GABAergic neurons regulate lateral ventricular development via transcription factor Pax5
The authors declare that there are no potential sources of conflict of interest.
This work was supported by NIMH MERIT AWARD R37 MH042261 Limbic lobe in Schizophrenia and Bipolar Disorder (Principal Investigator: F.M.B.), and an E-STAR fellowship funded by the EC's FP6 Marie Curie Host fellowship for Early Stage Research Training (MEST-CT-2004–504640) to S.B. M.B.'s research was supported by Boehringer Ingelheim.
Summary
Postmortem studies have revealed a downregulation of the transcription factor Pax5 in GABAergic neurons in bipolar disorder, a neurodevelopmental disorder, raising the question whether Pax5 in GABAergic neurons has a role in normal brain development. In a genetic approach to study functions of Pax5 in GABAergic neurons, Pax5 was specifically deleted in GABAergic neurons from Pax5 floxed mice using a novel Gad1-Cre transgenic mouse line expressing Cre recombinase in Gad1-positive, that is, GABAergic neurons. Surprisingly, these mice developed a marked enlargement of the lateral ventricles at approximately 7 weeks of age, which was lethal within 1–2 weeks of its appearance. This hydrocephalus phenotype was observed in mice homozygous or heterozygous for the Pax5 conditional knockout, with a gene dosage-dependent penetrance. By QTL (quantitative trait loci) mapping, a 3.5 Mb segment on mouse chromosome 4 flanked by markers D4Mit237 and D4Mit214 containing approximately 92 genes including Pax5 has previously been linked to differences in lateral ventricular size. Our findings are consistent with Pax5 being a relevant gene underlying this QTL phenotype and demonstrate that Pax5 in GABAergic neurons is essential for normal ventricular development. genesis 51:234–245. © 2013 Wiley Periodicals, Inc.
INTRODUCTION
Pax5 is a paired box gene, which functions as a transcription factor in canonical Wnt signaling and determines cellular fate in the cerebral cortex and hippocampus (Machon et al., 2007). In the mid-gestation stage, a dynamic gradient of Wnt signaling controls neural tube patterning (Ciani and Salinas, 2005), which is controlled by an organizing center located at the midbrain/hindbrain boundary where Pax5 is highly expressed (Bouchard et al., 2000). Glutamic acid decarboxylase 67 (GAD67), encoded by the Gad1 gene, is responsible for synthesis of GABA from glutamate and is widely distributed in the brain (Tamamaki et al., 2003). It is expressed in postmitotic and proliferating cells, suggesting that using this gene promoter to drive Cre will allow recombination of floxed genes at these stages to study fundamental cellular activities such as differentiation and migration during developmental stages (Ma et al., 1992; Tashiro et al., 2007). A Pax5 global knockout is postnatally lethal (Urbanek et al., 1994). To explore the role of Pax5 in inhibitory neurons on brain development and function, we have generated GAD67-specific conditional Pax5-deficient mice (Gad1-Pax5KO).
RESULTS
Generation and Analysis of Gad1-Cre Mice
To achieve expression of the Cre recombinase in GAD67-expressing GABAergic neurons, we generated a Gad1-Cre transgenic mouse using chromosomal Gad1 sequences from a BAC clone (Fig. 1a). Gad1-Cre transgenic mice did not show gross abnormalities (Fig. 1b) and were indistinguishable from their wild-type (WT) littermates in open-field tests (Supporting Information Fig. S1).
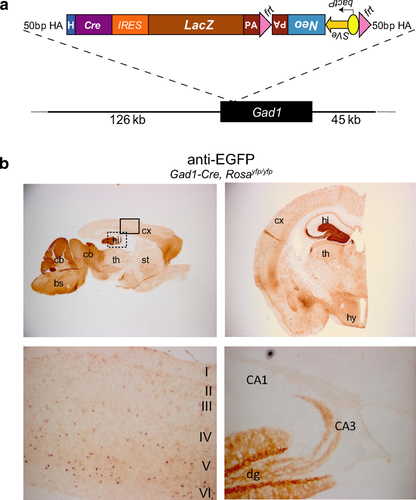
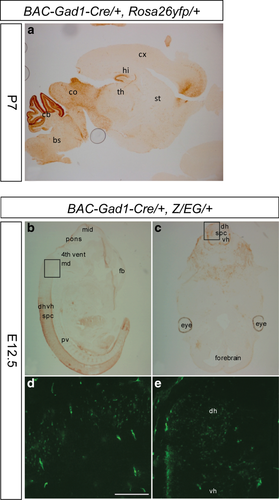
The recombination profile of Gad1-Cre transgenic mice was assessed using the reporter lines Rosa26-EYFP (Srinivas et al., 2001) and Z/EG (Novak et al., 2000) in which EYFP (Enhanced Yellow Fluorescent Protein) or EGFP (Enhanced Green Fluorescent Protein) is expressed upon Cre-mediated recombination of a loxP-flanked STOP-cassette. We observed EYFP-immunoreactive (IR) cells in various brain areas that contain GABAergic neurons including the brainstem, cerebellum, inferior and superior colliculus, hypothalamus, and dentate gyrus of hippocampus and neocortex (mainly layers IV–VI) (Fig. 1b). However, in some brain areas that are rich in GABAergic neurons (Supporting Information Fig. S2) only few cells showed recombination, for example, in the striatum (Fig. 1b) and the olfactory bulb (data not shown). Gad1-Cre-mediated recombination was already present at E12.5 (Fig. 2b) and in P7 pups its pattern was similar to that seen in adult animals (Fig. 2a).
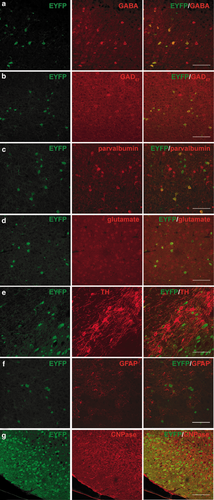
To examine the specificity of recombination events in Gad1-Cre mice, co-immunostaining studies were performed in the lower layers (layers IV–VI) of the somatosensory cortex. In the mice examined, 100% and 81.8% of EYFP-IR cells were colabeled with GABA, indicating that the recombination is specific for GABAergic cells (Fig. 3a, Supporting Information Table S1, Fig. S4). Conversely, 42.1% and 40.9% of GABA-positive cells were positive for EYFP, indicating that less than half of the GABAergic cells can be targeted by the Gad1-Cre recombinase. Some EYFP-IR cells were labeled using an antibody against GAD67 (Fig. 3b). However, the GAD67 staining is punctated and it is not easy to distinguish signal from background and cell bodies from neurites. We think that this could be the reason why not all EYFP-IR cells are recognized as GAD67-positive. There are multiple populations of GABAergic interneurons, which are defined by the presence of marker proteins. To assess which interneuronal population expresses the Gad1-Cre transgene, double-labeling experiments were performed, which revealed that 25.6%–39.8% of the EYFP-positive cells were positive for parvalbumin, and 23.0%–30.9% of parvalbumin-positive cells were colabeled with EYFP (Fig. 3c, Supporting Information Table S1, Fig. S5a). No EYFP-positive cells were colabeled with calretinin or neuropeptide Y (Supporting Information Table S1, Fig. S5b). None of the EYFP-positive cells was colocalized with the excitatory neurotransmitter glutamate (Fig. 3d, Supporting Information Table S1). Furthermore, Gad1-Cre-mediated recombination does not occur in dopaminergic neurons as shown by the absence of reporter signal from tyrosine hydroxylase (TH)-positive neurons (Fig. 3e). Recombination was absent in astrocytes and oligodendrocytes (Fig. 3f,g). Collectively, this initial analysis shows that the Gad1-Cre transgene is expressed in parvalbumin-positive interneurons and as yet unidentified interneuron classes, excluding the calretinin or neuropeptide Y-expressing subtypes.
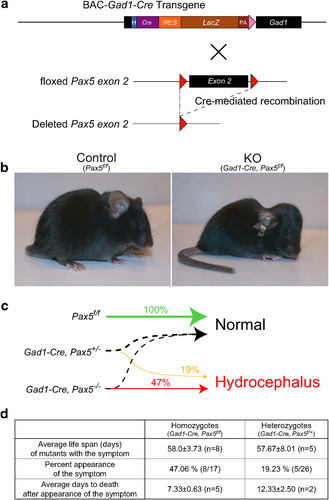
Gad1-Pax5KO Mice Develop Hydrocephalus in a Gene Dosage-Dependent Manner
The Gad1-Cre line was crossed with Pax5-floxed mice (Horcher et al., 2001) to generate Gad1-Pax5KO mice with a conditional deletion of Pax5 in GAD67-expressing neurons (Fig. 4a). About 47% of homozygous mutant mice (Gad1-Cre, Pax5f/f) and 19% of heterozygous mutant mice (Gad1-Cre, Pax5f/+) developed a dome-shaped skull around postnatal day 50 (Fig. 4b), indicating that the development of hydrocephalus is dependent on the Pax5 gene dosage (p = 0.0021, Spearman's rank correlation; Fig. 4c and d). Gad1-Pax5KO mice died approximately 1–2 weeks after the appearance of macroscopic changes (Fig. 4d), and displayed lethargy and emaciation. Both homozygous and heterozygous mutants developing a dome-shaped head had a life expectancy of 58 days (Fig. 4d), whereas animals not developing this phenotype by P60 were able to survive for at least 12 months and were fertile. Abnormalities were not observed in littermates with control genotypes (Gad1-Cre; Pax5f/+; Pax5f/f).
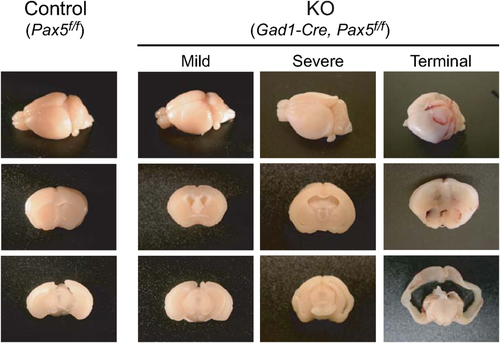
Gad1-Pax5KO Mice Have Enlarged Lateral Ventricles
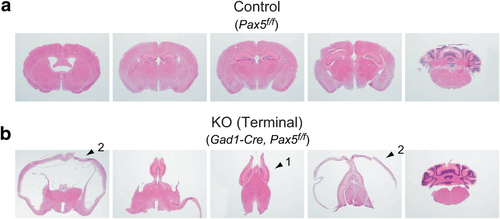
The ventriculomegaly of Gad1-Pax5KO was prominent in the lateral ventricles (Fig. 5, Supporting Information Fig. S6). In the most severe cases, the medial structures between lateral ventricles were deformed and compressed as represented by vertically relocated hippocampi (Fig. 6b, arrow 1). The cerebral cortex surrounding the lateral ventricles was also markedly thinner (Fig. 6b, arrow 2). The ventricular dilation may be caused by obstruction of interventricular connections, increased production of cerebrospinal fluid by ependymal cells of the choroid plexus or decreased resorption by arachnoid villi. As our present analysis provided no evidence for the enlargement of other ventricles, the extensive enlargement of the lateral ventricles indicates an obstruction of interventricular connections, while other possibilities cannot be excluded. The potential contribution of other mechanisms, for example, that Gad1-Cre-induced recombination in the choroid plexus (Supporting Information Fig. S3) may have affected ependymal cell function, remains to be elucidated.
Gad1-Pax5KO Mice Show Increased Locomotion and Anxiogenic-Like Behavior in the Open Field Test
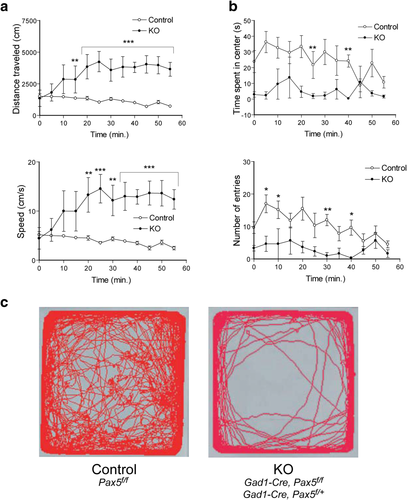
Locomotor activity of Gad1-Pax5KO mice with dome-shaped heads was assessed in a novel open field. All four measures taken, that is, distance traveled, speed, time spent in center, and number of entries into center showed significant differences in genotype, (F1,77 = 20.94, p = 0.0026; F1,77 = 18.94, p = 0.0033; F1,77 = 9.1, p = 0.0196; F1,77 = 9.3, p = 0.0187 by two-way repeated measures ANOVA). Bonferroni posthoc analysis confirmed these differences (Fig. 7a). Because the mutants appeared to be lethargic in their home cages, the increased locomotion is likely induced by the novel environment. The observation that mutant mice spent less time in the center and entered the center less frequently (Fig. 7b, c) is potentially indicating an anxiogenic-like behavior.
DISCUSSION
The major phenotype of these mice, that is, enlargement of the lateral ventricles has been found to be associated with neurodevelopmental disorders such as schizophrenia (Meduri et al., 2010; Rosa et al., 2010) and bipolar disorder (Kempton et al., 2008) [not in the first episode (Rosa et al., 2010), only when psychosis was present (Edmiston et al., 2011)]. Thus, reduced Pax5 expression in GABAergic neurons in patients with bipolar disorder (Benes et al., 2007) might contribute to changes of ventricular size observed in these patients.
Pax5 is strongly expressed in the mid–hindbrain boundary (MHB) at embryonic stages, whereas in adults the expression level is low and expression appears to be widespread (Allen Developing Mouse Brain Atlas, http://developingmouse.brain-map.org). It is tempting to speculate that Pax5-deficiency in the MHB of the embryo may be responsible for the hydrocephalus phenotype in Gad1-Pax5KO mice. However, the time window for the appearance of hydrocephalus is quite narrow, potentially suggesting a role of the Pax5-deficit in the adult animals in the pathogenesis of hydrocephalus in the Gad1-Pax5KO mice. To address this question, temporal control of Pax5 expression in GAD67-positive neurons would be required. Our finding that GABAergic neurons can play a role in determining ventricular size is consistent with the observation of a linear correlation between degree of ventricular enlargement and reduction of GAD-immunoreactive neurons in a kaolin-induced hydrocephalus model in rats (Tashiro et al., 1997).
It is interesting to note that one functional copy of Pax5 is not sufficient to maintain a normal development, as a sizable percentage of Gad1-Pax5+/− mice develop hydrocephalus (Fig. 4c). Although global Pax5+/− have not been examined in this study, such mice have been generated in other contexts (e.g., as breeders to generate Pax5−/− mice), and development of an obvious hydrocephalus like the one observed in this study has not been noticed, which does not exclude the possibility that a more subtle ventricular enlargement may be present. This raises the question why heterozygous loss of Pax5 in GABAergic neurons leads to hydrocephalus, while global heterozygous loss of Pax5 is so far not known to do so. Although we do not know the answer to this question, one speculative possibility is that in global Pax5+/− mice loss of Pax5 in other cell types helps to compensate for loss of Pax5 in GABAergic neurons. Another noteworthy aspect is that not all Gad1-Pax5−/− mice develop hydrocephalus (Fig. 4c), indicating the presence of modifier genes. This is in line with previous observations that several genes have an influence on ventricle size in the mouse (Zygourakis and Rosen, 2003).
Gad1 is thought to be expressed exclusively in neurons, and our analysis supports the notion that the Gad1-Cre transgene is expressed specifically in GABAergic neurons. However, we cannot completely rule out the possibility that Gad1-Cre is also expressed in non-neuronal tissues and that Gad1-Cre-mediated recombination in non-neuronal cells might lead to hydrocephalus. Gad1 knockout mice (and also Gabrb3 knockout mice that lack the GABAA receptor β3 subunit or mice lacking the vesicular inhibitory amino acid transporter Viaat) have been found to have a cleft palate phenotype (Asada et al., 1997; Condie et al., 1997; Homanics et al., 1997; Wojcik et al., 2006). The developmental mechanism of cleft palate in these mice are unknown, but it is not unreasonable to assume that non-neuronal cells are involved in the development of cleft palate. However, it has been shown recently that CNS-specific inactivation of Gad1 was sufficient to disrupt palate development (Oh et al, 2010), suggesting that the primary defect is in the CNS. We assume that the same is true for the hydrocephalus phenotype in the Gad1-Pax5−/− mice.
Pax2 and Pax5 arose by gene duplication and have maintained equivalent biochemical functions (Bouchard et al., 2000). It has been suggested that there is functional substitution between Pax2 and Pax5 in brain regionalization (Schwarz et al., 1997). Pax2 expression in the MHB positively regulates Pax5 expression through a Pax-binding site of a minimal MHB-specific enhancer in the 5′-flanking region of Pax5 (Pfeffer et al., 2000). Combinatorial interactions between the transcription factors Pax2, En1, Grg4, Otx2, Gbx2, and the signaling molecule Wnt1 control the expression of FGF8, which is required to maintain mid-hindbrain identity by supporting development of multiple neurons (Ciani and Salinas, 2005; Ye et al., 2001). FGF8, Wnt, En1, En2, Pax2, and Pax5 depend on each other for stable expression; if one is missing, the expression of the remaining genes is extinguished over time, and these genes can induce and maintain each other's expression at ectopic locations (Funahashi et al., 1999; Wurst and Bally-Cuif, 2001; Ye et al., 2001). These interactions establish a positive regulatory loop, which governs brain development as an organizer located at the MHB. The direct interactions between Pax2 and the MHB-specific enhancer in the 5′-flanking region of Pax5 mentioned above are directly involved in this feedback loop (Pfeffer et al., 2000). Hydrocephalus has also been observed in other mutants of genes in this regulatory network, for example, Otx2+/− mice (Makiyama et al., 1997), Wnt1 null mutants, transgenic mice expressing En1 ectopically under the control of the Wnt1 enhancer, and Wnt1sw/sw (swaying) point mutants (Louvi and Wassef, 2000; Rowitch et al., 1999; Thomas and Capecchi, 1990), demonstrating the essential role of the MHB region for ventricular development. All previous loss-of-function studies have been performed with global mutants, so that no conclusion could be drawn as to the relevant neuronal cell types required for normal ventricular development. Our data clearly implicate that GABAergic neurons and, in particular, Pax5-dependent functions in these neurons play a pivotal role in this process. The closely related gene, Pax6, is expressed in the subcomissural organ (SCO) from which neuroepithelial cells originate, and mice lacking Pax6 fail to develop the SCO (Estivill-Torrus et al., 2001). In the H-Tx rats that develop congenital hydrocephalus, Pax6 is downregulated (Miller et al., 2006). Thus, Pax6 could play a role in the development of hydrocephalus. Recently, it has also been reported that deregulation of the p57-E2F1-p53 axis in the SCO and Pax2-positive interneurons results in nonobstructive hydrocephalus and cerebellar malformation in mice (Matsumoto et al., 2011).
QTL mapping revealed a 3.5 Mb locus on chromosome 4 containing 92 genes including Pax5, which interacts with another as yet uncharacterized locus on chromosome 7 to modulate ventricle size of the mouse brain (Zygourakis and Rosen, 2003). Our data suggest that Pax5 is likely a major gene at this locus responsible for modulating ventricle size.
METHODS
Generation of Gad1-Cre Mouse Line
The plasmid pIZKeoX1 containing the Cre-IRES-lacZ construct (Fig. 1) was generated in Escherichia coli. The Neo cassette with flanking frt sequences was inserted downstream of the Cre-IRES-lacZ in reversed orientation. Homologous arms of 50 bp were added for subsequent specific recombination into the first coding ATG of the Gad1 gene in the BAC clone, RPCI23–118N13 (BACPAC resources, Children's Hospital, Oakland, California) by ET recombination (Muyrers et al., 2004). Specifically, the cassettes inserted into the BAC clone consist of a hybrid intron (HI) followed by the Cre-recombinase encoding gene, an internal ribosomal entry site (IRES), the beta-galactosidase gene (lacZ) for identification of transgene expression by X-gal staining, and a neomycin/kanamycin cassette (neo) for selection of recombined BAC clones in E. coli. The Neo cassette was removed by Flp-mediated recombination at the frt sites after the selection. The consequent transgenic construct was validated also by DNA sequencing at the borders of insertion sites using primer 1 5′-ACGGCCGGAGTGGACACCTGTGGAGAG-3′ for the boundary between the first coding ATG of Gad1 and Cre, and primer 2 5′-AGATCAGCAGCCTCTGTTCCACAT-3′ at the boundary between frt and Gad1. BAC DNA was prepared using a CsCl gradient centrifugation-based method or a QIAGEN kit for large constructs. Circular BAC DNA was injected at 1–2 ng/µl into pronuclei of oocytes (CBA/C57 hybrid). Offspring of founder mice was screened for transgene expression by X-gal staining, showing that the LacZ-cassette is functional in the cerebellum and superior colliculus (regions with many Gad1-Cre- and LacZ-expressing cells). However, LacZ expression was relatively weak presumably because of low efficiency of the IRES site. Cre recombinase function was tested in crosses with reporter mice, which express EYFP or EGFP after Cre-mediated deletion of a STOP-cassette (Rosa26-EYFP reporter line, Srinivas et al. 2001; Z/EG reporter line, Novak et al. 2000).
Generation of Gad1-Pax5KO Mice
Pax5 floxed mice had been backcrossed to C57BL/6 at least eight generations and Gad1-Cre mice for at least four generations. After these backcrossings, Gad1-Cre mice were crossed with floxed Pax5 mice.
Experiments involving mice at the Mailman Research Center of the McLean Hospital were approved by the Institutional Animal Care and Use Committee of McLean Hospital (protocols #07–1/2–1 and #08–1/2–2 to UR), and all animal procedures at the European Biology Molecular Laboratory (Mouse Biology Unit) conformed to National and International laws and policies (EEC Council Directive 86/609, OJ L 358, 1, December 12, 1987; NIH Guide for the Care and Use of Laboratory Animals, NIH Publication No. 85–23, 1985 revised in 1995), personal project number: 7.1; at the Centre for Neuroregeneration, conformed to UK legislation (Scientific procedures) ACT 1986 and the University of Edinburgh ethical review committee policy, personal project license: 60/4049. Mouse reagents will be made available to the research community.
Histology and Immunostainings
Adult mice and pups were anesthetized by intraperitoneal injection of 3% avertin in PBS, and perfused with PBS, followed by 4% PFA in PB buffer (pH7.4) for fixation. Embryos were dissected out of the uterus of the mother (euthanized by cervical dislocation). Dissected brains and embryos were postfixed in 4% PFA at 4°C overnight, then rinsed twice with cold PBS, and stored in cold 30% sucrose in Tris-azide buffer for 2–3 days at 4°C followed by cryopreservation in embedding medium (OCT Sakura TissueTek). Cryosections (30 µm) were obtained using a Leica cryostat CM3050S and for DAB stainings were incubated in 0.1 M PB buffer (pH7.4), quenched with 2% H2O2, washed in TBS, blocked in 10% serum in histo solution (0.3% carrageenan, 0.5% Triton X-100, in TBS), incubated with the primary antibody in 1% serum in histo solution at 4°C for 24–72 h, washed in wash solution (1% serum in 0.5% Triton X-100 in TBS), incubated with the secondary antibody in 1% serum in histo solution at 4°C overnight, rinsed in wash solution, incubated in vectastain ABC solution for 1 h, washed in TBS and TB, and incubated in 0.5 mg/ml DAB and 0.03% H2O2, followed by cold TB washes, drying and mounting in DEPEC-Eukitt. For immunofluorescent stainings after the primary antibody incubation, sections were washed in TBS and incubated in secondary antibody in TBS at room temperature for 2 h, washed in TBS, stained with DAPI, dried, and mounted in vectashield mounting medium. Stained sections were analyzed using a dissecting microscope (Leica MZ12 and Zeiss Schott KL1500 light source and Leica FireCam 1.2.0 imaging software) or a Leica fluorescent microscope (Leica, Wetzlar, Germany DMR and camera DC500 and imaging software IM500 or a Leica confocal microscope LASAF TCS SP5). For histological analyses, Gad1-Cre and Pax5 mutant brains were fixed as described above except that animals were anesthetized using Isoflurane and sliced using a coronal brain dice at AP: Bregma +0.38 and −4.16, approximately for macro photography shown in Figure 5 and Supporting Information Figure S6. A series of 25 µm coronal sections were prepared after cryoprotection procedure at 100 µm interval, and were subjected to hematoxylin/eosin staining.
Antibodies
EGFP (for DAB:1:2000, ab290 Abcam; for IF:1:3000 ab10980 Abcam), GABA (1:1000, A2052 Sigma), glutamate (1:1000, G9282 Sigma), GAD67 (for DAB: 1:, for IF: 1:1000, MAB5406 Chemicon, Temecula, CA), parvalbumin (1:1000, P3088 Sigma), calretinin (1:2000, #7699/4 Swant, Marly, Switzerland), NPY (1:8000, ab10980 Abcam, Cambridge, MA), TH (1:200, #Mab318 Chemicon), GFAP (1:1000, #sc9065 Santa Cruz Biotechnology, Dallas, TX), CNPase (1:100, #Mab326 Chemicon), biotinylated anti-chicken, anti-mouse, anti-rabbit (1:200, Vector Labs, Burlingame, CA), Alexa488-anti-chicken (1:1000, Moleular Probes (Life Technologies), Grand Island, NY), Alexa555-anti-mouse, anti-rabbit (1:1000, Molecular Probes).
Behavioral Analysis
All behavioral studies were performed on male mice, littermates being housed together. Mice were moved to the behavioral facility at least a week before starting the tests in order to allow for habituation to the new environment.
Novelty-Induced Locomotor Activity in Gad1-Cre Mice
After habituation to the test room, four mice at a time were placed into four different open field arenas (50 × 50 × 28 cm) at light conditions of 100 lux in the arena and video tracked for 30 min. The time spent and distance traveled at the border and at the center of the arena were recorded using VideoMot2, version 5.62 (TSE). A 34 cm × 34 cm square inside the open field was arbitrarily defined as center of the field.
Novelty-Induced Locomotor Activity in Gad1-Cre Pax5 Mutants
Mice were placed in 42 × 42 × 31 cm plexiglas chambers lit at 160 lux. Mice were videorecorded for 60 min. Five minutes time bins were applied to process activity measurements by the aid of computer software EthoVision XT (Noldus Information Technology, Wageningen, The Netherlands). A 20 cm × 20 cm square inside the open field was arbitrarily defined as center of the field.
Statistics
Data in Figure 4d, “Percent appearance of the symptom,” were analyzed using Spearman's rank correlation for number of deleted Pax5 alleles by appearance of the symptom. Data in Figure 7 and Supporting Information Figure S1 are expressed as mean ± SEM. Data in Figure S1a and b were analyzed using two-way ANOVA (genotype × area). Data in Figure S1c were analyzed using unpaired 2-sample t-test. Data in Figure 7a,b were analyzed using two-way repeated-measures ANOVA (genotype × time) and the mutant group and control group were compared by two-sided Student's t-test.
ACKNOWLEDGMENTS
We thank the EMBL-Monterotondo gene-expression and transgenic services for the production of the BAC-transgenic mouse and thank Ms. Hew Mun Lau (Laboratory of Genetic Neuropharmacology, McLean Hospital) for technical assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health. MB contributed study material. NO, SB, FMB, LM, and UR contributed to experimental design. NO and SB collected, assembled, and analyzed data. NO, SB, LM, and UR contributed to data interpretation and composed the manuscript. All authors approved the final version of the manuscript.
COMPETING FINANCIAL INTERESTS
UR has provided professional services to Sunovion Pharmaceuticals and to Concert Pharmaceuticals. The other authors declare no competing financial interests.




