (10 µg/mL), stained as described in Materials and methods and run on the ImageStream imaging cytometer. Data were analyzed to determine the percentage of pDC with nuclear localization of IRF-7 (D, E) or NF-κB (F) using the IRF-7/DRAQ5 or NF-κB/DRAQ5 similarity scoring following CpG A stimulation. Kinetics of translocation for one representative adult versus cord blood donor is shown (D, F). Scatter plot shows the fold change of the percentage of translocated cells, calculated as ratios between the percentage of translocated cells in treated versus untreated pDC for each donor after a 6-h incubation with CpG A (E). This was done for six adult donors and six cord blood samples; **p<0.01 cord blood versus adult pDC.
Upon TLR7/9 signalling, IRF-7 becomes phosphorylated and then translocates into the nucleus where it can activate type I IFN genes. We first analyzed the basal expression of IRF-7. IRF-7 mRNA levels, assessed by real-time RT-PCR, and IRF-7 protein levels, measured by intracellular cytoplasmic staining technique, were comparable in both adult and cord blood pDC (Fig. 4B, C). Furthermore, the basal expression of several molecules implicated in the TLR7/9 signalling pathway such as MyD88, TNF receptor-associated factor (TRAF)6, IRAK4 and IRAK1 was comparable between resting adult and cord blood pDC as assessed by the microarray analysis (Supporting Information Table 2).
To quantify nuclear translocation of IRF-7 in activated pDC, we used the ImageStream technology developed by Amnis Corporation. This technique combines high-resolution digital imaging with flow cytometry technology, enabling quantitative image-based analysis of nuclear translocation in rare subpopulations 9.
BDCA-2/4+ cells with high IRF-7/DRAQ5 similarity scores were gated to quantify the percentage of cells with nuclear-localized IRF-7. Fig. 4D shows the kinetics of the nuclear translocation of IRF-7 in pDC following CpG incubation. We observed that the highest percentage of adult pDC showing a nuclear localization of IRF-7 was reached after 6 h of stimulation. In contrast, IRF-7 nuclear translocation was not induced in cord blood pDC upon CpG stimulation at any time point tested.
We next compared adult and cord blood samples following 6 h of incubation with CpG A. The results presented in Fig. 4E are expressed as fold changes of percentage of translocated cells, calculated as ratios between percentage of translocated cells in treated versus untreated pDC. Our data indicate that the mean of the fold changes was 3.3 after CpG treatment in adult pDC versus 1.3 for cord blood pDC. In those conditions, IRF-7 nuclear translocation was significantly reduced in cord blood pDC as compared to their adult counterparts (Fig. 4E). Of note, we did not observe IRF-7 nuclear translocation in response to R-848 using the ImageStream technique.
We also used this technique to quantify the nuclear translocation of NF-κB in pDC following CpG stimulation. Indeed, this transcription factor has been shown to control essential functions in human pDC, such as induction of their maturation, survival and inflammatory cytokine production. As shown in Fig. 4F, NF-κB rapidly translocated to the nucleus in adult pDC with a peak of translocation after 2 h of CpG treatment, followed by a reduction in nuclear localization after 6 h. Fig. 4F shows that NF-κB translocation occurred in CpG-stimulated cord blood pDC but appeared weaker after 2 and 4 h of incubation as compared to adult pDC. However, after 6 h of CpG stimulation, cord blood pDC reached adult levels in terms of percentage of translocated cells.
These results indicate that although IRF-7 is present in resting cord blood pDC, its translocation is profoundly compromised in response to TLR9 ligation. In contrast, NF-κB translocation seems to be less affected in cord blood pDC under the same conditions.
Impaired cytokine secretion and defective IRF-7 translocation in cord blood pDC exposed to viruses
pDC have been largely described as the most potent cell type endowed with the unique property to secrete large amounts of type I IFN following viral invasion 6. In contrast to other cells types, viruses do not need to be replication-competent to activate pDC to produce IFN-α/β 10. Until now, TLR are considered as the major receptors for the initiation of type I IFN production in pDC upon virus recognition. More particularly, viral particles containing double-stranded DNA genomes, such as HSV-1 and murine cytomegalovirus are recognized by TLR9 10, 11.
To investigate the ability of cord blood pDC to produce IFN-α following virus exposure, we first incubated pDC from one adult and one cord blood donor with increasing doses of live human CMV (HCMV). Fig. 5A shows that as expected, adult pDC secreted large quantities of IFN-α following wild-type HCMV stimulation in a dose-dependent manner. In contrast, cord blood pDC were profoundly impaired in their capacity to mount IFN-α production whatever the MOI used. To confirm this result, we incubated pDC from a larger number of adult donors and cord blood samples with HCMV at MOI 10, showing that cord blood pDC produced significantly less IFN-α as compared to their adult counterparts in response to HCMV (Fig. 5B). The defect in IFN-α production by cord blood pDC could be observed at any time of our kinetics, between 24 and 72 h (data not shown).

Impaired IFN-α production and IRF-7 translocation in cord blood pDC exposed to viruses. (A) Purified pDC were cultured at 2×105 cells/mL with graded doses (MOI) of wild-type HCMV. Cell-free supernatants were collected at 72 h and IFN-α levels were assayed by ELISA. One representative adult and cord blood sample out of three is shown for the dose response. (B) Purified pDC were cultured at 2×105 cells/mL with wild-type HCMV (MOI 10). Cell-free supernatants were collected at 72 h and IFN-α levels were assayed by ELISA. Scatter plot represents the IFN-α levels of 14 adult and nine cord blood samples. (C, D) Purified pDC (2×105 cells/mL) were exposed to HSV-1 for 24 h, and IFN-α (C) and TNF-α (D) levels were assayed by ELISA. Data represent the means ± SEM of five adult and five cord blood samples. (E) Adult and cord blood enriched pDC were stimulated for the indicated times with HSV-1 (MOI 1). After viral exposure, cell samples were stained as described in Materials and methods and run on the ImageStream machine. Data were analyzed to determine the percentage of pDC with nuclear localization of IRF-7 using the IRF-7/DRAQ5 similarity score (Supporting Information Fig. 2) following HSV-1 stimulation. Kinetics of translocation for one representative adult versus cord blood donor is shown. (F) Scatter plot shows the fold changes, calculated as ratios between the percentages of translocated cells in treated versus untreated pDC for each individual donor after 6 h incubation with HSV-1. This was done for six adult donors and six cord blood samples; **p<0.01 cord blood versus adult pDC.
Furthermore, similar dose-response experiments were performed with live HSV-1, a double-stranded DNA virus. As shown in Fig. 5C, live HSV-1 induced large amounts of IFN-α in adult pDC whereas IFN-α levels were significantly reduced from cord blood pDC for each MOI. Our data indicate that cord blood pDC are thus profoundly defective in IFN-α production upon HCMV and HSV-1 exposure. We also compared TNF-α production by adult and cord blood pDC and we could observe that cord blood pDC produce less TNF-α after HSV-1 exposure (Fig. 5D).
We next analyzed nuclear translocation of IRF-7 in response to HSV-1 using the ImageStream technology as described as above 9 (Supporting Information Fig. 2). The kinetic study of the nuclear translocation following HSV-1 incubation indicated that the highest percentage of adult pDC showing a nuclear localization of IRF-7 was reached after 6 h of stimulation (Fig. 5E). In contrast, IRF-7 nuclear translocation was not induced in cord blood pDC upon HSV-1 stimulation at any time of the kinetics. The analysis of a larger number of samples indicates that the mean of the fold changes for IRF-7 translocation in adult pDC reached the value of 1.9 after HSV-1 incubation whereas it was 1.1 for cord blood pDC in the same conditions (Fig. 5F). IRF-7 nuclear translocation in cord blood pDC was significantly impaired as compared to their adult counterparts following HSV-1 treatment. Our results thus show that nuclear translocation of the transcription factor IRF-7 is reduced in cord blood pDC after TLR9 ligation as well as after HSV-1 exposure.
Discussion
Human newborns are more susceptible than adults to infections with viruses, including human immunodeficiency virus, respiratory syncytial virus, HCMV and HSV 12–15. The immaturity of the cord blood immune system is often proposed as the main underlying factor for such limited anti-viral immune responses in early life 16. More precisely, functional alterations of T lymphocytes and antigen-presenting cells are critically involved in such compromised neonatal immune responses 17, 18. Here, we show that highly purified pDC from cord blood exhibit a profound defect in their capacity to produce IFN-α/β in response to TLR7/9 ligation as well as to CMV or HSV-1 exposure. This defect is detected both at the protein and mRNA levels. Interestingly, long-term resistance to HSV-1 could be induced in neonatal mice following IFN-α treatment at birth, highlighting the decisive role of IFN-α and pDC for viral resistance in early life 19. The fact that TLR7-dependent type I IFN synthesis is impaired in cord blood pDC suggests that cord blood pDC responses to single-stranded RNA viruses would be also compromised.
Through the activation of TLR7/9, robust induction of IFN-α/β was found to be regulated by the assembly of a multi-component complex MyD88/IRAK1/IRAK4/TRAF3/TRAF6 allowing direct recruitment and activation of IRF-7. After being phosphorylated by IRAK1 and activated by TRAF6, IRF-7, which is highly expressed by resting pDC, translocates to the nucleus to initiate IFN-α/β transcription and thus serves as a key mediator for transcription of IFN-α/β genes 20, 21. Recent data suggest that IRF-5 is also important for TLR7-dependent induction of type I IFN 22. On the other hand, IRF-5 nuclear translocation, together with NF-κB and MAPK activation, is crucial for the production of inflammatory cytokines and chemokines and the maturation of the pDC 23. Our results showed that TNF-α production and phenotypical maturation are also partially affected in cord blood pDC stimulated with R-848.
Beside TLR, a separate nucleic acid recognition pathway for type I IFN production during anti-viral responses involves RNA helicases RIG-1-MDA-5 and their adaptor IPS-1 21. Although the RIG-1-MDA-5-IPS-1 signalling is essential for type I IFN production in response to viruses by other immune cells, the implication of this pathway in type I IFN production by pDC will need more investigations 24. Until now, TLR are considered as the major receptors for the initiation of type I IFN production in pDC upon virus recognition.
Herein, we found that the defect of IFN-α/β production by cord blood pDC could not be attributed to a lower expression of TLR7 and TLR9 in those cells. Importantly enough, we demonstrate an impaired nuclear translocation of IRF-7 in cord blood pDC following CpG and HSV stimulation. In contrast, NF-κB translocation following TLR9 ligation using CpG treatment is less affected as compared with IRF-7 translocation in cord blood pDC. This is in concordance with our findings that NF-κB-dependent responses such as TNF-α induction as well as CD80, CD40 and HLA-DR up-regulation were less altered in cord blood pDC following TLR7 ligation. The fact that NF-κB activation was partially affected in cord blood pDC suggests that a perturbation in the assembly of the multi-component complex could account for our observation. Of note, we did not detect any difference in MyD88, IRAK and TRAF expression between adult and cord blood resting pDC. Whether the recruitment and/or activation of critical signalling molecules involved in IRF-7 activation is impaired remains to be determined. Altogether, we propose that impaired nuclear translocation of IRF-7 provides a molecular basis for deficient type I IFN production by cord blood pDC.
IRF-3-dependent responses to lipopolysaccharide were recently shown to be defective in cord blood cells. Indeed, deficient IRF-3 activity in lipopolysaccharide-stimulated cord blood monocyte-derived DC is associated with a reduced ability to produce IFN-β, IL-12p70 and IFN-inducible chemokines 25. We suggest that defective IRF-3 activation in cord blood monocyte-derived DC and impaired IRF-7 translocation in cord blood pDC represent two key molecular signatures underlying the immaturity of neonatal DC.
In conclusion, we suggest that deficient IRF-7 translocation in cord blood pDC blunts their ability to elicit anti-viral responses by limiting their production of type I IFN. This might be relevant to the increased susceptibility of human newborns to viral infections but also their inability to mount efficient immune responses to vaccines. Indeed, although CpG and R-848 molecules have been shown to display promising adjuvant activities in adults 26, 27, the use of these TLR agonists as neonatal vaccine adjuvants needs to be revisited in light of the present findings. Finally, defective IRF-7/type I IFN axis in cord blood pDC could also have important implications for the understanding of higher susceptibility to microbial infections encountered after cord blood cell transplantation 28, 29.
Materials and methods
Blood samples
Adult fresh blood was obtained from healthy volunteers with informed consent and umbilical cord blood samples were collected from healthy full-term neonates at the departments of obstetrics of four hospitals with parental agreement (Clinique Notre Dame-Charleroi, Notre Dame de Grâce-Gosselies, CHU-Charleroi and CHU-Tivoli-La Louvière). All procedures were approved by the ethics committee of the faculty of medicine at the Université Libre de Bruxelles.
pDC purification, culture and IFN-α production
PBMC and cord blood mononuclear cells (CBMC) were obtained from the blood samples by centrifugation on Ficoll-Hypaque (Axis-Shield PoC AS, Oslo, Norway). pDC were isolated by MACS, first by a negative depletion using a cocktail of antibodies (CD16, CD19, CD14, CD15, CD33, CD3 and Glycophorin-A) directly coupled to magnetic beads (Miltenyi Biotec) passed on an LD column, followed by a positive selection using the anti-BDCA-4-conjugated magnetic microbeads passed over two MS columns. The purity of the pDC fraction was checked by staining with the anti-BDCA-2-FITC (Miltenyi Biotec) and the CD123-PE (BD Biosciences, Erembodegem, Belgium) mAb and was >95%.
Freshly isolated pDC were cultured at 2.5×105 cells/mL in complete medium (RPMI 1640) (Biowhittaker Europe, Verviers, Belgium) supplemented with 10% FCS (Biowhittaker Europe), 2 mM L-glutamine (Life Technologies, Paisley, UK), 20 µg/mL gentamicin, 50 µM 2-mercaptoethanol and 1% nonessential amino acids (Life Technologies), and stimulated with resiquimod (R-848; Pharma Technologies, China) or CpG A (2216; Tib Molbiol, Berlin, Germany) (5′-ggGGGACGATCGTCgggggG-3′) for 48 h.
Viability of purified pDC after 48 h of culture in presence or absence of TLR ligand was determined by propidium iodide staining (10 µg/mL final concentration). In five independent experiments, we observed that the fractions of viable cells (mean ± SD) were 59 ± 10%, 76 ± 10%, 60 ± 3% for adult pDC, and 47 ± 8%, 68 ± 9%, 46 ± 10% for cord blood pDC in untreated, R-848 or CpG A conditions, respectively.
HCMV strain TB40/E (1×108 PFU/mL) was originally provided by Z. Tabi (College of Medicine, Cardiff, UK) and then propagated in our institute as described by Tabi et al. 30. pDC (2.5×105 cells/mL) were exposed to the virus at the indicated MOI. After 3 h medium was replaced by fresh medium supplemented with IL-3 (4 ng/mL; R&D Systems, Abingdon, UK). HSV-1 strain 2931 was provided by P. Fitzegerald-Bocarsly (Newark, NJ); pDC were exposed to the virus for 24 h at the indicated MOI. Cell-free supernatants were collected and stored at –20°C until quantification by ELISA for IFN-α, IFN-β (Biosource, Nivelles, Belgium), TNF-α and IL-6 (R&D Systems, Abdingdon, UK) following manufacturer's instructions.
Flow cytometry
For immunophenotyping analysis, pDC were membrane-stained during 20 min at 4°C with the following conjugated antibodies: anti-CD80-FITC, anti-CD86-FITC, anti-CD40-PE, anti-HLA-DR-FITC, anti-CD54-PE and anti-CCR7-PE (BD Biosciences). Intracellular staining of IRF-7 was done as described before 5 with the following modifications: fixation occurs in 1× CellFix (BD Biosciences) for 10 min at room temperature and the cells were permeabilized with Perm2 (BD Biosciences) for 10 min at room temperature. Finally, the cells were fixed in 1× CellFix (BD Biosciences). Data were obtained on a CyAnTM ADP LX9 flow cytometer and analyzed using the summit 4.2. software (DakoCytomation).
RNA purification and quantitative real-time RT-PCR
mRNA were extracted from purified pDC (>95%) using the MagNa Pure LC mRNA Isolation Kit following manufacturer's instructions (Roche Diagnostics, Brussels, Belgium). RT and real-time PCR were then carried out using LightCycler-RNA Master Hybridization Probes (one-step procedure) on a Lightcycler device (Roche Diagnostics). Primers and sequences are listed in Supporting Information Table 3.
Microarray experiments
RNA preparation
pDC from three adult and three cord blood donors were left untreated or stimulated with R-848 (10 µg/mL) for 2 h. The RNA was isolated using TRIzol reagent (Invitrogen, Merelbeke, Belgium) and RNAeasy Micro Kit (Qiagen, The Netherlands).
Affymetrix GeneChip hybridization
The RNA quality was checked with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). All RNA used in our experiments present the quality criteria defined by Agilent, and processed according to Affymetrix's instructions. Briefly, 150 ng of total RNA were reverse-transcribed using a T7-(dT) 24 primer and Superscript II reverse transcriptase. Double-stranded cDNA was synthesized and purified with GeneChip® Sample Cleanup Modules. cRNA was then synthesized using the Megascript T7 kit (Ambion) and purified. Six-hundred nanograms of this cRNA was reverse-transcribed using random hexamer primers, and Superscript II RT and double-stranded cDNA was subsequently synthesized and purified. Biotinylated cRNA was synthesized from the double-stranded cDNA using T7 RNA polymerase and a biotin-conjugated pseudo-uridine containing nucleotide mixture provided in the Affymetrix kit.
Prior to hybridization, the cRNA was purified and fragmented. Fifteen micrograms of each experimental sample and 15 µg of each Affymetrix eukaryotic hybridization control were hybridized for 16 h to human U133 Plus 2.0 GeneChips. Arrays were washed and stained with streptavidin-PE (Molecular Probes, Eugene, OR), biotinylated anti-streptavidin (Vector Laboratories, Burlingame, CA) and scanned with an Affymetrix GeneChip® Scanner 3000. The Affymetrix eukaryotic hybridization control kit and Poly-A RNA control kit were used to ensure efficiency of hybridization and cRNA amplification. All cRNA were synthesized at the same time. Experiments were done by DNAVision® (Gosselies, Belgium). Array images (“.DAT” files) were visually screened to discount for signal artefacts, scratches or debris and Affymetrix quality metrics were analyzed.
Data analysis
Various Bioconducter (www.bioconducter.org) packages for R (http://www.r-project.org) were used for data analysis. In brief, the Simpleaffy package was used to pre-process the data using the RMA algorithm 31. Differentially expressed genes were identified using linear models and empirical Bayesian methods 32 as implemented in the LIMMA package.
Three statistical approaches, all part of BioConductor software, were used to identify differentially expressed genes: (1) significance of analysis for microarrays 33, (2) a paired t-test controlling the false discovery rate using Benjamini and Hochberg correction 34, (3) the moderated t-test in linear models for microarray data 32 also enconcompassing the Benjamini and Hochberg correction. Data were first filtered based on “MA plot”, where M = log2(fluorescent dye intensity of infected DC/uninfected DC) and A = log2√fluorescent dye intensity of (treated pDC) × (untreated pDC), as described by Yang et al. [35] for cDNA microarray. In the analysis between treated and untreated pDC or untreated adult versus cord blood pDC, a gene was called differentially expressed if the M value was more than 2 (up-regulated) or less than –2 (down-regulated) and the p value was less or equal to 0.001. A gene is called differentially regulated between adult pDC and cord blood pDC if the M value was more than 1 or less than –1 and the p value was less or equal to 0.05.
Determination of nuclear translocation using the ImageStream
pDC were enriched from PBMC or CBMC by negative depletion as described above. Autologous PBMC or CBMC were added back to the enriched pDC fraction to reach the number of 2×106 cells per condition. Cells were stimulated for the indicated time with CpG A (10 µg/mL) or HSV-1 (MOI 1) at 37°C. After incubation, cells were surface-stained with both anti-BDCA-2-PE and anti-BDCA-4-PE antibodies, fixed in 1% formaldehyde and stored overnight at 4°C. The following day, samples were permeabilized with 0.1% Triton in 1× PBS and incubated with anti-IRF-7 or anti-NF-κB (p65) antibodies (SantaCruz Biotechnology, Santa Cruz, CA) 30 min at room temperature. For the secondary antibodies, goat anti-rabbit IgG-FITC (Jackson Immunoresearch, Suffolk, UK) was added and the cells were incubated for 15 min at room temperature. Following washing, cells were fixed in 1% formaldehyde and shipped to Amnis. Nuclei were stained with DRAQ5 immediately before acquisition. Samples were acquired using the ImageStream imaging flow cytometer. Images were captured in five channels: darkfield, brightfield, FITC, PE and DRAQ5, and analyzed using IDEAS software (Amnis) as recently described 9.
Briefly, single BDCA-2/4+ cells were gated and then analyzed for nuclear translocation of IRF-7 or NF-κB using similarity scoring of the transcription factor and DRAQ5 image pair on a per-cell basis. The similarity score is a log-transformed Pearson's correlation coefficient of the pixel values of the DRAQ5 and transcription factor images. If the transcription factor is nuclear-localized within a cell, the two images will appear similar and the cell will have a large positive value.
Statistical analysis
Data were compared using unpaired Mann–Whitney U–test.
Acknowledgements
We thank the staff from the obstetric departments of CHU Tivoli-La louvière, CHU-Charleroi, Clinique Notre Dame from Charleroi and Clinique Notre Dame de Grâce from Gosselies. We thank Nguyen Muriel and Thomas Séverine for their kind technical support. This work was supported by a grant from the Fonds pour la formation à la Recherche dans l'Industrie et dans l'Agriculture (F.R.I.A.) and the Télévie-Fonds National de la Recherche Scientifique program. S.G. is a post-doctoral Researcher of the Fonds National de la Recherche Scientifique. The Institute for Medical Immunology is supported by GSK Biologicals and the government of the Walloon Region. Part of these results have been presented at the 13th International Congress of Immunology (ImmunoRio, August 2007).
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
Appendix
Conflict of interest: The authors declare no financial or commercial conflict of interest.




