Transcriptomic changes during caste development through social interactions in the termite Zootermopsis nevadensis
Abstract
One of the most striking examples of phenotypic plasticity is the different phenotypes (i.e., castes) within a same nest of social insects. Castes usually derive from a single genotype initially by receiving social cues among individuals during development. Specific gene expression changes may be involved in caste differentiation, and thus, the regulatory mechanism of these changes should be clarified in order to understand social maintenance and evolution. The damp-wood termite Zootermopsis nevadensis is one of the most important model termite species, due to not only the availability of genomic, transcriptomic, and epigenomic information but also evidence that soldier- and worker-destined individuals can be identified in natural conditions. Given that the nutritional intakes via social interactions are crucial for caste differentiation in this species, there is a possibility that transcriptomic changes are influenced by the nutritional difference among these individuals. Here, whole body RNA-seq analysis of 3rd-instar larvae with biological replications and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were conducted. We found the drastic expression differences during caste developments between soldier- and worker-destined individuals. The results indicated that there are several key signaling pathways responsible for caste formations, which are involved in developments and social interactions. Particularly, the nutritional sensitive signaling was upregulated in soldier-destined individuals, while some metabolic pathways were identified in worker-destined individuals. These bioinformatic data obtained should be utilized to examine the molecular mechanisms of caste determination in social insects.
1 INTRODUCTION
One of the major goals in ecological developmental biology is to reveal the molecular mechanisms underlying phenotypic plasticity, which allows organisms to produce different phenotypes from the same genomic background depending on environmental cues (Gilbert & Epel, 2009). Phenotypic plasticity is particularly widespread in insects (Simpson, Sword, & Lo, 2011), and for social insects (e.g., bees, wasps, ants, and termites), it is a crucial for their social organization and evolutionary success (Wilson, 1971). In a colony of social insects, there are multiple phenotypes (i.e., castes) sharing the same genomic background, and they have specialized morphologies and behaviors to achieve their respective roles. Such species are an excellent resource to study the molecular mechanisms of phenotypic plasticity, because the complex social signals as well as environmental cues usually impact on caste differentiation during postembryonic development (Linksvayer, Fewell, Gadau, & Laubichler, 2012).
Termite is one of the most successful social insects, and their social organization was acquired independently from hymenopteran insects. Termite castes are normally composed of reproductives, workers, and soldiers, and unlike the social Hymenoptera, the soldier caste was first acquired as a permanently sterile caste during social evolution (Nalepa, 2011; Tian & Zhou, 2014). Soldiers are differentiated from workers via an intermediate presoldier stage. The new production of soldiers is promoted by the presence of reproductives (Bordereau & Han, 1986; Maekawa, Nakamura, & Watanabe, 2012) and suppressed by existing soldiers (Mitaka, Mori, & Matsuura, 2017; Watanabe, Gotoh, Miura, & Maekawa, 2011). Caste differentiation is generally determined in an environmentally sensitive period during postembryonic development, probably by pheromonal substrates transmitted via inter-individual interactions (Noirot, 1991; Watanabe, Gotoh, Miura, & Maekawa, 2014). Consequently, changes in gene expression caused by the social interactions should be clarified to understand the molecular mechanisms underlying caste differentiation.
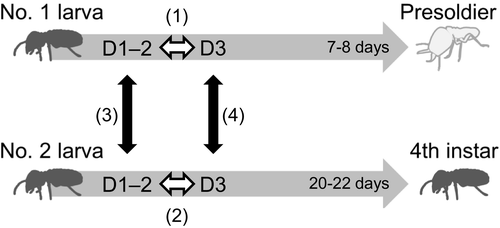
The damp-wood termite Zootermopsis nevadensis is useful taxon for sociogenomic studies, because genomic, transcriptomic, and epigenomic information is available (Glastad, Gokhale, Liebig, & Goodisman, 2016; Terrapon et al., 2014). Importantly, in this species soldier-destined individuals can be identified in natural conditions (Maekawa et al., 2012; Masuoka, Yaguchi, Suzuki, & Maekawa, 2015; Yaguchi, Inoue, Sasaki, & Maekawa, 2016). In an incipient colony, the first soldier was differentiated from the first-molted 3rd-instar larva (hereafter No. 1 larva). On the other hand, workers were molted from the 2nd (No. 2) or subsequent molted 3rd-instar larva. The No. 1 larva always differentiated into a presoldier after 7–8 days, whereas the No. 2 larva normally molted into a 4th instar after approximately 20 days and functioned as a worker. The No. 2 larvae could differentiate into a soldier, when the No. 1 larva was artificially removed from the incipient colony, suggesting that soldier differentiation was determined by environmental conditions (Maekawa et al., 2012). Importantly, proctodeal trophallaxis (i.e., anal feeding) from the reproductives to the No. 1 larvae was more frequently observed than to the No. 2 larvae, but allogrooming of the No. 2 larvae was more frequently observed than that of the No. 1 larvae. These behavioral differences initially occurred at Day 1 or 2 after their appearance (Maekawa et al., 2012). These observations suggested that the nutritional conditions were apparently different between these larvae. Developments of the No. 1 larvae typically depended on the unique food intakes from reproductives, but those of the No. 2 larvae were required mainly for self-feeding of woods. Therefore, there is a possibility that the caste formations are influenced by the intrinsic factors in response to the nutritional differences between these larvae. There are, however, no molecular evidences to support these developmental changes.
Generally, soldier differentiation in termites requires an increase in juvenile hormone (JH) titer in workers (Miura & Scharf, 2011). In Z. nevadensis, JH biosynthetic genes in the No. 1 larvae were highly expressed from the period beyond 3 days after their appearance (Yaguchi, Masuoka, Inoue, & Maekawa, 2015). Moreover, a specific expressed lipocalin gene (Neural Lazarillo; NLaz) was identified in the No. 1 larva at day 3 after the appearance (Yaguchi et al., 2018). A lipocalin gene ZnNLaz1 was a crucial regulator for solder differentiation through the regulation of trophallactic interactions with a queen. These results suggested that there was a cross talk between lipocalin and the intrinsic factors to integrate the social signals including unique nutrient intakes from a queen. Thus, soldier- and worker-destined larvae, that is, the No. 1 and No. 2 larvae, especially within 3 days after their appearance, are important materials to examine the gene expression patterns for the behavioral and physiological changes involved in caste differentiation.
Here, we revealed the transcriptomic changes in response to the nutritional differences underlying caste differentiation in Z. nevadensis. By performing RNA-seq analysis, large numbers of differentially expressed genes (DEGs) were identified in both the No. 1 and No. 2 larvae. Functional significance of DEGs was inferred using Gene Ontology (GO) database, and Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Based on the results obtained using these bioinformatic analyses, we discuss the molecular basis underlying caste differentiation in termites.
2 MATERIALS AND METHODS
2.1 Incipient colony foundation of Zootermopsis nevadensis
Mature colonies (total 11) of the damp-wood termite Z. nevadensis were collected from Kawanishi-shi, Hyogo Prefecture, Japan, in April and June 2015. The collected colonies were placed in plastic containers and maintained in consistent darkness at room temperature in the laboratory until the emergence of alates (winged adults). Alates were collected from these colonies, and the sexes of individuals were confirmed using the morphology of abdominal sternites (Weesner, 1969). The incipient colonies were established with unrelated alates from 11 different mature colonies in accordance with previous reports (Maekawa et al., 2012; Yaguchi et al., 2016, 2015). The constructed 48 incipient colonies (Supporting Information Table S1) were kept in consistent darkness at 25°C in an incubator. After the appearance of the 2nd-larval instar, the colonies were checked carefully every 24 hr to confirm whether a 3rd-instar larva had appeared. The oldest 3rd-instar larva (No. 1 larva = soldier-destined larva) and the second 3rd-instar larva (No. 2 larva = worker-destined larva) at Day 0 after their appearance were marked with different colored ink spots to discriminate each larva. The two respective larvae were collected at Day 1 (n = 6), 2 (n = 6), and 3 (n = 12), all of which were individually sampled from the different colonies (Supporting Information Table S1). The No. 2 larvae were collected at the periods when the presoldier exists in the colony, and thus, they were definitely worker-destined larvae. The collected larvae were immersed immediately in liquid nitrogen and stored in −80°C for the following experiments.
2.2 Total RNA extraction, RNA-seq library preparation, and sequencing by HiSeq1500
Total RNA was extracted from whole body (4 individuals were used for each library) of the No. 1 and No. 2 larvae using a SV Total RNA extraction kit (Promega, Madison, WI, USA). Extracted total RNA from larvae at Day 1–2 was mixed equally (2 individuals from each day; Supporting Information Table S1). The amounts of RNA and DNA in each sample were quantified using a Qubit fluorometer (Life Technology, Eugene, OR, USA), and the qualities of those were confirmed using an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Total RNA (500 ng) was used for cDNA synthesis and purification based on a low-throughput protocol with a TruSeq Stranded RNA LT kit (Illumina, San Diego, CA, USA). A half-scale reaction of the standard protocol was applied for library preparation. The quality and quantity of cDNA were validated using an Agilent 2100 bioanalyzer and a KAPA qPCR SYBR green PCR kit (GeneWorks, Thebarton, Australia). RNA-seq analysis was performed by single-end sequencing (66 bp) using Hiseq1500 (Illumina, San Diego, CA, USA). Three replicates (i.e., biological triplicates) were prepared for four different developmental stages (No. 1 larvae at Day 1–2, No. 2 larvae at Day 1–2, No. 1 larvae at Day 3, and No. 2 larvae at Day 3), and a total of 12 libraries were sequenced (Supporting Information Table S1). All reads have been deposited in the DDBJ Sequence Read Archive (DRA) database under accession numbers DRA007363.
2.3 Identification of differentially expressed genes between soldier- and worker-destined larvae
Prior to mapping of the transcriptomic data, all libraries were processed in order to perform expression analyses as follows. Firstly, the read quality of the obtained sequence reads was checked using FastQC (Andrews, 2010), and the adaptor sequences were removed from all libraries using cutadapt 1.4.2 (Martin, 2011) with default parameters. The trimming of low-quality reads was performed using SolexaQA v2.5 (Cox, Peterson, & Biggs, 2010) with a Phred score cutoff of 28 (-h 28) in DynamicTrim.pl and a minimum trimmed read length of 23 (-l 23) in LengthSort.pl. These reads were mapped to a Z. nevadensis reference genome (gene model OGSv2.2; Terrapon et al., 2014) using TopHat v2.0.9 (Kim et al., 2013) with default parameters. Counting of reads was performed using featureCounts v1.5.2 (Liao, Smyth, & Shi, 2014). Differential gene expression levels were compared using a generalized linear model (GLM) approach implemented using the edgeR 3.18.1 Bioconductor package (Robinson, McCarthy, & Smyth, 2010). Normalization factors for each library were calculated using the trimmed mean of M-values (TMM) method (Robinson & Oshlack, 2010). A multidimensional scaling (MDS) plot was described using the normalized count data of all libraries in R v.3.4.0 (R Core Team, 2017). Four pairwise comparisons were performed: (a) Day 1–2 × Day 3 in the No. 1 larva, (b) Day 1–2 × Day 3 in the No. 2 larva, (c) the No. 1 larva × No. 2 larva at Day 1–2, and (d) the No. 1 larva × No. 2 larva at Day 3 (Figure 1). MA plots were used to represent the difference in counts in each developmental stage by edgeR. A false discovery rate (FDR) <0.05 was used to the cutoff of differentially expression. Furthermore, ANOVA-like comparisons were performed to know the caste-biased genes with age effects using edgeR with GLM analysis (McCarthy, Chen, & Smyth, 2012).
2.4 Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis
To understand the different functional profiles between the No. 1 and No. 2 larvae, GO and KEGG enrichment analyses were performed. For those analyses, we assigned an ortholog of Drosophila melanogaster for each Z. nevadensis gene, based on reciprocal best hits between Z. nevadensis genes (gene model OGSv2.2; Terrapon et al., 2014) and D. melanogaster genes (available from FlyBase, version: FB2018 r6.23). BlastP searches were carried out for 15,876 Z. nevadensis genes and 30,506 D. melanogaster protein sequences (version: FB2018 r6.23) with default parameters. The reciprocal best hit genes showing less than e-value 1.0e−50 were assigned as the orthologs. Of the assigned 5,424 genes, DEGs with more than twofold changes (total 800 genes) were used for GO and KEGG enrichment analyses performed using an R package clusterProfiler (Yu, Wang, Han, & He, 2012) with fly annotations (Carlson, Pagès, Arora, Obenchain, & Morgan, 2016). The GO enrichment analysis was performed focused on biological process. p-Values were adjusted using the multiple test correction method of Benjamini and Hochberg (Benjamini & Hochberg, 1995).
3 RESULTS
3.1 Overall transcriptomic profiles
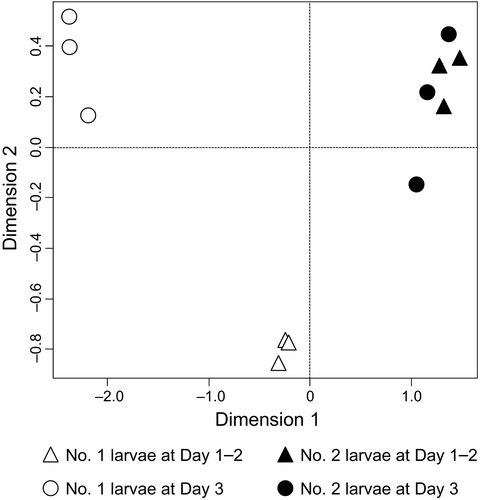
We investigated overall gene expression profiles from whole bodies of soldier-destined (No. 1) and worker-destined (No. 2) larvae at Day 1–2 and Day 3. Sequencing using Illumina HiSeq1500 platform yielded 138.9 million 66-bp single-end sequence reads (Supporting Information Table S2). To evaluate the transcriptomic differences among the No. 1 and No. 2 larvae at both time points after their appearance, the MDS plot was described using normalized count data calculated for each library. Each replication was clustered and similar to each other (Figure 2). The MDS plot showed that the overall transcriptome was differentially plotted between Day 1–2 and Day 3 in the No. 1 larvae. However, no obvious differences were observed between Day 1–2 and Day 3 in the No. 2 larvae relative to those of the No. 1 larvae.
3.2 Differentially expressed genes between each developmental stage
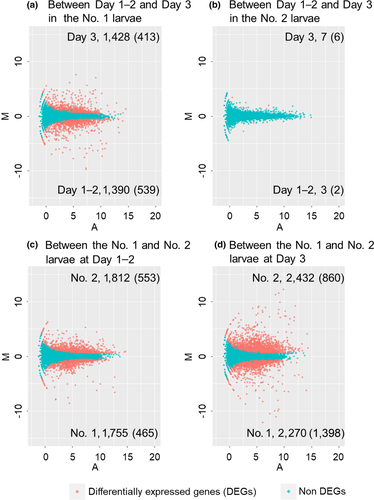
We obtained different numbers of DEGs for the four pairwise comparisons (Figure 3). The number of DEGs between Day 1–2 and Day 3 in soldier-destined (No. 1) larvae was 2,818. Of these genes, 1,390 and 1,428 genes were upregulated in Day 1–2 and Day 3, respectively (Figure 3a and Supporting Information Tables S3 and S4). By contrast, there were only 10 DEGs between Day 1–2 (three genes were upregulated) and Day 3 (seven genes were upregulated) in worker-destined (No. 2) larvae (Figure 3b and Supporting Information Tables S5 and S6). The expression levels of 3,567 genes were significantly different between the No. 1 and No. 2 larvae at Day 1–2. Of these 3,567 DEGs, 1,755 and 1,812 genes were upregulated in the No. 1 and No. 2 larvae, respectively (Figure 3c and Supporting Information Tables S7 and S8). The expression levels of 4,702 genes were significantly different between the No. 1 and No. 2 larvae at Day 3. Of these 4,702 DEGs, 2,270 and 2,432 genes were upregulated in the No. 1 and No. 2 larvae, respectively (Figure 3d and Supporting Information Tables S9 and S10). The ZnNLaz1 (gene ID: Znev_05665) was highly expressed in the No. 1 larvae at Day 3 relative to the No. 2 larvae at Day 3 (Supporting Information Table S9). In the ANOVA-like comparisons, caste-biased genes with age effects were detected between the No. 1 and No. 2 larvae (Supporting Information Figure S1). In the No. 1 larvae, 932 and 1,723 genes were highly expressed at Day 1–2 and Day 3, respectively (Supporting Information Figure S1 and Tables S11 and S12). On the other hands, 1,859 and 905 genes were highly expressed at Day 1–2 and Day 3 in the No. 2 larvae, respectively (Supporting Information Figure S1 and Tables S13 and S14). Again, the ZnNLaz1 (Znev_05665) was highly expressed in the No. 1 larvae at Day 3 (Supporting Information Table S12).
3.3 Gene Ontology enrichment analysis in DEGs
GO enrichment analyses (Yu et al., 2012) were performed using the identified DEGs with more than twofold changes for each pairwise comparison (Figure 3) in order to reveal the molecular signatures for caste differentiation. The enriched GO terms (biological process) were identified among the DEGs between each developmental stage (Supporting Information Figure S2, Tables S15–S20).
Using 539 DEGs upregulated in soldier-destined (No. 1) larvae at Day 1–2 relative to Day 3 (Figure 3a), there were 28 enriched GO terms, which were related to metabolic process as well as cuticle development (Supporting Information Figure S2, Table S15). In particular, GO terms related to nutritional substrates were identified, such as lipid (GO:0006629, 0008610, and 0055088) and fatty acids (GO:0006631, 0006633, 0030497, and 0000038). On the other hand, using 413 DEGs upregulated in soldier-destined (No. 1) larvae at Day 3 compared with Day 1–2 (Figure 3a), there were 40 enriched GO terms (Supporting Information Figure S2, Table S16). These included the GO terms related to organ morphogenesis accompanying cell proliferations (e.g., GO:0002009 and 0048729) and those related to carbohydrate metabolisms as well as cuticle development (e.g., GO:0005975, 0035017, and 0042335). There were no GO terms in DEGs between the two periods in worker-destined (No. 2) larvae (Supporting Information Figure S2).
The characterized GO terms were identified between the No. 1 and No. 2 larvae. Using 465 DEGs at Day 1–2 in soldier-destined (No. 1) larvae relative to worker-destined (No. 2) larvae (Figure 3c), 135 enriched GO terms were identified (Supporting Information Figure S2, Table S17). Of the 135 enriched GO terms, there were biological processes related to the cell cycle with chromosome organizations, nuclear division, and organelle fission (e.g., GO:0022402, 0051276, and 0048285). Using 553 DEGs at Day 1–2 in worker-destined (No. 2) larvae relative to soldier-destined (No. 1) larvae (Figure 3c), 29 enriched GO terms were identified (Supporting Information Figure S2, Table S18). Of the 29 terms observed in the No. 2 larvae, cuticle development and nutritional metabolism (e.g., GO:0042335 and 1901071) were mainly enriched. These enriched GO terms were also identified in the comparison between the No. 1 and No. 2 larvae at Day 3 using 1,398 and 860 DEGs; a total 221 and 32 GO terms were enriched in the No. 1 and No. 2 larvae, respectively (Supporting Information Figure S2, Tables S19 and S20).
3.4 Kyoto Encyclopedia of Genes and Genomes enrichment analysis in differentially expressed genes
Finally, KEGG enrichment analysis (Yu et al., 2012) was performed using DEG data for each gene to reveal the different key metabolic and signaling pathways between soldier- and worker-destined larvae. The results showed enriched KEGG pathways involved in amino acid and sugar metabolism (KEGG ID: dme00350, dme00260, and dme00040) were identified in highly expressed genes in soldier-destined (No. 1) larvae at Day 1–2 compared with Day 3 (Table 1). However, no pathways were enriched at the two time points in worker-destined (No. 2) larvae. In the No. 1 larvae, nutritional pathways, such as amino sugar and sucrose metabolism (KEGG ID: dme00520 and dme00500), and an insect hormone biosynthetic pathway (KEGG ID: dme00981), especially ecdysone synthesis genes, were enriched at Day 3 relative to Day 1–2 (Table 1). In the No. 1 larvae compared to the No. 2 larvae, DNA replication (dme03030) and nucleotide synthesis related pathways (e.g., dme03430, dme03420, and dme03410) were upregulated at both Day 1–2 and 3 (Table 1). Moreover, the Hippo signaling pathways (KEGG ID: dme04391 and dme04392) were upregulated at Day 3 (Table 1). On the other hand, in the No. 2 larvae relative to the No. 1 larvae, several metabolic pathways (e.g., KEGG ID: dme00520, dme04142, and dme00350) were enriched at Day 1–2 and Day 3 (Table 1).
| KEGG ID | Description | % in caste-DEG | % in all | p Value | p.adjust | q Value | Count |
|---|---|---|---|---|---|---|---|
| (a) Day 1–2 compared with Day 3 in the No. 1 larva | |||||||
| dme00350 | Tyrosine metabolism | 10.53 | 0.68 | 8.60E−05 | 3.09E−03 | 2.26E−03 | 4 |
| dme00260 | Glycine, serine, and threonine metabolism | 10.53 | 1.11 | 6.41E−04 | 1.15E−02 | 8.44E−03 | 4 |
| dme00040 | Pentose and glucuronate interconversions | 7.89 | 0.74 | 2.32E−03 | 2.78E−02 | 2.03E−02 | 3 |
| (b) Day 3 compared with Day 1–2 in the No. 1 larva | |||||||
| dme00520 | Amino sugar and nucleotide sugar metabolism | 13.89 | 1.84 | 3.97E−04 | 1.51E−02 | 1.29E−02 | 5 |
| dme00981 | Insect hormone biosynthesis | 8.33 | 0.63 | 1.23E−03 | 2.33E−02 | 2.00E−02 | 3 |
| dme00500 | Starch and sucrose metabolism | 8.33 | 0.79 | 2.44E−03 | 3.09E−02 | 2.65E−02 | 3 |
| (c) The No. 1 larva compared with the No. 2 larva at Day 1–2 | |||||||
| dme03030 | DNA replication | 23.21 | 1.32 | 1.19E−14 | 4.52E−13 | 3.25E−13 | 13 |
| dme00240 | Pyrimidine metabolism | 16.07 | 3.00 | 2.64E−05 | 3.35E−04 | 2.41E−04 | 9 |
| dme03430 | Mismatch repair | 10.71 | 0.79 | 2.05E−06 | 3.90E−05 | 2.81E−05 | 6 |
| dme03420 | Nucleotide excision repair | 10.71 | 1.58 | 1.73E−04 | 1.64E−03 | 1.18E−03 | 6 |
| dme00983 | Drug metabolism—other enzymes | 8.93 | 1.32 | 6.32E−04 | 4.62E−03 | 3.33E−03 | 5 |
| dme03410 | Base excision repair | 7.14 | 0.79 | 7.29E−04 | 4.62E−03 | 3.33E−03 | 4 |
| dme03440 | Homologous recombination | 5.36 | 0.74 | 7.03E−03 | 3.82E−02 | 2.75E−02 | 3 |
| (d) The No. 2 larva compared with the No. 1 larva at Day 1–2 | |||||||
| dme00520 | Amino sugar and nucleotide sugar metabolism | 18.92 | 1.84 | 2.68E−06 | 8.86E−05 | 8.20E−05 | 7 |
| dme04142 | Lysosome | 16.22 | 2.79 | 4.25E−04 | 7.02E−03 | 6.49E−03 | 6 |
| dme00531 | Glycosaminoglycan degradation | 8.11 | 0.63 | 1.33E−03 | 1.46E−02 | 1.35E−02 | 3 |
| (e) The No. 1 larva compared with the No. 2 larva at Day 3 | |||||||
| dme03030 | DNA replication | 12.87 | 1.32 | 3.90E−11 | 2.26E−09 | 1.72E−09 | 13 |
| dme04391 | Hippo signaling pathway—fly | 9.90 | 2.11 | 2.60E−05 | 7.53E−04 | 5.74E−04 | 10 |
| dme03420 | Nucleotide excision repair | 5.94 | 1.58 | 4.09E−03 | 3.39E−02 | 2.58E−02 | 6 |
| dme04392 | Hippo signaling pathway—multiple species | 4.95 | 0.68 | 3.54E−04 | 6.84E−03 | 5.21E−03 | 5 |
| dme03430 | Mismatch repair | 4.95 | 0.79 | 7.58E−04 | 1.10E−02 | 8.38E−03 | 5 |
| dme00511 | Other glycan degradation | 3.96 | 0.58 | 1.87E−03 | 2.17E−02 | 1.65E−02 | 4 |
| dme00531 | Glycosaminoglycan degradation | 3.96 | 0.63 | 2.69E−03 | 2.60E−02 | 1.98E−02 | 4 |
| dme03410 | Base excision repair | 3.96 | 0.79 | 6.55E−03 | 4.75E−02 | 3.62E−02 | 4 |
| (f) The No. 2 larva compared with the No. 1 larva at Day 3 | |||||||
| dme00040 | Pentose and glucuronate interconversions | 6.90 | 0.74 | 3.39E−07 | 2.27E−05 | 1.68E−05 | 8 |
| dme00350 | Tyrosine metabolism | 5.17 | 0.68 | 5.53E−05 | 1.85E−03 | 1.37E−03 | 6 |
4 DISCUSSION
4.1 Transcriptomic changes between soldier- and worker-destined larvae
Our results showed that the transcriptome profiles were extraordinarily different between soldier-destined (No. 1) and worker-destined (No. 2) larvae, as shown by the MDS plot (Figure 2) and diagrams obtained by ANOVA-like test (Supporting Information Figure S1). Consequently, there is a possibility that transcriptomic differences might be immediately occurred at early periods (within 3 days) after their 3rd larval molts. The No. 2 larvae always molt into the 4th instar when a particular individual (the No. 1 larva, presoldier or soldier) exists in a colony (Maekawa et al., 2012). Therefore, the transcriptomic differences between the No. 1 and No. 2 larvae are related to the differentiation of each caste (i.e., soldier or worker). There are two previous reports on the transcriptome changes during developments of these larvae, both of which were conducted with no biological replications. The first one indicated that transcriptome profiles were essentially similar among each molting process using only head samples of these larvae and presoldiers (Masuoka, Yaguchi, Toga, Shigenobu, & Maekawa, 2018). The second report showed that a homolog of the lipocalin gene NLaz (ZnNLaz1) was involved in the soldier differentiation by the regulation of social interaction between the No. 1 larva and a queen (Yaguchi et al., 2018). The present in-depth study suggests that the different transcriptome profiles at the early periods (within 3 days) during each molt are important for the determination of caste developmental fates. We provide the excellent data to understand the possible genetic networks to integrate social signals and intrinsic factors during soldier differentiation.
Several transcriptome analyses have been performed during caste differentiation in hymenopteran species, all of which suggested that social cues were crucial modulators of gene expression changes (Toth & Rehan, 2017). For example, huge transcriptomic differences were observed among different social contexts in the ants Pogonomyrmex californicus and Cardiocondyla obscurior (Helmkampf, Mikheyev, Kang, Fewell, & Gadau, 2016; Schrader, Simola, Heinze, & Oettler, 2015). Moreover, the expression levels of nutrition-sensitive genes involved in caste determination changed in different conditions with/without reproductive castes in the ant Diacamma sp. (Okada, Watanabe, Tin, Tsuji, & Mikheyev, 2017). In termites, caste differentiation can occur in response to social signals through inter-individual interactions, and drastic morphological changes are observed especially during presoldier differentiation (Watanabe et al., 2014). Consequently, there is a possibility that the dynamic transcriptomic differences between Day 1–2 and Day 3 in soldier-destined (No. 1) larvae observed here (Figure 2) may be triggered by different social interactions. Although the molecular mechanisms underlying social behaviors observed in the No. 1 larvae were poorly understood, neuronal alterations should be required, because the brain levels of biogenic amines (e.g., tyramine and dopamine) are involved in the regulation of social behaviors in termites (Ishikawa, Aonuma, Sasaki, & Miura, 2016; Yaguchi et al., 2016). We suggest that transcriptomic differences between the No. 1 and No. 2 larvae are attributed not only to physiological changes for soldier-specific traits but also to behavioral changes in a context-dependent environment.
4.2 The possible transcriptomic changes in response to the nutritional differences
In social insects, nutritional signals are involved in caste-specific developments and behaviors through endocrine regulations (Smith, Toth, Suarez, & Robinson, 2008). The nutritional differences were observed between the soldier and worker developmental processes in an incipient colony of Z. nevadensis (Maekawa et al., 2012). The ZnNLaz1 was crucial factor for soldier differentiation through the regulation of trophallactic interactions with a queen (Yaguchi et al., 2018). In D. melanogaster, NLaz was assigned as one of the GO terms such as lipid metabolic process (GO:0006629). When NLaz was overexpressed in flies, lipid contents were significantly changed in adult individuals (Hull-Thompson et al., 2009). In the present study, lipid metabolic and biosynthetic processes (GO:0006629 and 0008610) were identified in the No. 1 larvae at Day 1–2 compared with Day 3 (Supporting Information Table S15), but ZnNLaz1 (Znev_05665) was highly expressed at Day 3 (Supporting Information Tables S4 and S9), as shown in the previous work (Yaguchi et al., 2018). Although the certain relationships between ZnNLaz1 and lipid contents are unknown in termites, there is a possibility that the nutritional contents (e.g., lipid and related compounds) are frequently received from a queen by the regulation of ZnNLaz1 expression. Alternatively, larval nutritional conditions may be enhanced by the highly ZnNLaz1 expression during the soldier developments. Further physiological cross talk between nutritional status and ZnNLaz1 expression should be analyzed to clarify the role of endocrine regulations during soldier differentiation.
Compared to soldier-destined (No. 1) larvae, GO terms involved in metabolic process were mainly upregulated in worker-destined (No. 2) larvae. No GO terms were enriched in the No. 2 larvae between Day 1–2 and Day 3, but 4 terms (GO:0006022, 0006030, 0006040, and 1901071) were commonly upregulated in the No. 2 larvae compared to the No. 1 larvae at both time points (Supporting Information Tables S18 and S20). There is a possibility that these metabolic pathways are related to the response to nutritional intakes in the No. 2 larvae. Interestingly, these four GO terms were also upregulated in the worker-biased genes in 16 ant species, even though these terms were not related to worker sterility (Morandin et al., 2016). These four GO terms were also enriched within nourishment-responsive genes in the paper wasp Polistes metricus (Berens, Hunt, & Toth, 2015). In addition, the pentose-related pathway (dme00040) was also upregulated in the No. 2 larvae at Day 3 (Table 1). This metabolic pathway was identified in the caste-biased genes in the ant Diacamma sp. (Okada et al., 2017). Consequently, these metabolic and nutritional pathways may be potentially conserved modules (“genetic toolkits”; Toth & Rehan, 2017) also in termites. Further analyses should be performed to know how these pathways are involved in the termite worker formation through nutritional intakes.
4.3 The potential molecular mechanisms of soldier differentiation
We identified potential genetic components underlying soldier differentiation. First, a KEGG pathway related to insect hormone biosynthesis (dme00981) was identified at Day 3 compared with Day 1–2 in soldier-destined (No. 1) larvae (Table 1). Especially, expression levels of three genes involved in ecdysone synthesis (shadow, shade, and spookier) were highly upregulated. This was consistent with the previous reports, which showed that ecdysone receptor signaling activity was important for presoldier and soldier formations in Z. nevadensis (Masuoka & Maekawa, 2016), and that the caste-specific expressions of ecdysone synthesis genes were broadly observed among three termite species including Z. nevadensis (Harrison et al., 2018).
Second, in the No. 1 larva relative to the No. 2 larva, GO terms involved in the cell cycle (e.g., GO:0000278 and 0022402) were upregulated (Supporting Information Tables S17 and S19). Outer cuticles, especially in the head and mandibles, developed rapidly after the gut-purged period of the No. 1 larvae (Masuoka et al., 2015), and the total body sizes were quite different between presoldiers and the 4th-instar larvae (Itano & Maekawa, 2008). These morphological changes may be regulated by genes involved in the cell cycle, chromosome organization, and tissue morphogenesis (e.g., GO:0000278, 0022402, 0051276, and 0048729). In the migratory locust Locusta migratoria, minichromosome maintenance (MCM) genes in DNA replication pathway were positively involved in the DNA synthesis of fat body cells through JH actions (Guo et al., 2014). These KEGG pathways including MCM genes were enriched in the No. 1 larvae relative to the No. 2 larvae at both time points (Table 1). During termite soldier differentiation, cell numbers and sizes in the fat body were increased in Hodotermopsis sjostedti (Cornette, Matsumoto, & Miura, 2007), and JH biosynthetic genes were highly expressed in Z. nevadensis (Yaguchi et al., 2015). Thus, it is possible that the genes involved in DNA replication and cell proliferation may be responsible for soldier development through JH actions.
Third, KEGG enrichment analysis showed that the Hippo signaling pathway (dme04391 and dme04392) were significantly upregulated in the No. 1 larvae at Day 3. The Hippo signaling pathway is broadly conserved among animals including mammals and insects (Yu & Guan, 2013). This pathway comprises a serine–threonine kinase cascade and mediates organ and tissue growth through the regulation of cell proliferation (Pan, 2010). For example, a mutant of warts, one of the members of this pathway, resulted in tumorous overgrowth of Notum in D. melanogaster adults (Staley & Irvine, 2012), and a transcriptional coactivator Yorkie, the downstream target of warts, was involved in tissue growth of D. melanogaster and the silkworm Bombyx mori (Huang, Wu, Barrera, Matthews, & Pan, 2005; Xu et al., 2018). In the rhinoceros beetle Trypoxylus dichotomus, knockdown of ds and ft in this pathway resulted in a decrease in head and thoracic horn size (Hust et al., 2018). Although the roles of Hippo signaling pathway in insects other than the holometabola (with a pupal stage) are still unknown, the present results suggest that soldier-specific exaggerated morphogenesis (e.g., mandibular elongation and head enlargement) is also regulated by this pathway in response to high nutritional conditions. The Hippo signaling pathway might play an important potential role in the integration of nutritional signals and endocrine factors, which resulted in specific organ morphogenesis in some beetles (Casasa, Schwab, & Moczek, 2017; Gotoh et al., 2015). Given that Hippo signaling pathway is broadly conserved among animals (Yu & Guan, 2013), they may also be crucial for termite soldier differentiation via the integration of nutrition and hormone signals. Because food intake from reproductives was frequently observed in the No. 1 larvae at Day 3 after their appearance (Maekawa et al., 2012), nutritional conditions may enhance the growth of these larval bodies. Moreover, recent work suggested that the presoldier molts were positively regulated by the TGF-beta signaling pathway through hormonal actions in Z. nevadensis (Masuoka et al., 2018). There is a possible cross talk between TGF and Hippo signaling pathways in D. melanogaster (Pan, 2010). Further expression and function analysis of components in both pathways are needed to clarify a possible role for soldier developments through nutritional intakes.
5 CONCLUSIONS
The complex social organization of termites can be achieved by social interactions among multiple phenotypes (i.e., castes). Importantly, social interactions contributed to the determination of larval developmental fates (Watanabe et al., 2014). Here, we could identify clear lists of DEGs between the soldier- and worker-destined individuals by focusing on larval development in an incipient colony of Z. nevadensis. The present findings suggest that huge transcriptomic changes are involved in the regulation of caste differentiation. The obtained lists of DEGs represent a cornerstone for our understanding of how termite individuals are determined for the development of each caste under the influence of the social signals through inter-individual interactions.
ACKNOWLEDGMENTS
We are grateful to T. Tsuchida, R. Kimura, and K. Yamahira for their valuable advice on this study, and K. Yamaguchi for technical support in Illumina sequencing. We thank Y. Masuoka, R. Suzuki, T. Hatano, M. Takizawa, N. Kanasaki, K. Kai, N. Hayase, and I. Kobayashi for their help during field and laboratory work. We also thank University of the Ryukyus Center for Research Advancement and Collaboration for use of the computer server. This study was supported in part by a grant for Scientific Research (Nos. 25128705 and JP16K07511 to KM) from the Japan Society for the Promotion of Science, by research grants promoted by University of the Ryukyus: Research Project Promotion Grant (Strategic Research Grant) (No. 16SP01302) and “the Spatiotemporal Genomics Project,” and by NIBB Collaborative Research Program (No. 17-202 and 18-422).
CONFLICT OF INTEREST
None declared.
AUTHORS’ CONTRIBUTIONS
H.Y., R.S., and K.M. designed the study; H.Y., R.S., and K.M. collected samples; H.Y., R.S., and M.M. performed the experiments; H.Y., R.S., M.M., and S.S. analyzed the data; H.Y., R.S., and K.M. drafted the manuscript; all authors contributed to the final version of the manuscript.
DATA ACCESSIBILITY
RNA-seq data obtained are available from the DDBJ Sequence Read Archive (DRA) database (accession no. DRA007363). All other relevant data are within the paper and its electronic supplementary material.




