Full spectrum of clonal haematopoiesis-driver mutations in chronic heart failure and their associations with mortality
Stefanie Dimmeler, Andreas M. Zeiher, and Michael A. Rieger shared last authorship.
Abstract
Aims
Somatic mutations in haematopoietic stem cells can lead to the clonal expansion of mutated blood cells, known as clonal haematopoiesis (CH). Mutations in the most prevalent driver genes DNMT3A and TET2 with a variant allele frequency (VAF) ≥ 2% have been associated with atherosclerosis and chronic heart failure of ischemic origin (CHF). However, the effects of mutations in other driver genes for CH with low VAF (<2%) on CHF are still unknown.
Methods and results
Therefore, we analysed mononuclear bone marrow and blood cells from 399 CHF patients by deep error-corrected targeted sequencing of 56 genes and associated mutations with the long-term mortality in these patients (3.95 years median follow-up). We detected 1113 mutations with a VAF ≥ 0.5% in 347 of 399 patients, and only 13% had no detectable CH. Despite a high prevalence of mutations in the most frequently mutated genes DNMT3A (165 patients) and TET2 (107 patients), mutations in CBL, CEBPA, EZH2, GNB1, PHF6, SMC1A, and SRSF2 were associated with increased death compared with the average death rate of all patients. To avoid confounding effects, we excluded patients with DNMT3A-related, TET2-related, and other clonal haematopoiesis of indeterminate potential (CHIP)-related mutations with a VAF ≥ 2% for further analyses. Kaplan–Meier survival analyses revealed a significantly higher mortality in patients with mutations in either of the seven genes (53 patients), combined as the CH-risk gene set for CHF. Baseline patient characteristics showed no significant differences in any parameter including patient age, confounding diseases, severity of CHF, or blood cell parameters except for a reduced number of platelets in patients with mutations in the risk gene set in comparison with patients without. However, carrying a mutation in any of the risk genes remained significant after multivariate cox regression analysis (hazard ratio, 3.1; 95% confidence interval, 1.8–5.4; P < 0.001), whereas platelet numbers did not.
Conclusions
Somatic mutations with low VAF in a distinct set of genes, namely, in CBL, CEBPA, EZH2, GNB1, PHF6, SMC1A, and SRSF2, are significantly associated with mortality in CHF, independently of the most prevalent CHIP-mutations in DNMT3A and TET2. Mutations in these genes are prevalent in young CHF patients and comprise an independent risk factor for the outcome of CHF, potentially providing a novel tool for risk assessment in CHF.
Introduction
Chronic ischemic heart failure (CHF) is a potentially fatal condition that is age-associated and increases globally.1 Although there are considerable advances in the treatment of CHF, the mortality rate remains high, and there is thus an increasing need to understand the mechanisms underlying the pathogenesis and development of CHF. Clonal haematopoiesis (CH) is the emergence of expanded mutated blood cell clones, originating from somatic mutations in haematopoietic stem cells (HSCs) in genes such as DNMT3A, TET2, JAK2, TP53, ASXL1, SF3B1, and PPM1D, known to be associated with haematologic malignancies.2 CH is present in the absence of haematologic disorders, and its prevalence increases with age.3 When the mutated blood cell clone reaches a size of at least 2% variant allele frequency (VAF), which corresponds to 4% mutated blood cells assuming the mutation is heterozygous, the technically defined term clonal haematopoiesis of indeterminate potential (CHIP) has been introduced.3 In recent years, it has been shown that CHIP is associated with increased risk of atherosclerosis, coronary artery disease (CAD), and coronary calcification and with poor outcome in patients with CHF as well as in patients with severe aortic stenosis undergoing transcatheter aortic valve implantation.4-10 The most frequently mutated genes code for the epigenetic modifiers DNMT3A and TET2. In CHF, patients with DNMT3A or TET2 mutations with a VAF ≥ 2% show an increased progression of the disease and increased mortality,9 which correlated in a dose-dependent manner with the size of the mutated blood cell clone (VAF). Elegant mechanistic studies in preclinical models of atherosclerosis and cardiac failure have demonstrated that TET2-deficiency and DNMT3A-deficiency in blood cells induce inflammation, atherosclerosis progression, and cardiac dysfunction.11-13 In contrast, the role of recurrent mutations in other CH-related genes, especially at low VAF (≥0.5%), in the development of cardiovascular diseases is still unknown.
In this study, we aimed to assess the potential prognostic significance of blood cell mutations in CH-genes other than DNMT3A and TET2 causing CH at low VAF (between 0.5% and 2%) in a cohort of patients with CHF due to post-ischemic origin.
Methods
Study cohort
Clinical data and biological specimens [bone marrow (BM)-derived or serum-free peripheral blood (PB)] were collected from a total of 399 patients suffering from CHF and participated in various controlled trials between January 2002 and November 2018 at the University Hospital of the Johann Wolfgang Goethe University, Frankfurt/Main, Germany.
Patients were eligible for inclusion into the study if they suffered from stable CHF-symptoms with New York Heart Association ≥ II, had a previous successfully revascularized myocardial infarction at least 3 months prior to inclusion, and had a well-demarcated region of left ventricular dysfunction on echocardiography. Exclusion criteria were the presence of acutely decompensated heart failure with New York Heart Association (NYHA) class IV, an acute ischemic event within 3 months prior to inclusion into the registry, a history of severe chronic diseases, documented haematological disease or cancer within the preceding 5 years or unwillingness to participate.
All patients provided written informed consent for one of the following registered clinical trials: TOPCARE-CHD (Crossover or Registry, n = 310, NCT00289822 or NCT00962364), Cellwave (n = 65, NCT00326989) or REPEAT (n = 24, NCT 01693042). All patients provided informed consent for genetic testing of BM and PB samples. The ethics review board of the Goethe University in Frankfurt, Germany, approved the protocols. The study complies with the Declaration of Helsinki.
Sample preparation and next generation sequencing
DNA was isolated with QIAamp DNA Mini Kit (Qiagen, Netherlands) from either deep-frozen BM mononuclear cells or serum-free PB cells. The custom panel based on the Illumina TruSeq Custom Amplicon Low Input assay including 594 amplicons in 56 genes and the next generation sequencing were applied as previously reported.9 The median coverage across all samples was 4354× before unique molecular identifier (UMI) family clustering and 675× with inclusion of UMIs.
Variant calling was performed as previously reported.9 Variants with a VAF of 0.45 to 0.55 and ≥0.95 were not considered to exclude potential germline variants. The identified variants were processed and filtered using the r programming language (Ver. 3.3.1). Common SNPs with a minor allele frequency ≥ 5% in either the 1000 Genome Project, Exome Variant Server or ExAC databases were excluded. In addition, variants with a low mapping quality (<20) and variants occurring in 5% or more of the subjects in the studied cohort were considered as technical artefacts and excluded. Furthermore, variants covered with less than 100 reads in at least one set of amplicons (CAT A or CAT B), variants called with only one of the set of amplicons (CAT A or CAT B), SNPs identified as common in the dbSNP database (≥1% in the human population), and variants with sequence ontology terms ‘LOW’ or ‘MODIFIER’ were filtered out.
Statistical analysis
Continuous variables are presented as median (interquartile range), unless otherwise noted. Gaussian distribution was tested through a consideration of Kolmogorov–Smirnov test, Shapiro–Wilk test, and Q–Q plot. Kruskal–Wallis H testing was used for comparison of continuous and categorical variables between the groups. Numerical and categorical variables were compared with the χ² test, correlations were tested with Spearman correlation as appropriate.
For survival analysis within the individual groups, Kaplan–Meier and Cox-regression analyses were used and both 95% confidence intervals (CI) and hazard ratios (HR) were calculated. Log rank (Mantel–Cox) testing was applied for comparison of event-free survival analysis.
Statistical significance was assumed if P < 0.05, all reported P values are two-sided. Statistical analysis was performed with spss (Version 26.0, SPSS Inc.). The mutation matrix was created with paplot-0.5.6.post3 (Python-2.7.10.0).
Results
High frequency of CH-mutations with a VAF ≥ 0.5% in CHF patients
We analysed mononuclear BM and PB cells of 399 CHF patients by error-corrected deep targeted amplicon sequencing of 56 genes associated with myeloid malignancies and CH.9, 14 Utilizing an advanced variant calling, we reached a sensitivity of the VAF of 0.5% in our cohort, meaning that clones representing 1% of heterozygote mutated blood cells could be detected. To avoid VAF quantification artefacts, we included UMI in our sequencing analysis and determined the presence of the mutation with independent primer pools on both DNA strands. Patients in our cohort had a median age of 63 years, NYHA class II, and a median follow-up time of 3.95 years (interquartile range, 2.4–4.9). The median left ventricular ejection fraction was 30% owing to a previous myocardial infarction.
We identified 1,113 mutations in 347 CHF patients (Supporting Information, Table S1), which result in non-synonymous alterations of the coding sequence or in alterations of the splicing sites. All synonymous mutations were not considered in this study. Eighty-seven per cent of the patients carried at least one mutation with a VAF ≥ 0.5%, and only 52 patients did not show any mutation above the detection threshold. Intriguingly, 82% of patients younger than 50 years old harboured at least one CH-mutation. The prevalence of CH even further increased with age (P = 0.026, Figure 1). We detected at least one mutation in 81.6% in patients between age 50–59 years, 90.7% in patients between age 60–69 years, 88.8% in patients between age 70–79 years, and 100% in patients older than 80 years (Figure 1).
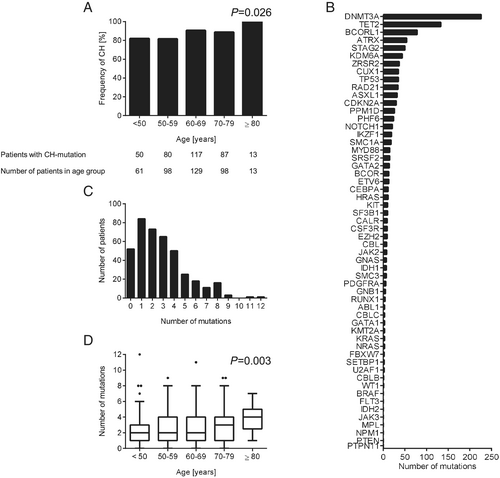
Mutations in the CH driver gene DNMT3A (226 mutations) were most frequent, followed by mutations in the genes TET2 (133 mutations), BCORL1 (78 mutations), ATRX (54 mutations), STAG2 (50 mutations), and in 43 other genes. (Figure 1B and Table S1). Twenty-one per cent of the patients carried 1 mutation, 47% showed 2 to 4 mutations, and 19% of the patients carried 5 to 12 CH-mutations. Only 13% CHF patients had no detectable mutations (Figure 1). The median number of CH-mutations per patient remained relatively stable for the age groups under the age of 70 but increased with age (P = 0.003) (Figure 1). Furthermore, the median VAF of CH-mutations increased significantly with age (P < 0.001, Figure S1). The median VAF of each individual gene is displayed in Figure S2. Among the single nucleotide substitutions, the appearance of mainly transitions from C to T (67%) and from A to G (23%) over transversions (in total less than 10%) has been described as age-associated mutation signature also being predominant in CH (Figure S3).15, 16
Prognostic significance of clonal haematopoiesis-mutations in chronic heart failure patients
During the follow-up of the study, 81 patients died (20%). In Figure 2, the death rate of CHF patients is shown in relation to mutations in different CH driver genes. Our results demonstrate that patients with mutations in genes such as SETBP1, KMT2A, CBL, CEBPA, and SRSF2, among other genes, showed an increased death rate compared with the average death rate of all patients (Figure 2). On the basis of these findings, we analysed the effects of 26 genes with a death rate of at least 25% to identify their relative contributions to increase death rate by employing a forest plot analysis (Figure 3). Mutations in the CH driver genes SETBP1, KMT2A, CBL, CEBPA, SRSF2, U2AF1, SMC1A, EZH2, GNB1, and PHF6 resulted in statistically significant increased HR in comparison with patients without CH mutations. Although mutations in SETBP1, U2AF1, and KMT2A had a large effect on CHF survival, as indicated by increased HR (Figure 3), the very low number of cases (two, two, and three patients, respectively) precludes any interpretation of the results, and these mutations were excluded for further analysis.
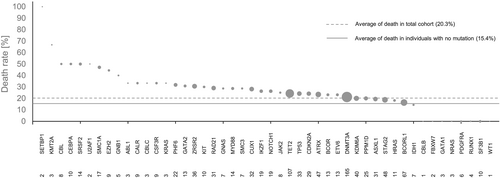
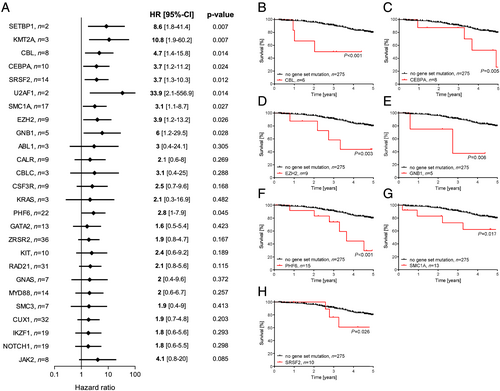
To eliminate confounding effects caused by the presence of DNMT3A and TET2 CHIP-mutations, we excluded patients with DNMT3A-related, TET2-related, and other CHIP-related mutations with a VAF ≥ 2% for further analysis. Kaplan–Meier analysis for overall survival showed that CHF patients with a CH driver mutation in CBL have a worse outcome of their disease than patients with no mutations in the genes CBL, CEBPA, EZH2, GNB1, PHF6, SMC1A, and SRSF2 (Figure 3). Similarly, Kaplan–Meier survival analyses revealed that mutations in CEBPA, EZH2, GNB1, PHF6, SMC1A, and SRSF2 are associated with increased mortality in CHF (Figure 3), independent from the presence of CHIP-driver mutations in DNMT3A, TET2, and other genes.
Recurrent mutations in a distinct set of genes are associated with reduced survival of chronic heart failure patients
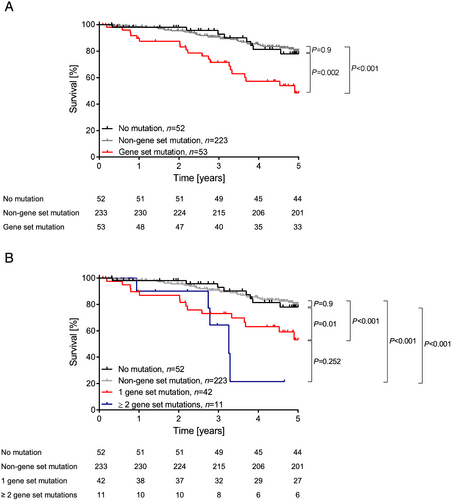
We restricted our further analyses on the effects of CBL, CEBPA, EZH2, GNB1, PHF6, SMC1A, and SRSF2, hereby called ‘risk gene set’, in survival of CHF patients. After excluding patients with other CHIP-related mutations with a VAF ≥ 2%, we could detect 66 mutations in the genes of the risk gene set carried by 53 patients (42 patients had 1 mutation, 9 patients 2 mutations, and 2 patients 3 mutations in genes of the risk gene set (Figures S4 and S5).
Chronic heart failure patients carrying mutations in this gene set showed significantly lower survival compared with patients with mutations in any other gene (P < 0.001) and patients without any mutations (P = 0.002) (Figure 4). The effects were independent of the number of mutations in the gene set (P < 0.001) (Figure 4). Additionally, the HR was significantly increased in patients carrying a mutation in the risk gene set compared with the patients without any mutation (HR, 3.3; 95%-CI, 1.5–7.5; P = 0.004), while there is no significant difference between the patients without any mutation and patients with mutations with a VAF < 2% in other genes than in the risk gene set (HR, 0.95; 95%-CI, 0.4–2.0; P = 0.9). To further demonstrate the independence of the increased risk of patients carrying a mutation in the gene set from the coexistence of mutations in DNMT3A and TET2, two prominent CH-driver genes with reported association with worse outcome in CHF,9 we excluded all patients in our cohort with any DNMT3A or TET2 mutation. Yet the Kaplan–Meier survival analysis demonstrated a significant decrease of survival when patients carry a mutation in the gene set (P = 0.0003), in comparison with patients without any mutation or mutations in other genes (Figure S6), independent on the presence of mutations in DNMT3A and TET2.
In Table 1, the baseline characteristics of patients with mutations in the gene set, patients carrying mutations in other genes, and patients without any mutations at all (with VAF ≥ 0.5% to <2%) are summarized. There were no significant differences in any parameter including the age of the patients, confounding diseases, severity of the CHF disease, heart failure risk assessments (according to the NYHA and Seattle Heart Failure Model) and blood cell parameters except for the number of platelets. Patients with mutations in the risk gene set presented slightly reduced platelet numbers in the PB. However, multivariable Cox regression analysis revealed that death remained independently associated with the presence of mutations in the risk gene set (HR, 3.1; 95% CI, 1.8–5.4; P < 0.001), whereas the number of platelets was no more significant (P = 0.084) (Table S2).
| Characteristic | Total cohort n = 328 | No mutation n = 52 | Non-gene set mutation n = 223 | Gene set mutation n = 53 | P value |
|---|---|---|---|---|---|
| Age (years, n = 328) | 61 (54; 69) | 57 (51; 64) | 62 (54; 69) | 62 (55; 69) | 0.219 |
| Sex (male/female, n = 328) | 275/53 | 43/9 | 190/33 | 42/11 | 0.554 |
| BMI (kg/m2, n = 323) | 27 (24; 30) | 28 (24; 30) | 27 (25; 30) | 27 (24; 31) | 0.752 |
| Hypertension (%, n = 328) | 71 | 69 | 71 | 72 | 0.960 |
|
Hypercholesterolemia (%, n = 318) |
77 | 73 | 79 | 72 | 0.344 |
| Diabetes mellitus (%, n = 325) | 30 | 29 | 30 | 30 | 0.986 |
| Smoking history (%, n = 317) | 71 | 79 | 68 | 78 | 0.624 |
| Family history of CAD (%, n = 312) | 53 | 43 | 55 | 54 | 0.151 |
| Extent of CAD 1/2/3 vessel disease (%, n = 315) | 32/24/44 | 30/36/34 | 34/22/44 | 25/23/52 | 0.082 |
| Atrial fibrillation (%, n = 321) | 80 | 82 | 77 | 90 | 0.081 |
| NYHA I/II/III/IV (%, n = 321) | 17/42/39/2 | 22/42/36/0 | 15/45/38/2 | 20/29/45/6 | 0.199 |
| Left ventricular ejection fraction (%, n = 302) | 30 (23; 40) | 30 (20; 40) | 30 (25; 40) | 30 (25; 40) | 0.511 |
| SHFM Score (n = 243) | 0.24 (−0.23;0.82) | 0.20 (−0.3;0.63) | 0.20 (−0.28;0.79) | 0.51 (−0.08;1.0) | 0.262 |
| Creatinine (mg/dL, n = 326) | 1.08 (0.91; 1.27) | 1.01 (0.88; 1.17) | 1.09 (0.92; 1.3) | 1.07 (0.9; 1.56) | 0.499 |
| C-reactive protein (mg/dL, n = 319) | 0.35 (0.2; 0.95) | 0.3 (0.16; 0.42) | 0.36 (0.23; 0.95) | 0.54 (0.21; 1.46) | 0.143 |
| Haemoglobin (g/dL, n = 326) | 14 (13; 15) | 14.4 (13.5; 15.2) | 14 (13; 15) | 14 (13; 15.2) | 0.969 |
| Haematocrit (%, n = 326) | 42 (40; 45) | 42 (40; 45) | 42 (40; 45) | 43 (39; 46) | 0.467 |
| Platelets (109/L, n = 276) | 211 (181; 249) | 233 (183; 262) | 211 (182; 247) | 199 (177; 229) | 0.038 |
| White blood cells (109/L, n = 328) | 8 (6; 9) | 8 (7; 9) | 8 (6; 9) | 7 (6; 9) | 0.814 |
- Patients with clonal haematopoiesis of indeterminate potential (CHIP)-mutations with a variant allele frequency ≥ 2% were excluded. Continuous variables are presented as median (interquartile range [IQR]). Continuous (not Gaussian distributed) and categorical variables were compared with the Kruskal–Wallis H test. Nominal variables were compared with the χ² test.
- BMI, body mass index; CAD, coronary artery disease; NYHA, New York Heart Association; SHFM Seattle Heart Failure Model.
Patients with mutations in the risk gene set did not harbour larger clones but had significantly more mutations than CHF patients with mutations in other genes (Figure S7A,B). There was no statistically relevant association of mutations in the gene set and co-operating mutations in other CH-driver genes in our cohort except of CSF3R (Table S3).
Interestingly, mutations in CBL, CEBPA, EZH2, GNB1, PHF6, SMC1A, and SRSF2 were abundant also in patients under the age of 60, and there was not a consistent increase of the prevalence of mutations in these genes in older individuals (P = 0.249; Figure S7C). These results suggest that accumulation of driver mutations in these specific risk genes may confer a profound increase in mortality even in younger CHF patients.
Discussion
We demonstrate here that recurrent mutations in a distinct set of genes, namely, in CBL, CEBPA, EZH2, GNB1, PHF6, SMC1A, and SRSF2, are significantly associated with increased mortality in patients with CHF of ischemic origin. Importantly, these effects were not confounded by co-occurring CHIP-mutations in DNMT3A, TET2, or other CHIP-driver genes with a VAF ≥ 2%. This is the first study implicating blood cell mutations with low VAF (0.5–2%) in the risk gene set in decreased long-term survival of CHF patients.
While first functional studies and correlative clinical patient data provide mechanisms of action for the association of DNMT3A and TET2 mutations in blood cells with the progression and outcome of cardiovascular diseases including CHF,4-14 the functional consequences of mutations in the risk set genes await further clarification.
CBL is an E3 ubiquitin protein ligase mediating the ubiquitination and degradation of activated receptor and non-receptor tyrosine kinases, and thereby regulating cytokine and growth factor cell signalling. CBL has been implicated as a negative regulator of immune responses. Genetic deletion of CBL augmented pro-inflammatory cytokine production and lung inflammation.17 CBL interferes with production of inflammatory cytokines by IgE-activated mast cells.18 CBL deficiency activates inflammatory cell recruitment19 and augmented microglia-mediated CNS inflammation after LPS stimulation.20 These results indicate that loss-of function mutations in CBL could be involved in sustaining chronic inflammation.
CEBPA is a myeloid transcription factor determining haematopoietic cell fate by inhibiting erythroid and inducing myeloid differentiation,21 and it is important for terminal differentiation and maturation of granulocytes.22-25 Mice deficient in CEBPA in HSCs are highly vulnerable to LPS-induced septic shock and CEBPA-deficient macrophages show altered functionality and produce less IL-10, an anti-inflammatory cytokine.26 CEBPA interferes with the production of pro-inflammatory cytokines.27, 28 These results suggest that CEBPA is a negative regulator of inflammation and in this way loss-of function mutations may contribute to CHF disease progression.
EZH2 is as a histone-lysine N-methyltransferase the enzymatic component of the polycomb repressive complex II and involved in such in gene repression, regulating cell cycle, proliferation, and differentiation.29-32 Mutations in EZH2 can lead to gain-of-function or loss-of-function and exert context-specific and cell-specific effects. Several studies have provided evidence suggesting EZH2 as an important regulator of immune responses. EZH2 regulates maintenance and proliferative capacity of B cells.33 EZH2 plays also important roles in CD4 + and CD8 + T cell function, TH1 and TH2 differentiation, and Treg differentiation.34-36 Moreover, EZH2 is highly expressed in activated lymphocytes,37 and it is an important regulator of macrophage activation and inflammation and its inhibition leads to suppressed activation of BM-derived macrophages and reduces inflammatory responses.38 These results suggest that the involvement of EZH2 in regulation of immune responses maybe a potential mechanism by which EZH2 contributes to the progression of CHF.
GNB1 encodes the β subunit of heterotrimeric G proteins, which are essential to the signalling function of G-protein-coupled receptors.39 GNB1 plays an important role during inflammation being involved in integrin activation and leukocyte recruitment and arrest.40 Conversely, GNB1 inhibits the NLRP3 inflammasome activation in macrophages.41 The exact role of GNB1 in CHF remains to be investigated.
PHF6 is a chromatin-binding transcriptional regulator of the self-renewal of HSCs by enhancing TNF-α mediated growth inhibition in HSCs.42 PHF6 inactivating mutations are commonly observed in human myeloid and T-cell malignancies.43 Interestingly, decreased numbers of granulocyte-monocytic progenitors but increased haematopoietic stem and progenitor cells (HSPCs) were found in the BM of PHF6 knockout mice.44 Moreover, PHF6 deficient mice have reduced numbers of CD4 + T and CD8 + T cells and increased numbers of B cells in the PB.44 These results suggest that alterations in PHF6 expression and function may result in changes in the composition of lymphocytes subsets and by this way contribute to disease progression in CHF.45
SMC1A is a core component of the cohesin complex that is required for proper cohesion and correct segregation of sister chromatids during cell division. Mutations in SMC1A have been also associated with Cornelia de Lange Syndrome characterized by cardiac malformations.46 Individuals with Cornelia de Lange Syndrome display decreased ratios of certain T cell subsets.47 Cohesin proteins control inflammatory gene expression in HSPCs,48 and their differentiation in response to inflammation and aging.49 It can be speculated that changes in SMC1A/cohesion function due to mutations may result in enhanced inflammatory responses and in this way contribute to adverse outcomes in CHF patients.
SRSF2 is a splicing factor involved in the proliferation and differentiation of HSPCs, and mutations in SRSF2 have been associated with many myeloid diseases. Additionally, SRSF2 has been associated with a variety of immune disorders including systemic lupus erythematosus,50 and human immunodeficiency virus infection.51 Furthermore, SRSF2 is known to regulate gene expression of many T-cell molecules,52 suggesting that SRSF2 may regulate T-cell functions and in this way inflammatory responses.
Future studies on the functional alterations and pathogenic mechanisms induced by distinct mutations in the risk gene set in blood cell lineages and their role in the development and progression of CHF must be conducted using relevant preclinical models.
The high prevalence of CH in our cohort of CHF patients was striking. Even at relatively young age (younger than 50 years), most patients carried at least one CH-mutation with a VAF ≥ 0.5%. Although a few other studies have reported on a high prevalence of CH cases in middle age people by increasing the sensitivity and accuracy of the detection method,53 they have not reported such a high frequency of CH with a VAF of at least 0.5% as in our CHF patients.54-56 Interestingly, CHF patients in our cohort carried increased numbers of CH mutations with low VAF ≥ 0.5% independently of their age. These results suggest that the presence of mutations in distinct CH-related genes in HSPCs, although it may not lead to haematological malignancies, may contribute to the development of other diseases, such as heart failure in the presence of other risk factors. Strikingly, mutations in the risk gene set were identified in all age groups, and they were abundant even in younger individuals. Taken together, these findings suggest that mutations in certain CH-related genes contribute to the development of cardiovascular diseases and heart failure independent of age, in contrast to previous reports, demonstrating a correlation between CHIP mutations, age, and cardiovascular disease progression.
Although the results of this study demonstrate that mutations in the risk gene set associate with poor survival in CHF patients, we provide no functional evidence demonstrating how the characterized mutations are involved in CHF. Functional studies in relevant preclinical models will enlighten the mechanisms by which these mutations contribute to the outcome in CHF. Due to the fact that the patients studied in this cohort are not a consecutive series of patients and the sample size is limited, future studies will have to validate our findings in larger CHF patient cohorts. Moreover, it would be of great importance to assess the potential prognostic value of genetic testing of the risk gene set in addition to, for example, N terminal pro brain natriuretic peptide (NT-proBNP) serum levels or the Seattle Heart Failure Model score in CHF patients in a larger cohort.
In conclusion, we show in this study that somatic mutations with low VAF in a distinct set of genes, namely, in CBL, CEBPA, EZH2, GNB1, PHF6, SMC1A, and SRSF2, are significantly associated with mortality in CHF, independently of mutations in the classical CHIP-driver genes DNMT3A and TET2. Mutations in the CH risk gene set are prevalent in young CHF patients and comprise an independent risk factor for the outcome of CHF, potentially providing a novel tool for cardiovascular risk assessment and precision medicine in CHF.
Acknowledgements
The study was supported by the German Research Foundation (SFB 834 project B6 to B.A. and A.M.Z and Z1 to L.D. and M.A.R., the Excellence Cluster Cardio-Pulmonary Institute, and project RI2462/1–1 (M.A.R.)), and by the German Center for Cardiovascular Research, DZHK, Berlin, Germany, partner site Frankfurt Rhine-Main. M.A.R received financial support by the Jose Carreras Leukämie-Stiftung (Grant 10R/2017), the Hessisches Ministerium für Wissen-schaft und Kunst (III L 5-519/03/03.001 – [0015]), and the Wilhelm Sander-Stiftung (Grant 2018.116.1).
Open access funding enabled and organized by Projekt DEAL.
Conflict of interest
All authors declare no competing interests.
Authorship contributions
S.D., A.Z., and M.A.R. initiated and supervised the study. M.A.R. wrote the manuscript with the help of E.P. and K.C.K. K.C.K. and E.P. performed the experiments, analysed the data. S.C., K.K., L.D., T.R., and B.A. established technologies and acquired data. K.A. performed the bioinformatic data analyses. K.C.K. and A.B. performed the statistical data analyses. H.S., S.D., and A.Z. supported the study and provided valuable comments on the manuscript. All authors discussed early versions of the manuscript and provided comments and suggestions for change. All authors have approved the final report for publication.




