Allotetraploid Origin and Putative Ancient Introgression in Plantago hakusanensis (Plantaginaceae)
Funding: This work was supported by Environmental Restoration and Conservation Agency, No. 4-2001 and Japan Society for the Promotion of Science (17K07527, 19K06805 and 24K09566).
ABSTRACT
Plantago hakusanensis (2n = 4x = 24) is an endangered endemic species that occurs in subalpine zones in Japan. To clarify the unresolved phylogenetic position of P. hakusanensis within subgenus Plantago, we constructed a phylogenetic tree based on the nuclear-encoded single-copy gene sucrose–proton symporter 1 (SUC1) using 60 previously reported alleles from 24 taxa in subgenus Plantago. We found that P. hakusanensis was closely related to Plantago asiatica var. densiuscula. The phylogenetic relationships between P. hakusanensis and P. asiatica var. densiuscula were further examined by analyses of the SUC1 nuclear regions and the internal transcribed spacer (ITS) of rDNA, genome-wide single-nucleotide polymorphism genotyping (via multiplexed inter-simple sequence repeat genotyping by sequencing), as well as by additional analyses of three chloroplast (cp) regions (trnL-F, ndhF-rpl32, and rpl32-trnL) in 25 individuals of P. hakusanensis and 53 individuals of P. asiatica var. densiuscula covering the species' geographical distribution. Monophyly of P. hakusanensis was suggested by the nuclear marker analyses, whereas the cp haplotypes of P. hakusanensis were shared with P. asiatica var. densiuscula and P. asiatica in China. The disparity between the nuclear and cp data may be explained by the introgression of the cp genome (cp capture). This research provides a phylogenetic tree showing the position of P. hakusanensis within subgenus Plantago and molecular evidence that implies complicated introgressions between P. hakusanensis and P. asiatica var. densiuscula.
1 Introduction
Plantago hakusanensis Koidz. (Plantaginaceae) (2n = 4x = 24) is a perennial herb endemic to Japan (Yamazaki 1993). P. hakusanensis is distributed around snow patches and wet fields in subalpine zones in Honshu (ca. 1500–2300 m above sea level; Koizumi 1930; Yamazaki 1992, 1993). The species has been found on 12 mountains located between Mt. Moriyoshi (Akita Prefecture) to the north and Mt. Hakusan (Ishikawa Prefecture) to the south (Yamada and Satomi 1975). P. hakusanensis and its hairless form, P. hakusanensis f. glabra T. Yamaz., have been recognized as category I endangered taxa in Akita and Nagano Prefectures, and as category II vulnerable taxa in Fukushima, Gunma, Ishikawa, and Gifu Prefectures (Japanese Red Data 2020). Based on morphological features, P. hakusanensis has been assigned to the subgenus Plantago, which comprises ca. 131 species in five sections (Rahn 1996). Rahn (1996) suggested that P. hakusanensis belongs to sect. Plantago and is related to P. asiatica L. However, molecular phylogenetic studies within this subgenus (Ishikawa et al. 2009) have shown that the sect. Plantago is paraphyletic and requires reclassification, and no molecular phylogenetic analyses have been conducted to identify the species most closely related to P. hakusanensis. In addition, the sectional placement of P. hakusanensis has been debated by some researchers. For example, Yamazaki (1992) suggested that P. hakusanensis is most closely related to P. gentianoides and placed it in section Gentianoides Pilg. because it possesses anthers with a slender acuminate projection at the apex and seeds with a concave ventral face, as observed in Plantago gentianoides Sibth. et Smith from central Asia.
P. hakusanensis has been identified as a conservation target on Mt. Hakusan because of concerns that it may be genetically polluted via hybridization with P. asiatica L. var. densiuscula Pilg. (2n = 4x = 24) (Pilger 1922; Nogami 2001; Nakayama et al. 2006, 2008; Sano et al. 2016, 2019). P. asiatica var. densiuscula is a perennial herb distributed from the Japanese Archipelago to Taiwan (Pilger 1922). It commonly grows in sunny locations, such as roadsides and unpaved parking lots, at low elevations (Pilger 1922; Yamazaki 1993; Ishikawa et al. 2006, 2009). P. asiatica var. densiuscula is morphologically similar to P. hakusanensis, but the two taxa differ in several respects, including the numbers of seeds per fruit, seed morphology, and leaf shape. P. hakusanensis has one to two seeds per fruit, whereas P. asiatica var. densiuscula has four to seven seeds per fruit (Pilger 1922; Yamazaki 1993; Nakayama et al. 2008; Ohashi 2017). P. asiatica var. densiuscula has invaded the native habitats of P. hakusanensis on Mt. Hakusan, such as Minami Ryugabanba Campsite (approximately 2080 m above sea level) (Nogami 2001, 2002, 2003; Nakayama et al. 2005, 2008; Sano et al. 2022). The sticky wet seeds of P. asiatica var. densiuscula may have been carried to the subalpine zone via the shoes of hikers or along with construction materials of mountain shelters (Nogami 2001, 2003). The two taxa are genetically compatible, and fertile F1 hybrids have been obtained by artificial pollination (Sano et al. 2016). The occurrence of protogynous and wind-pollinated flowers in the two taxa increases their outcrossing rate; moreover, they had overlapping flowering periods on Mt. Hakusan during 3 out of the 4 years between 2011 and 2014 (Sano et al. 2019). Putative hybrids with an intermediate leaf shape have been found on Mt. Hakusan at locations where the taxa are sympatric (Nakayama et al. 2008). The government ministries of Japan (Ministry of Agriculture, Forestry and Fisheries of Japan et al. 2015) led efforts to remove P. asiatica var. densiuscula from Mt. Hakusan, where it is regarded as an exotic taxon. Genetic pollution is also a concern in other locations. Invasions of P. asiatica var. densiuscula have been reported on Mt. Gassan (Yokoyama 2015). Although P. hakusanensis is (i) an endemic species with limited distribution and (ii) threatened by hybridization with P. asiatica var. densiuscula, its phylogenetic relationships and evolutionary origins have not been investigated adequately.
Polyploidy is frequent in the subgenus Plantago (67% of species, as shown by chromosome counts, Rahn 1996), and allopolyploidy has been identified by molecular phylogenetic evidence (Ishikawa et al. 2009). Hence, we aimed to determine the phylogenetic position of tetraploid P. hakusanensis within the subgenus Plantago via phylogenetic analyses based on the nuclear-encoded single-copy gene sucrose–proton symporter 1 (SUC1) using 60 previously reported alleles from 24 representative taxa in the subgenus Plantago. We obtained the DNA sequence of SUC1 in P. hakusanensis using either cloning or allele (homoeolog)-specific PCR amplification. Furthermore, although Mt. Chokai is one of the popular destinations among hikers during the summer, there have been no previous reports of an invasion by P. asiatica var. densiuscula. To investigate the potential invasion of P. asiatica var. densiuscula into subalpine areas and to assess possible hybridization between Plantago hakusanensis and P. asiatica var. densiuscula on Mt. Chokai, we conducted phylogenetic analyses of the nuclear-encoded rDNA internal transcribed spacer (ITS) regions and performed genome-wide single-nucleotide polymorphism (SNP) genotyping using multiplexed inter-simple sequence repeat genotyping by sequencing (MIG-seq) (Suyama and Matsuki 2015; Suyama et al. 2022) and three chloroplast (cp) regions (trnL-F, ndhF-rpl32, and rpl32-trnL).
2 Materials and Methods
2.1 Taxon Sampling and DNA Isolation
To determine the phylogenetic position of Plantago hakusanensis within the subgenus Plantago, we conducted a phylogenetic analysis using a nuclear-encoded, single-copy SUC1 gene region spanning from exon 1 to exon 2. We newly sequenced this region from two individuals of P. hakusanensis collected from Mt. Hakusan and incorporated 60 previously reported SUC1 alleles from 24 taxa (Ishikawa et al. 2009; Table 1). These taxa represent all five sections (Micropsyllium, Mesembrynia, Virginica, Oliganthos, and Plantago) of the globally distributed subgenus Plantago (Rahn 1996). Alleles were isolated from one individual of each of these 24 taxa, except in the case of diploid P. major L., from which we collected two individuals. The numbers of alleles obtained from each individual varied from one to seven, depending on the levels of ploidy and/or heterozygosity (Ishikawa et al. 2009). We selected P. tenuiflola Waldst. & Kit, and one of the 24 taxa evaluated by Ishikawa et al. (2009) as an outgroup based on previously determined phylogenetic relationships within subgroup Plantago (Rønsted et al. 2002; Ishikawa et al. 2009; Iwanycki Ahlstrand et al. 2019). Note that Plantago formosana Tateishi & Masam. is considered a synonym of P. asiatica subsP. asiatica and has been discussed as a synonym of Plantago major L. (Hatusima 1971; Shimabuku 1997). The phylogenetic analysis showed that P. hakusanensis is closely related to P. asiatica var. densiuscula. Therefore, to investigate differences between P. hakusanensis and P. asiatica var. densiuscula further, we conducted additional analyses using genetic polymorphisms obtained from the same SUC1 region (extending from exon 1 to exon 2), the nuclear-encoded rDNA ITS regions, and three cp regions (trnL-F, ndhF-rpl32, and rpl32-trnL). We also used MIG-seq to identify genome-wide SNPs (Suyama and Matsuki 2015; Suyama et al. 2022). We used 25 individuals of P. hakusanensis and 53 individuals of P. asiatica var. densiuscula, including P. asiatica var. densiuscula f. yakusimensis (Masam) N. Ishikawa et al. The samples of P. hakusanensis were collected from Mt. Hakusan (4 individuals), Mt. Chokai (3 populations, 17 individuals), Mt. Gassan (1 individual), and Mt. Asahi (3 individuals), thereby covering the main distribution range of the taxon. Mt. Hakusan is the lectotype locality, and both Mt. Chokai and Mt. Gassan were listed as known localities of P. hakusanensis specimens in the original description (Koizumi 1930). The samples of P. asiatica var. densiuscula comprised 46 individuals collected from Taiwan, Cheju Island in Korea, and a broad geographical range across the Japanese Archipelago. We also included seven individuals from the subalpine zone on Mt. Chokai to investigate hybridization between P. hakusanensis and P. asiatica var. densiuscula invaders in the subalpine zone. Although P. hakusanensis was primarily distributed around snow patches and P. asiatica var. densiuscula was observed near both former and current mountain huts (Table 2), the distributions of the two taxa overlapped along a mountain trail on Mt. Chokai at elevations of 1320–1540 m.
| No. | Section | Species | Number of allele (s) | References (sample name) |
|---|---|---|---|---|
| 1 | Plantago | P. cornuti Gouan. | 1 | Ishikawa et al. (2009) |
| 2 | P. major L. | 1 | Ishikawa et al. (2009), (Japan) | |
| P. major | 1 | Ishikawa et al. (2009), (Germany) | ||
| 3 | P. major var. japonica (Franch. et Sav.) Miyabe | 1 | Ishikawa et al. (2009) | |
| 4 | P. maxima Jacq. | 2 | Ishikawa et al. (2009) | |
| 5 | P. reniformis Beck. | 1 | Ishikawa et al. 2009 | |
| 6 | P. asiatica L. var. densiuscula Pilg. | 2 | Ishikawa et al. (2009) | |
| 7 | P. asiatica var. densiuscula f. yakusimensis (Masam.) N. Ishikawa et al. | 2 | Ishikawa et al. (2009) | |
| 8 | P. palmata Hook. f. | 2 | Ishikawa et al. (2009) | |
| 9 | P. rugelii Decne. | 2 | Ishikawa et al. (2009) | |
| 10 | P. formosana Tateishi et Masam. | 3 | Ishikawa et al. (2009) | |
| 11 | P. media L. | 4 | Ishikawa et al. (2009) | |
| 12 | Mesembrynia | P. debilis R. Br. | 1 | Ishikawa et al. (2009) |
| 13 | P. depressa Willd. | 1 | Ishikawa et al. (2009) | |
| 14 | P. camtschatica Link. | 1 | Ishikawa et al. (2009) | |
| 15 | P. stauntoni Reichardt. | 2 | Ishikawa et al. (2009) | |
| 16 | P. spathulata Hook. f. | 7 | Ishikawa et al. (2009) | |
| 17 | P. raoulii Decne. | 4 | Ishikawa et al. (2009) | |
| 18 | Virginica | P. tomentosa Lam. | 2 | Ishikawa et al. (2009) |
| 19 | P. trinitatis Rahn. | 2 | Ishikawa et al. (2009) | |
| 20 | P. virginica L. | 2 | Ishikawa et al. (2009) | |
| 21 | P. australis Lam. | 4 | Ishikawa et al. (2009) | |
| 22 | Oliganthos | P. uniglumis Walp. | 5 | Ishikawa et al. (2009) |
| 23 | P. rigida Kunth. | 5 | Ishikawa et al. (2009) | |
| 24 | Micropsyllium | P. tenuiflola Waldst. et Kit | 2 | Ishikawa et al. (2009) |
| Population no. | Sample size | ITS genotype (n) | Cp haplotype (n) | Latitude, longitude | Altitude (m) | Locality | Reference |
|---|---|---|---|---|---|---|---|
| P. hakusanensis | |||||||
| 1 | 3 | H (3) | H1 (1), H11 (2) | Unknown | Unknown | Mt. Asahi, Asahi-cho, Nishimurayama-gun, Yamagata pref., Japan | |
| 2 | 9 | H (9) | H1 (4), H2 (5) | 39°04′ N, 140°02′ E | 1556 | Mt. Chokai, Yusa-cho, Akumi-gun, Yamagata pref., Japan | |
| 3 | 5 | H (5) | H1 (3), H2 (1) | 39°04′ N, 140°02′ E | 1517 | Mt. Chokai, Yusa-cho, Akumi-gun, Yamagata pref., Japan | |
| 4 | 3 | H (3) | H1 (3) | 39°04′ N, 140°02′ E | 1282 | Mt. Chokai, Yusa-cho, Akumi-gun, Yamagata pref., Japan | |
| 5 | 1 | H (1) | H11 (1) | 38°32′ N, 139°59′ E | 1388 | Mt. Gassan, Tsuruoka, Yamagata pref., Japan | |
| 6 | 4 | G (4) | H10 (4) | 36°08′ N, 136°46′ E | 2072 |
Mt. Hakusan, Shiramine, Hakusan city, Ishikawa pref., Japan |
|
| P. asiatica var. densiuscula | |||||||
| 7 | 4 | A (4) | H3 (4) | 39°04′ N, 140°02′ E | 1539 | Mt. Chokai, Yusa-cho, Akumi-gun, Yamagata pref., Japan | |
| 8 | 3 | A (3) | H4 (2), H8 (1) | 39°04′ N, 140°02′ E | 1282 | Mt. Chokai, Yusa-cho, Akumi-gun, Yamagata pref., Japan | |
| 9 | 3 | A (3) | H3 (2), H4 (1) | 39°06′ N, 139°52′ E | 4 | Fukuura Misaki, Yusa-cho, Akumi-gun, Yamagata pref., Japan | |
| 10 | 2 | A (1) | H8 (1) | 34°40′ N, 135°50′ E | 112 | Nara park, Nara pref., Japan | Ishikawa et al. (2006)a |
| 11 | 1 | ND | H8 (1) | 34°39′ N, 135°50′ E | 107 | Nara University of Education, Nara pref., Japan | |
| 12 | 1 | A (1) | H1 (1) | Unknown | Unknown | Shirakawa, Gifu pref., Japan | Ishikawa et al. (2006)a |
| 13 | 2 | A (1) | H8 (1) | 35°00′ N, 135°51′ E | 123 | Otu city, Shiga pref., Japan | Ishikawa et al. (2006)a |
| 14 | 1 | A (1) | ND | 35°00′ N, 135°47′ E | 64 | Sakyo-ku, Kyoto pref., Japan | Ishikawa et al. (2006)a |
| 15 | 1 | A (1) | H4 (1) | 34°27′ N, 136°43′ E | 18 | Ise city, Mie pref., Japan | Ishikawa et al. (2006)a |
| 16 | 1 | A (1) | H8 (1) | 34°14′ N, 133°00′ E | 6 | Ohmi-shima Island, Ehime pref., Japan | Ishikawa et al. (2006)a |
| 17 | 1 | A (1) | H6 (1) | 33°53′ N, 133°11′ E | 44 | Saijyo city, Ehime pref., Japan | Ishikawa et al. (2006)a |
| 18 | 1 | A (1) | ND | 33°35′ N, 130°24′ E | 3 | Fukuoka city, Fukuoka pref., Japan | Ishikawa et al. (2006)a |
| 19 | 2 | A (2) | H8 (1) | 33°31′ N, 130°32′ E | 50 | Dazaifu city, Fukuoka pref., Japan | Ishikawa et al. (2006)a |
| 20 | 1 | A (1) | H9 (1) | Unknown | Unknown | Mt. Yuwan, Amamiohshima Island, Kagoshima pref., Japan | Ishikawa et al. (2006)a |
| 21 | 1 | D (1) | H8 (1) | 30°44′ N, 130°59′ E | 22 | Tanega-shima Island, Kagoshima pref., Japan | Ishikawa et al. (2006)a |
| 22 | 1 | A (1) | ND | 38°18′ N, 141°33′ E | 62 | Kinkazan Island, Miyagi pref., Japan | Ishikawa et al. (2006)a |
| 23 | 2 | A (1) | H8 (1) | 34°17′ N, 132°18′ E | 22 | Miyajima Island, Hiroshima pref., Japan | Ishikawa et al. 2006)a |
| 24 | 1 | A (1) | H4 (1) | 43°03′ N, 141°18′ E | 34 | Sapporo city, Hokkaido pref., Japan | Ishikawa et al. (2006)a |
| 25 | 1 | A (1) | H4 (1) | Unknown | Unknown | Akita pref., Japan | Ishikawa et al. (2006)a |
| 26 | 1 | A (1) | H4 (1) | 38°20′ N, 141°30′ E | 364 | Mt. Hikariyama, Miyagi pref., Japan | Ishikawa et al. (2006)a |
| 27 | 1 | A (1) | H1 (1) | 35°42′ N, 139°45′ E | 23 | Hongo campus of the University of Tokyo, Tokyo, Japan | Ishikawa et al. (2006)a |
| 28 | 1 | B (1) | H1 (1) | 35°18′ N, 139°32′ E | 8 | Kamakura, Kanagawa pref., Japan | Ishikawa et al. (2006)a |
| 29 | 1 | C (1) | ND | 36°35′ N, 137°26′ E | 474 | Toyama pref., Japan | Ishikawa et al. (2006)a |
| 30 | 1 | E (1) | H5 (1) | Unknown | Unknown | Mt. Ontake, Nagano pref., Japan | Ishikawa et al. (2006)a |
| 31 | 1 | A (1) | ND | Unknown | 2200 | Nagano pref., Japan | Ishikawa et al. (2006)a |
| 32 | 1 | A (1) | H1 (1) | 36°35′ N, 137°27′ E | 977 | Toyama pref., Japan | Ishikawa et al. (2006)a |
| 33 | 1 | A (1) | H7 (1) | 34°57′ N, 137°09′ E | 48 | Okazaki, Aichi pref., Japan | Ishikawa et al. (2006)a |
| 34 | 1 | A (1) | ND | 34°50′ N, 137°32′ E | 140 | Mt. Maruyama, Aichi pref., Japan | Ishikawa et al. (2006)a |
| 35 | 1 | A (1) | ND | 34°29′ N, 136°51′ E | 66 | Sakatejima Island, Mie pref., Japan | Ishikawa et al. (2006)a |
| 36 | 1 | A (1) | H8 (1) | 34°32′ N, 134°59′ E | 2 | Awajishima Island, Hyogo pref., Japan | Ishikawa et al. (2006)a |
| 37 | 1 | A (1) | ND | Unknown | 800 | Mt. Daisen, Tottori pref., Japan | Ishikawa et al. (2006)a |
| 38 | 1 | A (1) | H8 (1) | 35°18′ N, 133°40′ E | 516 | Hiruzen, Okayama pref., Japan | Ishikawa et al. (2006)a |
| 39 | 1 | A (1) | ND | Unknown | 1270 | Mt. Karasugasen, Tottori pref., Japan | Ishikawa et al. (2006)a |
| 40 | 1 | A (1) | H8 (1) | 33°37′ N, 130°25′ E | 3 | Kyushu University, Fukuoka pref., Japan | Ishikawa et al. (2006)a |
| 41 | 1 | D (1) | ND | 31°35′ N, 130°36′ E | 7 | Sakurajima Island, Kagoshima pref., Japan | Ishikawa et al. (2006)a |
| 42 | 1 | A (1) | H8 (1) | 31°33′ N, 130°42′ E | 33 | Sakurajima Island, Kagoshima pref., Japan | Ishikawa et al. (2006)a |
| 43 | 1 | A (1) | H1 (1) | Unknown | Unknown | Tawulun Fort, Keelung, Taiwan | Ishikawa et al. (2006)a |
| 44 | 1 | A (1) | H1 (1) | Unknown | Unknown | Ilan, Taiwan | Ishikawa et al. (2006)a |
| 45 | 1 | A (1) | ND | Unknown | 1700 | Mt. Halla, Cheju Island, Korea | Ishikawa et al. (2006)a |
| P. asiatica var. densiuscula f. yakusimensis | |||||||
| 46 | 2 | F (1) | H8 (1) | 30°20′ N, 130°30′ E | 1911 | Mt. Miyanoura, Yakushima Island, Kagoshima pref., Japan | Ishikawa et al. (2006)a |
| 47 | 1 | F (1) | H8 (1) | 30°19′ N, 130°30′ E | 1789 | Mt. Kuromi, Yakushima Island, Kagoshima pref., Japan | Ishikawa et al. (2006)a |
| P. asiatica | |||||||
| 48 | 1 | J (1) | H11 (1) | Unknown | Unknown | China, cult. | Rønsted et al. (2002)a and Iwanycki Ahlstrand et al. (2019), Kew DNA bank ID = 9585, K |
| P. major | |||||||
| 49 | 1 | I (1) | Not included | Unknown | Unknown | Sachsen-Anhalt, Germany | Rønsted et al. (2002)a |
| P. major var. japonica | |||||||
| 50 | 1 | J (1) | ND | 39°06′ N, 139°52′ E | 5 | Yusa-cho, Akumi-gun, Yamagata pref., Japan | |
| P. camtschatica | |||||||
| 51 | 1 | K (1) | H12 (1) | 38°43′ N, 139°40′ E | 1 | Tsuruoka city, Yamagata pref., Japan | |
| 52 | 1 | L (1) | H13 (1) | Unknown | Unknown | (Rahn 684, C) | Iwanycki Ahlstrand et al. (2019)b |
- Abbreviations: n, number of samples; ND, not determined.
- a Nucleotide sequence of the ITS region was determined in the indicated reference.
- b Nucleotide sequences of the ITS and chloroplast regions were determined in the indicated reference.
Data for the phylogenetic analysis based on the SUC1 region were obtained from 11 individuals of P. hakusanensis, 1 individual of P. asiatica var. densiuscula, and 1 individual of P. asiatica var. densiuscula f. yakusimensis. The North American putative tetraploid P. rugelii Decne. was added as an outgroup. Data for the phylogenetic analysis based on ITS sequences were obtained from 25 individuals of P. hakusanensis, 48 individuals of P. asiatica var. densiuscula, and P. asiatica var. densiuscula f. yakusimensis (including 38 previously reported individuals; Ishikawa et al. 2006, Table 2), and 1 individual of P. asiatica from China (Rønsted et al. 2002). We also included P. camtschatica Link., P. major, and Plantago major var. japonica (Franch. et Sav.) Miyabe as related taxa. Phylogenetic analysis of the MIG-seq data was performed using five representative individuals of P. hakusanensis, five of P. asiatica var. densiuscula, and one of P. asiatica var. densiuscula f. yakusimensis. The selected samples spanned the geographical range of each taxon. MIG-seq is a PCR-based method that concentrates and isolates inter-simple sequence repeat regions located mainly in the nuclear genome (Suyama and Matsuki 2015; Suyama et al. 2022). The cp phylogenetic analysis included 24 individuals of P. hakusanensis from the four mountains and 37 specimens of P. asiatica var. densiuscula that included plants collected from the subalpine zone on Mt. Chokai (Table 2). We also included the cp sequences of one individual of P. asiatica from China (Rønsted et al. 2002; Iwanycki Ahlstrand et al. 2019; Kew DNA bank ID = 9585, K) and two individuals of P. camtschatica as related taxa (Iwanycki Ahlstrand et al. 2019). Although the ITS analysis included P. major, the species was excluded from the cpDNA analysis because excessive numbers of polymorphisms were found in P. major relative to P. hakusanensis (compared with the other taxa included in this analysis).
Total genomic DNA was extracted from fresh or dried leaves using a slightly modified cetyltrimethylammonium bromide method (Murray and Thompson 1980).
2.2 Chromosome Observations
We checked the chromosome numbers of P. hakusanensis to confirm previous reports (Yamazaki 1993; Ohashi 2017). Three individuals of P. hakusanensis were transplanted from Mt. Chokai to a nursery at the University of Tokyo for cytological observations. Fresh root tips were pretreated in 2 mM 8-hydroxyquinoline solution for 1 h at 20°C and then stored at 4°C for 15 h. We subsequently fixed them in Newcomer's fluid (6:3:1:1:1 isopropanol, propionic acid, petroleum ether, acetone, 1,4-dioxane). The root tips were macerated in 1 N HCl at 60°C for 10 min, then stained with 2% lacto-propionic orcein, and squashed for cytological observation.
2.3 PCR Amplification and DNA Sequencing of the SUC1 Region
Preliminary analysis indicated that the determination of the nucleotide sequence of the SUC1 region by direct sequencing of the PCR product would be difficult, presumably because of the allotetraploid origin of P. hakusanensis. Thus, we applied two methods to efficiently determine two homoeologs of SUC1 (i.e., Homoeolog L and Homoeolog I) of P. hakusanensis. First, the nucleotide sequences of two individuals collected from Mt. Hakusan were determined by the cloning and sequencing method described by Ishikawa et al. (2009). In this procedure, the PCR conditions were optimized to avoid over-amplification, which potentially produces artificial recombinants among multiple alleles (homoeologs) of the polyploid (Bradley and Hillis 1997; Judo et al. 1998; Kanagawa 2003). Second, the SUC1 sequences of nine P. hakusanensis individuals were obtained by homoeolog-specific PCR amplification and direct sequencing because the cloning and sequencing method is excessively laborious. Primers for the specific PCR were designed using polymorphic sites between the homoeologs obtained by the cloning and sequencing method. Each of the two SUC1 homoeologs (Homoeolog L and Homoeolog I) was amplified as two overlapping regions and assembled into a continuous sequence (Figure 1). For example, Homoeolog L was amplified using two primer sets: (i) SUC1-F11 (5′-ATGGGTGAATTGTCAGGAATTGAA-3′) and SUC1-hJ-R1 (5′-TCAAACAAATTCTGAAGTC-3′) and (ii) SUC1-hJ-F1 (5′-GATCCGTTCAATACTGATAGATCCA-3′) and SUC1-R4 (5′-GAGCCACCATGTCTTAG-3′). Homoeolog I was amplified using the following primer sets: (iii) SUC1-F11 and SUC1-hM-R2 (5′-CGATGTATACCTCTTCTATG-3′) and (iv) SUC1-hM-F2: CATGGTACGGACATGGAAATGG and SUC1-R4. The homoeolog-specific PCR parameters were as follows: incubation at 94°C for 1 min, 20 cycles of touchdown PCR (denaturation at 94°C for 30 s, annealing at 60°C for 30 s with a temperature reduction of 0.5°C per cycle, and extension at 72°C for 1 min), 20 cycles of non-touchdown PCR (denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min), and final extension at 72°C for 7 min. PCR was performed in a 20 μL volume using TaKaRa Ex Taq polymerase (Takara Bio Inc., Shiga, Japan). The PCR products were purified with ExoSAP-IT reagent (Thermo Fisher Scientific K. K., Tokyo, Japan) following the manufacturer's instructions. The nucleotide sequences of the PCR products were sequenced by Fasmac Co. Ltd. (Kanagawa, Japan) using the Sanger method. The GenBank accession numbers of the SUC1 alleles are listed in Table 3. All sequencing chromatograms obtained by the Sanger method have been visually checked for quality and heterozygous sites using CLC Genomics Workbench v10.0.1 software (Filgen, Nagoya, Japan).
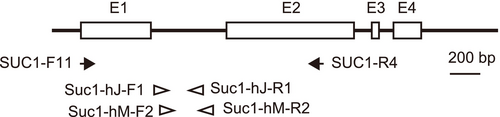
| SUC1 | Genbank accession No. (Length) | |
|---|---|---|
| Subclade I | Subclade L | |
| Mt. Asahi_h1 (1) | LC549233 (1271 bp) | LC549245 (1250 bp) |
| Mt. Asahi_h2 (1) | LC549234 (1271 bp) | LC549246 (1250 bp) |
| Mt. Asahi_h3 (1) | LC549235 (1271 bp) | LC549247 (1250 bp) |
| Mt. Chokai_h4 (2) | LC549236 (1271 bp) | LC549248 (1250 bp) |
| Mt. Chokai_h8 (4) | LC549237 (1271 bp) | LC549249 (1250 bp) |
| Mt. Chokai_h19 (4) | LC549238 (1271 bp) | LC549250 (1250 bp) |
| Mt. Chokai_h20 (4) | LC549239 (1271 bp) | LC549251 (1250 bp) |
| Mt. Gassan_ (5) | LC549240 (1271 bp) | LC549252 (1250 bp) |
| Mt. Hakusan_h24 | LC549241 (1251 bp) | ND |
| Mt. Hakusan_h25 | LC549242 (1252 bp) | LC549253 (1237 bp) |
| Mt. Hakusan_16–6 | LC549243 (1251 bp) | LC549255 (1250 bp) |
| Mt. Hakusan_5–10 | LC549244 (1251 bp) | LC549254 (1250 bp) |
- Abbreviation: ND, not determined.
2.4 PCR Amplification and DNA Sequencing of the ITS and Cp Regions
The ITS region was amplified using AB101 and AB102 primers (Douzery et al. 1999). The PCR conditions were as follows: incubation at 94°C for 1 min, 25 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 1 min, followed by final extension for 7 min. Three cp regions (trnL-trnF, ndhF-rpl32, and rpl32-trnL) were chosen based on a previous study showing that they are useful for phylogenetic studies of the subgenus Plantago (Dunbar-Co et al. 2008; Iwanycki Ahlstrand et al. 2019). The cp regions were amplified using universal primers (Taberlet et al. 1991; Shaw et al. 2007). The PCR and sequencing procedures were identical to those used for the SUC1 homoeolog-specific PCR.
2.5 Phylogenetic Analysis of the SUC1 Region
We assembled forward and reverse reads using CLC Genomics Workbench v10.0.1 software (Filgen, Nagoya, Japan). All sequence alignments were performed using MAFFT v7 software (Katoh et al. 2019) and manually edited using CLC Genomics Workbench v10.0.1 software. The phylogenetic relationships of the SUC1 region were inferred using the maximum parsimony (MP), neighbor-joining (NJ; Saitou and Nei 1987), and maximum likelihood (ML; Felsenstein 1981) procedures using PAUP* 4.0a software (Swofford 2003). In the MP analysis, parsimony informative indels were coded as binary (present or absent) characters, “gapmode” was set as missing, and all characters were weighted equally. The analysis was performed via a heuristic search using the tree bisection–reconnection branch-swapping option. One hundred rounds of random additions were performed to identify multiple islands of equally most parsimonious trees (Maddison 1991). The search setting used to find MP trees was applied to 1000 bootstrap replications (Felsenstein 1985). The distance option in our NJ analysis was set to ML; the substitution rate classes and gamma shape parameter were estimated following the PAUP* procedures manual (model correspondence = GTR + G) (Swofford and Sullivan 2009; Swofford and Bell 2017). This search setting was applied to 1000 bootstrap replications. In our ML tree search, the nucleotide evolution model was selected using PAUP* 4.0a software (model correspondence = GTR + I + G) (Swofford and Sullivan 2009), and the analysis was performed via a heuristic search using the tree bisection–reconnection branch-swapping option (addseq = random, nreps = 10). This search setting was applied to 100 bootstrap replications.
2.6 Median-Joining (MJ) Network Analysis of the ITS and Cp Regions
The MJ network was constructed using PopART v1.7 software (Bandelt et al. 1999; Leigh and Bryant 2015) to determine the relationships of ITS genotypes and cp haplotypes with ε = 0. All sites with only ambiguous base states (e.g., Y, R, and K) were excluded, and indels were coded as binary (present or absent) characters. A mononucleotide repeat region detected in rpl32-trnL (from 1844 bp to 1865 bp in a concatenated alignment) was removed before data analysis because of its significant homoplasy. There was a sequencing gap between 1947 bp and 1948 bp in the rpl32-trnL region of the concatenated alignment because sequencing of the middle part of the rpl32-trnL region was not feasible for either P. hakusanensis or P. asiatica var. densiuscula due to the poor quality of the raw data. The GenBank accession numbers of the ITS genotypes and cp haplotypes are listed in Tables 4 and 5, respectively.
| Genotype of ITS | Genbank accession No. |
|---|---|
| A | AB223151.1 |
| B | AB223153.1 |
| C | AB223154.1 |
| D | AB223156.1 |
| E | AB223160.1 |
| F | AB223162.1 |
| G | LC549256 |
| H | LC549257 |
| I | AY101861 |
| J | AY101862 |
| K | LC549258 |
| L | AJ548971.1 |
| Chloroplast haplotype (n) | Genbank accession no. | |||
|---|---|---|---|---|
| ndh-rpl32 | trnL-trnF | rpl32-trnL | rpl32-trnL | |
| 5′ | 3′ | |||
| 1 | LC549259 | LC549271 | LC549283 | LC549295 |
| 2 | LC549260 | LC549272 | LC549284 | LC549296 |
| 3 | LC549261 | LC549273 | LC549285 | LC549297 |
| 4 | LC549262 | LC549274 | LC549286 | LC549298 |
| 5 | LC549263 | LC549275 | LC549287 | LC549299 |
| 6 | LC549264 | LC549276 | LC549288 | LC549300 |
| 7 | LC549265 | LC549277 | LC549289 | LC549301 |
| 8 | LC549266 | LC549278 | LC549290 | LC549302 |
| 9 | LC549267 | LC549279 | LC549291 | LC549303 |
| 10 | LC549268 | LC549280 | LC549292 | LC549304 |
| 11 | LC549269 | LC549281 | LC549293 | LC549305 |
| 12 | LC549270 | LC549282 | LC549294 | LC549306 |
| 13 | MK487875 | MK487976 | MK487925 | |
2.7 Preparation of the MIG-seq Library, High-Throughput Sequencing, and Phylogenetic Inference
In brief, we used a two-step amplification procedure based on the protocol of Suyama and Matsuki (2015), but we changed the annealing temperature from 48°C to 38°C in the first PCR of this protocol. The amplicons were purified and sequenced on an Illumina MiSeq sequencer (Illumina, San Diego, CA, USA). Primer regions, anchors, and low-quality reads were removed using the FASTX Toolkit package (http://hannonlab.cshl.edu/fastx_toolkit/). To remove reads derived from extremely short library entries, we searched for the sequences of the primer-targeted regions within the sequences of reads 1 and 2, and reads containing the searched sequences were removed using TagDust software (Lassmann et al. 2009).
To obtain genotypes at SNPs, we used the Universal Network Enabled Analysis Kit (UNEAK) pipeline (Lu et al. 2013) to assemble 80-bp clean reads. UNEAK is a non-reference, network-based pipeline that has been successfully used for genotype calling in polyploid species (e.g., Clark et al. 2015; Li et al. 2014). A default setting was used in the read assembly by UNEAK. SNPs were exported in HapMap format and then filtered using TASSEL 5.0 software (Bradbury et al. 2007) with the following parameters: siteMinCount 10, MinAlleleFreq 0.05, and maxHetero 0.5. The loci genotyped in all samples were retained in the final dataset.
We used RAxML v.8.2.10 software (Stamatakis 2014) to infer a ML phylogenetic tree. In this analysis, we specified the GTRGAMMA model as the nucleotide evolution model and performed 1000 bootstrap iterations to assess the node support values. The GenBank accession numbers of the MIG-seq raw data are listed in Table 6.
| BioProject | BioProject submission | DRA submission | BioSample | BioSample submission | Sample name | Experiment | Experiment alias | Experiment title | Library source | Library strategy | Library layout | Run | Run alias | Run title | Run files |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224523 | SSUB014994 | Plantago hakusanensis_NI-1_Mt.Gassan | DRX217532 | pancheri54-0022_Experiment_0001 | Illumina MiSeq sequencing of SAMD00224523 | GENOMIC | AMPLICON | SINGLE | DRR227268 | pancheri54-0022_Run_0001 | Illumina MiSeq sequencing of SAMD00224523 | NI-1.fastq.gz |
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224524 | SSUB014994 | Plantago hakusanensis_NI-2_Mt.Hakusan_h24 | DRX217533 | Pancheri54-0022_Experiment_0002 | Illumina MiSeq sequencing of SAMD00224524 | GENOMIC | AMPLICON | SINGLE | DRR227269 | Pancheri54-0022_Run_0002 | Illumina MiSeq sequencing of SAMD00224524 | NI-2.fastq.gz |
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224525 | SSUB014994 | Plantago hakusanensis_NI-3_Mt.Hakusan_h25 | DRX217534 | Pancheri54-0022_Experiment_0003 | Illumina MiSeq sequencing of SAMD00224525 | GENOMIC | AMPLICON | SINGLE | DRR227270 | Pancheri54-0022_Run_0003 | Illumina MiSeq sequencing of SAMD00224525 | NI-3.fastq.gz |
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224526 | SSUB014994 | Plantago hakusanensis_NI-4_Mt.Chokai_h4 | DRX217535 | Pancheri54-0022_Experiment_0004 | Illumina MiSeq sequencing of SAMD00224526 | GENOMIC | AMPLICON | SINGLE | DRR227271 | Pancheri54-0022_Run_0004 | Illumina MiSeq sequencing of SAMD00224526 | NI-4.fastq.gz |
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224527 | SSUB014994 | Plantago hakusanensis_NI-5_Mt.Chokai_h10 | DRX217536 | Pancheri54-0022_Experiment_0005 | Illumina MiSeq sequencing of SAMD00224527 | GENOMIC | AMPLICON | SINGLE | DRR227272 | Pancheri54-0022_Run_0005 | Illumina MiSeq sequencing of SAMD00224527 | NI-5.fastq.gz |
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224528 | SSUB014994 | Plantago asiatica var. densiuscula_NI-6_Mt.Chokai_29 | DRX217537 | Pancheri54-0022_Experiment_0006 | Illumina MiSeq sequencing of SAMD00224528 | GENOMIC | AMPLICON | SINGLE | DRR227273 | Pancheri54-0022_Run_0006 | Illumina MiSeq sequencing of SAMD00224528 | NI-6.fastq.gz |
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224529 | SSUB014994 | Plantago asiatica var. densiuscula_NI-7_Nara-1 | DRX217538 | Pancheri54-0022_Experiment_0007 | Illumina MiSeq sequencing of SAMD00224529 | GENOMIC | AMPLICON | SINGLE | DRR227274 | Pancheri54-0022_Run_0007 | Illumina MiSeq sequencing of SAMD00224529 | NI-7.fastq.gz |
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224530 | SSUB014994 | Plantago asiatica var. densiuscula_NI-8_Miyajima-1 | DRX217539 | Pancheri54-0022_Experiment_0008 | Illumina MiSeq sequencing of SAMD00224530 | GENOMIC | AMPLICON | SINGLE | DRR227275 | Pancheri54-0022_Run_0008 | Illumina MiSeq sequencing of SAMD00224530 | NI-8.fastq.gz |
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224531 | SSUB014994 | Plantago asiatica f. yakusimensis_NI-9_Mt.Miyanoura-1 | DRX217540 | Pancheri54-0022_Experiment_0009 | Illumina MiSeq sequencing of SAMD00224531 | GENOMIC | AMPLICON | SINGLE | DRR227276 | Pancheri54-0022_Run_0009 | Illumina MiSeq sequencing of SAMD00224531 | NI-9.fastq.gz |
| PRJDB9828 | PSUB008116 | DRA010171 | SAMD00224532 | SSUB014994 | Plantago asiatica var. densiuscula_NI-10_Nara-2 | DRX217541 | Pancheri54-0022_Experiment_0010 | Illumina MiSeq sequencing of SAMD00224532 | GENOMIC | AMPLICON | SINGLE | DRR227277 | Pancheri54-0022_Run_0010 | Illumina MiSeq sequencing of SAMD00224532 | NI-10.fastq.gz |
3 Results
3.1 Chromosome Numbers
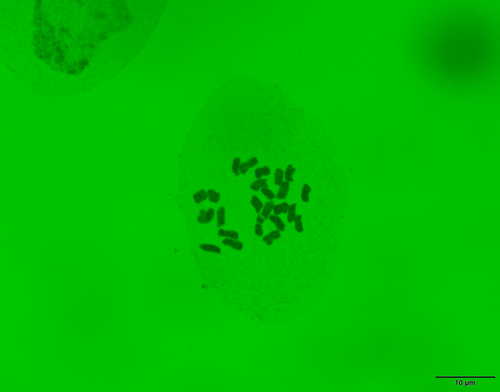
We counted chromosome numbers of 2n = 24 in three individuals of P. hakusanensis collected on Mt. Chokai (Figure 2). This number was reported previously by Yamazaki (1993) and Ohashi (2017), but we were unable to locate original data from either study showing the collection site or photographs of chromosomes. We confirmed that the chromosome number of P. hakusanensis was 2n = 24, and that the species was a tetraploid with a basic chromosome number x = 6 (Rahn 1996).
3.2 Phylogenetic Analysis Based on SUC1 Showed That P. hakusanensis is a Close Relative of P. asiatica var. densiuscula Within Sect. Plantago
The PCR-amplified SUC1 region of P. hakusanensis was ca. 1.25 kb long. Two distinct SUC1 alleles were obtained from each of the two individuals from Mt. Hakusan. After removing the entire intron 1 from the ambiguous alignment, the aligned matrix of all 63 unique alleles from P. hakusanensis and 24 taxa representing all five sections of subgenus Plantago (2 diploids and 17 polyploids, including P. tenuiflora as an outgroup) was 835 bp long, with 180 variable and 111 parsimony-informative sites. In the MP analysis, we obtained 59 most parsimonious trees with 311 steps. The overall consistency index (Kluge and Farris 1969) was 0.723, the overall retention index (Farris 1989) was 0.855, and the overall rescaled index (Farris 1989) was 0.618. The topologies produced from the MP, NJ, and ML procedures were mostly similar. The MP tree is shown in Figure 3. The NJ and ML trees are shown in Figures S1 and S2, respectively.
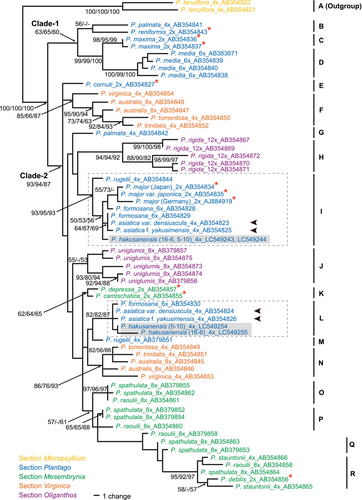
We compared our results with those of Ishikawa et al. (2009) and found that the addition of three P. hakusanensis alleles and a change in outgroup identity had little effect on the major topologies of the trees. Thus, we present below a brief summary of our results with a particular focus on P. hakusanensis. The alleles of the ingroups from four sections of the subg. Plantago fell into two sister clades: clade 1 (MP/NJ/ML support values = 63/65/60) and clade 2 (support values = 93/94/87). Three subclades (B–D) were recognized in clade 1. Three alleles (E, G, and M) and 11 subclades (F, H–L, and N–R) were found in clade 2 (Figure 3). Two alleles obtained from a P. hakusanensis individual were represented in the two distantly separated subclades I (support values = 93/95/93) and L (support values = 82/82/87) (Figure 3). The alleles found in the distinct subclades were considered homoeologs originating from allotetraploidization that occurred through interspecific hybridization between two diploids followed by full duplication of the hybrid genome (Ramsey and Schemske 1998; Glover et al. 2016). A set of two homoeologs in subclades I and L was also found in P. asiatica var. densiuscula and P. asiatica var. densiuscula f. yakusimensis; hence, P. hakusanensis is an allotetraploid originating from ancestral lineages shared with P. asiatica var. densiuscula and P. asiatica var. densiuscula f. yakusimensis.
3.3 Monophyly of P. hakusanensis Inferred by Analyses Based on SUC1, the ITS Region, and MIG-seq
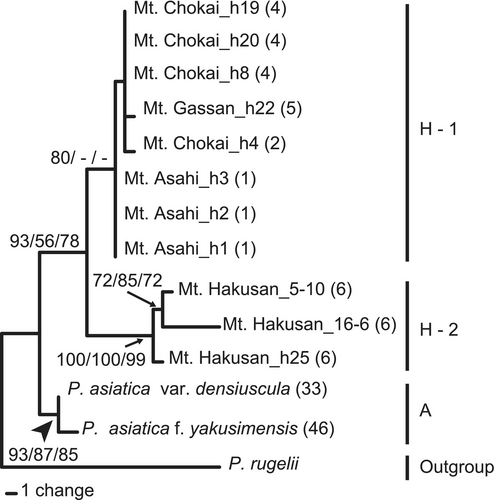
Two homoeologs of SUC1 were successfully amplified by homoeolog-specific PCR amplification in all 10 individuals of P. hakusanensis that we investigated. The homoeologs from 1250 to 1271 bp in length were successfully sequenced with one exception: one homoeolog in subclade L in an individual collected from Mt. Hakusan could not be sequenced, likely due to the presence of indels between two biparentally inherited alleles. The topologies of the trees inferred from each homoeolog of the I and L lineages were consistent (data not shown). We therefore concatenated the two homoeologs in the phylogenetic analysis. The aligned matrix of all 14 sequences comprised 2589 characters with 40 variable and 20 parsimony-informative sites. In our MP analysis, we obtained nine most parsimonious trees after 63 steps. The overall consistency index was 0.952, the overall retention index was 0.935, and the overall rescaled index was 0.89. The topologies produced by MP, NJ, and ML were nearly identical, although there were a few slight differences in resolution at the branch tips. Thus, we present only the MP tree in Figure 4. P. hakusanensis formed a monophyletic clade with high to moderate bootstrap support values (MP/NJ/BI support values = 93/56/78). This clade was sister to a clade comprising P. asiatica var. densiuscula and P. asiatica f. yakusimensis (clade A). The P. hakusanensis clade was divided into two sister clades: subclade H-1 (Mt. Chokai, Mt. Gassan, and Mt. Asahi) and subclade H-2 (Mt. Hakusan).
Among 78 individuals examined, substitutions and indels in the ITS regions were recognized at 44 and 4 sites in 742 characters, respectively. There were 25 parsimony-informative sites. The variable positions indicated a total of 12 genotypes (Table 7). The MJ network showed that those genotypes were divided into three taxon-specific groups (I, II, and P. camtschatica); the remaining members of the group belonged to P. asiatica from China, P. major, and P. major var. japonica (Figure 5 and Table 2). Group I contained six genotypes of P. asiatica var. densiuscula, and group II contained two genotypes of P. hakusanensis; these two groups were distinctly separated by six mutation steps. Within the MJ network, group I had a star-like configuration in which one major genotype was surrounded by several low-frequency genotypes distinguished by one or two mutation steps.
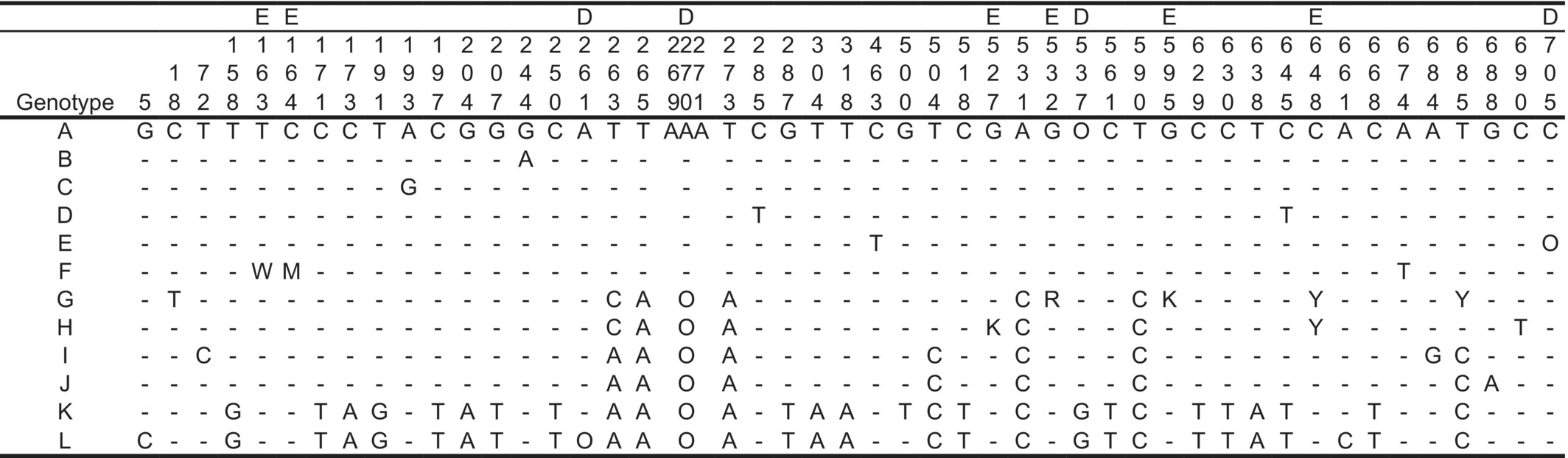
- Note: Sequences are numbered from the 5′ end to the 3′ end. —: Same as genotype A, O: absence, D: site with indel, E: polymorphic site consisted of only an ambiguous base (e.g., W and M), and the site was excluded from the analysis.
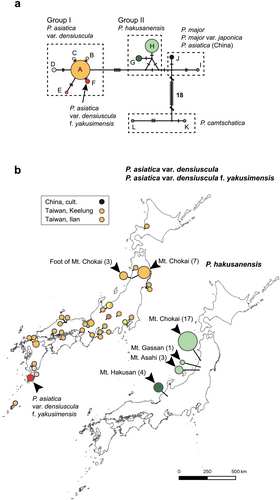
Since the distributions of both taxa overlap along a mountain trail on Mt. Chokai at elevations between 1320 and 1540 m, we hypothesized that recent hybridization between P. hakusanensis and P. asiatica var. densiuscula might be detectable in ITS sequences due to the biparental inheritance of nuclear alleles. However, sequencing chromatograms indicated no sign of hybridization in the 17 individuals of P. hakusanensis and 7 individuals of P. asiatica var. densiuscula sampled from Mt. Chokai.
MIG-seq analysis obtained an average of 2,561,487 reads from 10 individuals. The genotype matrix consisted of 420 SNPs, and the genotyping rate at these markers was 1.0. Phylogenetic analyses revealed the monophyly of P. hakusanensis (Figure 6, bootstrap value = 100). The P. hakusanensis clade was divided into two sister clades (H1 and H2, bootstrap value = 100). Clade H1 comprised individuals from Mt. Chokai and Mt. Gassan. Clade H2 comprised individuals from Mt. Hakusan, similar to the SUC1 tree (Figures 4 and 6). Branch lengths from the tips to the nodes at the last common ancestor for each taxon were shorter in P. asiatica var. densiuscula, even though it has a wide distribution across the Japanese Archipelago.
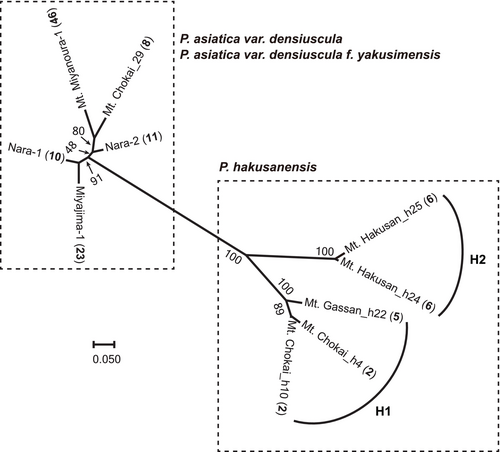
3.4 Cp Haplotypes Shared Among P. hakusanensis, P. asiatica var. densiuscula, and P. asiatica From China
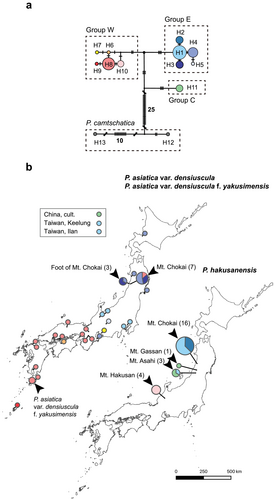
Among 64 individuals examined, substitutions and indels were recognized at 46 and 14 sites in 2175 characters, respectively. There were 36 parsimony-informative sites. The variable positions indicated a total of 13 haplotypes (Table 8). Eleven haplotypes were taxon-specific, but two haplotypes were shared by different taxa. The first shared haplotype, H1, was shared by P. hakusanensis and P. asiatica var. densiuscula. The second haplotype, H11, was shared by P. hakusanensis and P. asiatica in China (Table 2). MJ network analysis showed that these haplotypes were divided into four groups (E, W, C, and P. camtschatica, Figure 7a). P. asiatica var. densiuscula contained groups E (haplotypes H1, H3–H5) and W (haplotypes H6–H9). The distributions of the two groups in P. asiatica var. densiuscula did not overlap, except in the case of one individual on Mt. Chokai (Figure 7). Individuals with group E haplotypes were found in Taiwan and in the eastern part of the Japanese Archipelago. Group W was distributed mainly within the western part of the Japanese Archipelago (Table 2). Haplotypes of P. hakusanensis consisted of groups E (H1 and H2), W (H10), and C (H11). Haplotypes H2 and H10 were specific to P. hakusanensis, although they were separated from haplotypes H1 and H8 of P. asiatica var. densiuscula, respectively, by only one substitution (H1 was shared by both P. hakusanensis and P. asiatica var. densiuscula, as indicated above).
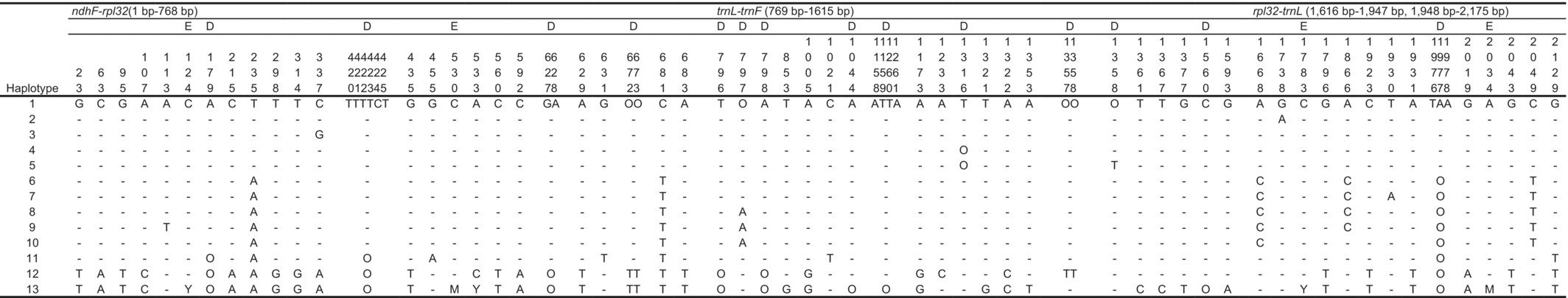
- Note: Sequences are numbered from the 5′ end to the 3′ end. —: Same as haplotype 1, O: absence, D: site with indel, E: polymorphic site consisted of only an ambiguous base (e.g., Y and M), and the site was excluded from the analysis.
4 Discussion
4.1 P. hakusanensis is an Allotetraploid Related to P. asiatica var. densiuscula
Our phylogenetic analyses based on the nuclear-encoded single-copy SUC1 region showed that P. hakusanensis and P. asiatica var. densiuscula (and P. asiatica var. densiuscula f. yakusimensis) are both allotetraploids with two distinctly related homoeologs. Each homoeolog was found to belong to the same subclade with high support values (subclade I and L in Figure 3). Thus, P. hakusanensis and P. asiatica var. densiuscula are closely related to one another, and P. hakusanensis is presumed to have originated either from the same ancestral allotetraploid as P. asiatica var. densiuscula, with subsequent differentiation into distinct taxa in different biomes, or from independent allopolyploidization via hybridization between the same or closely related parental species of P. asiatica var. densiuscula.
A diploid P. major (or P. major var. japonica) in subclade I and a diploid species related to P. depressa and P. camtschatica in sect. Mesembrynia (subclade K, Figure 3) have been proposed to be the parental species in the allopolyploidization of P. asiatica var. densiuscula, based on their phylogenetic positions and current distributions (Ishikawa et al. 2009). The assumptions made for P. asiatica var. densiuscula would also be applicable to the parental species of P. hakusanensis. In contrast to our SUC1 analysis results, only one lineage of ITS sequences was obtained from both P. asiatica var. densiuscula and P. hakusanensis; these sequences were closely related to P. major and P. major var. japonica (Figure 5). This result may be explained by the failure of PCR to amplify ITS sequences related to Plantago camtschatica, or biparentally inherited homoeologous ITS regions might have been homogenized by both inter-locus and intra-locus concerted evolution biased toward one of the two parental lineages (P. major or P. major var. japonica), as reported in other allopolyploids (Wendel et al. 1995; Kovarik et al. 2005). The former possibility may be less likely because the ITS region of P. camtschatica was successfully amplified in the present study, and the primer set used in this analysis has been shown to be appropriate for a wide taxonomic range (Douzery et al. 1999; Sonboli et al. 2011; Kokubugata et al. 2011; Koecke et al. 2013).
4.2 Disparities Between Phylogenies Based on Nuclear- and Cp-Encoded Genes in P. hakusanensis and P. asiatica var. densiuscula
Phylogenetic analyses of the two nuclear coding regions (SUC1 and ITS) and MIG-seq data revealed the monophyly of P. hakusanensis, at least in the samples examined in our study (Figures 3-6). In contrast, cp haplotypes of P. hakusanensis (haplotypes H1, H2, H10, and H11) were shared or phylogenetically close to P. asiatica var. densiuscula and P. asiatica in China. In general, shared genetic diversity between closely related taxa is explained by incomplete lineage sorting of ancestral polymorphisms and/or introgression (Rieseberg and Soltis 1991; Comes and Abbott 2001; Dixon et al. 2007). Although it is difficult to rule out the possibility of incomplete lineage sorting completely, the shared and related haplotypes in P. hakusanensis and P. asiatica var. densiuscula and P. asiatica may be best explained by introgressions of the cp genome (cp capture) for the three reasons outlined below. First, incomplete lineage sorting tends to occur shortly after speciation, but we consider the two taxa to be distinctly differentiated from one another based on the results of nuclear marker analyses (Figures 4-6). Second, major haplotype groups (W, E, and C; Figure 5) were also distinctly differentiated from one another (W, E, and C; Figure 5). Third, ancestral polymorphisms in incomplete lineage sorting are more readily fixed in the cp genome, which has a smaller effective population size than that of the nuclear genome (Schaal et al. 1998), although we detected shared genetic diversity between taxa in the cp markers examined in the present study.
We found the cp haplotypes of P. asiatica var. densiuscula groups W and E in the western and eastern parts of the Japanese Archipelago, respectively (Figure 7). A similar geographic structure also occurred in P. hakusanensis. Although the direction of cp genome introgression between P. asiatica var. densiuscula and P. hakusanensis is unclear, we consider two possible hypotheses below. First, the geographic structure was originally established in P. asiatica var. densiuscula, and two independent cp genome introgressions from P. asiatica var. densiuscula to P. hakusanensis occurred on Mt. Hakusan (group W) and in the Tohoku area (group E). Similar geographic distribution patterns of nuclear genotypes and/or cp haplotypes have been reported to be common in many other plants in temperate forests (Iwasaki et al. 2012), and the geographic patterns are assumed to have been formed by the isolation of ancestral populations into different refugia during the Quaternary glacial climate and postglacial expansions (Aoki et al. 2019; Sakaguchi et al. 2021). Alternatively, the second hypothesis proposes that haplotypes in groups W and E originated from P. asiatica var. densiuscula and P. hakusanensis, respectively. Both empirical and simulation studies have reported that neutral gene introgression tends to occur from locally established species to invading taxa (Currat et al. 2008; Excoffier et al. 2009). Studies of the genera Veratrum and Cercidiphyllum also hypothesized ancient introgression of the cp genome from local species in subalpine cool-temperate forest habitats to invading species in low- to mid-elevation warm-temperate locations (Kikuchi et al. 2010; Qi et al. 2012). Under this hypothesis, P. hakusanensis may have differentiated in central Honshu, with cp genome (group E) introgression subsequently occurring from local P. hakusanensis to invading P. asiatica var. densiuscula. P. asiatica var. densiuscula would have rapidly expanded its distribution range from the southwestern Japanese Archipelago toward the northeast because, within the median-joining network (Figure 5), ITS group I had a star-like configuration, in which one major genotype was surrounded by several low-frequency genotypes distinguished by one or two mutation steps, which is indicative of rapid population expansion. Moreover, the MIG-seq maximum parsimony tree shows shorter branch lengths from the tips to the last common ancestor for P. asiatica var. densiuscula (Figure 6), despite its broad distribution. This result also implies that P. asiatica var. densiuscula expanded more rapidly than P. hakusanensis. These findings support the second hypothesis; however, under this scenario, an additional cp genome introgression (group W) from P. asiatica var. densiuscula to P. hakusanensis on Mt. Hakusan would be required to account for the current haplotype distribution at least (Figure 7).
Author Contributions
Naoko Ishikawa: conceptualization (equal), formal analysis (lead), funding acquisition (lead), writing – original draft (lead). Shota Sakaguchi: formal analysis (supporting), writing – original draft (supporting), writing – review and editing (equal). Chikako Hasekura: investigation (equal). Alexey Shipunov: writing – review and editing (equal). Ayumi Matsuo: investigation (equal). Yoshihisa Suyama: writing – review and editing (equal). Hirokazu Tsukaya: writing – review and editing (equal). Hiroshi Ikeda: conceptualization (equal), writing – review and editing (equal). Motomi Ito: writing – review and editing (equal).
Acknowledgments
The authors are grateful to Mr. K. Sawa and Dr. Y. Nakayama for providing the plant materials. This research was approved by the Ministry of Environment, Japan (for investigations in Ishikawa, Yamagata, Toyama, and Kagoshima Prefectures). Funding was provided by the Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) (Nos. 17K07527, 19K06805 and 24K09566) and the Environmental Restoration and Conservation Agency of Japan for the Environmental Research and Technology Development Fund (No. 4-2001). Computation was performed with the assistance of the Research Center for Computational Science, Okazaki, Japan (National Institute for Basic Biology project no.: 24-IMS-C357).
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The DNA sequences generated in this study have been deposited in the National Center for Biotechnology Information, and the accession numbers are listed in Tables 3–6.




