Analysis of human chromosomes by imaging flow cytometry
Abstract
Chromosomal analysis is traditionally performed by karyotyping on metaphase spreads, or by fluorescent in situ hybridization (FISH) on interphase cells or metaphase spreads. Flow cytometry was introduced as a new method to analyze chromosomes number (ploidy) and structure (telomere length) in the 1970s with data interpretation largely based on fluorescence intensity. This technology has had little uptake for human cytogenetic applications primarily due to analytical challenges. The introduction of imaging flow cytometry, with the addition of digital images to standard multi-parametric flow cytometry quantitative tools, has added a new dimension. The ability to visualize the chromosomes and FISH signals overcomes the inherent difficulties when the data is restricted to fluorescence intensity. This field is now moving forward with methods being developed to assess chromosome number and structure in whole cells (normal and malignant) in suspension. A recent advance has been the inclusion of immunophenotyping such that antigen expression can be used to identify specific cells of interest for specific chromosomes and their abnormalities. This capability has been illustrated in blood cancers, such as chronic lymphocytic leukemia and plasma cell myeloma. The high sensitivity and specificity achievable highlights the potential imaging flow cytometry has for cytogenomic applications (i.e., diagnosis and disease monitoring). This review introduces and describes the development, current status, and applications of imaging flow cytometry for chromosomal analysis of human chromosomes.
1 INTRODUCTION
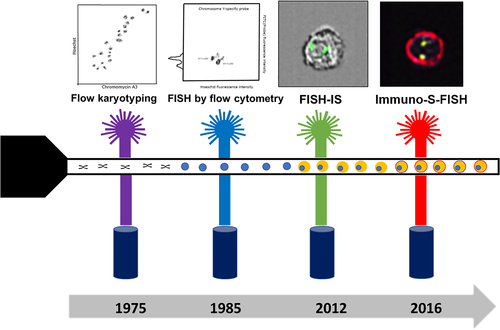
Chromosomal analysis plays a pivotal role in the assessment and understanding of cellular biology in health and disease. Since first introduced in the 1950s, a range of methods has been utilized to assess chromosomes in the life sciences and human disease (Ferguson-Smith, 2015; Hsu, 1952; Tjio & Levan, 1956). This includes using cell cultures and colchicine followed by chromosome banding methods. Chromosome size, type and shape can be determined and defects in constitutional disorders and malignancies (i.e., leukemia, lymphoma) identified (Martin & Warburton, 2015; O'Connor, 2008). These include changes in chromosome number (aneuploidy) such as whole chromosome loss (e.g., monosomy) or gain (e.g., trisomy) (Ferguson-Smith, 2015). Structural alterations, such as translocations between chromosomes, inversions, insertions, rings, deletions, isochromosomes, and duplications, can also be identified. Other methods were subsequently introduced including fluorescent in situ hybridization (FISH), chromosome painting and comparative genomic hybridization. FISH is a molecular-based method that utilizes hybridization of fluorescent-labeled probes to complementary DNA sequences to detect a specific region on a chromosome (Carter, 1994; Guo et al., 2014; Sinclair, 2002; Weiss et al., 1999). FISH is typically performed on metaphase spreads and interphase cells, including on tissue sections, on a slide, to detect numerical or structural alterations, and assessed by fluorescent microscopy (Cui et al., 2016; Van Der Logt et al., 2015). Flow cytometry was first applied to analyze chromosomes in the mid-1970s (Gray, Carrano, Moore, et al., 1975; Gray, Carrano, Steinmetz, et al., 1975; Stubblefield et al., 1975). Although not widely applied in human disease, it did offer rapid and high-throughput analysis as well as the ability to flow sort chromosomes. Subsequent developments in flow cytometric instrumentation, in particular the addition of digital cameras and visual imagery, is now leading to a reawakening of flow cytometry for the analysis of chromosomes (Figure 1). The new instrumentation is now being applied to assess ploidy and structural chromosome changes using FISH probes. Such is the capability of this new technology that it has been described as an “unexplored frontier” in cytogenomics and the molecular testing of cytological samples (Weissleder & Lee, 2020). In this review, the development and current status of imaging flow cytometry for chromosomal analysis in human disease are presented.
2 FLOW CYTOMETRY FOR CHROMOSOME ANALYSIS
Flow cytometry of chromosomes in suspension is a high-throughput, rapid and accurate method to assess chromosome number and structure (Gray, Carrano, Moore, et al., 1975; Gray, Carrano, Steinmetz, et al., 1975; Stubblefield et al., 1975). It has been applied for karyotyping, FISH analysis of telomeres and centromeres, as well as chromosome sorting for downstream analysis. Although neither flow karyotype nor flow FISH has been widely adopted for analysis of human chromosomes, an overview of each technique is presented to give some background to the field (Stepanov et al., 1996).
2.1 Karyotyping
Flow karyotyping, pioneered by Gray et al. was the first chromosomal analytical technique developed for flow cytometry (Gray, Carrano, Moore, et al., 1975; Gray, Carrano, Steinmetz, et al., 1975). This technique enabled the detection of metaphase chromosomes in suspension by utilizing two DNA stains: Hoechst 33342 (HO) and Chromomycin A3 (CA3), which bind to the A-T rich and G-C rich regions of the DNA respectively (Chazotte, 2011; Zihlif et al., 2010). Subsequent improvements in instrumentation and DNA dyes allowed better discrimination between chromosomes (e.g., 4′,6-diamidino-2-phenylindole [DAPI], HO, CA3, and propidium iodide) resulting in the ability to separate chromosome 9 from the chromosome 9–12 cluster (Ng et al., 2019). However, it is still not possible to clearly separate all chromosomes with chromosomes 10–12 forming a cluster, due to their similarity in size and base pair composition (Langlois et al., 1982; Ng et al., 2019). Chromosomes are assessed for the fluorescence profile and abnormalities detected by deviations compared to normal (Gray et al., 1988). Numerical changes are seen as changes in the volume or bivariate mean of each peak, whereas translocations cause changes in the DNA composition of chromosomes resulting in the presence of a new peak (Gray et al., 1988; Lebo et al., 1986). Flow karyotyping can be performed on standard high throughput flow cytometers (e.g., BD LSRFortessa and Beckman Coulter MoFlo Legacy) (Ng et al., 2019). It is not widely utilized to study human disease but is applied to animal and plant cells (Frohlich et al., 2017; Popescu et al., 1993; Schmitz et al., 1998; Stanyon & Stone, 2008).
2.2 FISH
Flow cytometers have also been used to analyze specific chromosomes and regions, including centromeres and telomeres, using FISH probes. Flow FISH is primarily performed on isolated chromosomes or nuclei of cells in interphase, but can also be achieved on preserved intact cells (Keyvanfar et al., 2012). The FISH probes used generally bind to the centromeres by hybridizing to specific alpha satellite sequences of individual chromosomes (Arkesteijn et al., 1995; Trask et al., 1985; Trask et al., 1988; van Dekken et al., 1990). Changes in the fluorescence intensity emitted by the probe can identify abnormalities in the number of chromosomes (Brind'Amour & Lansdorp, 2011; Keyvanfar et al., 2012). For example, a chromosome gain (e.g., trisomy) increases the fluorescence intensity compared to normal due to the presence of an extra chromosome; whereas with a chromosome loss (monosomy) there is a decrease. Examples of the clinical applicability have been demonstrated for the analysis of chromosomes 1, 8, and Y in leukemia (Arkesteijn et al., 1995).
Telomeres and their length have also been studied by flow cytometry by measuring the fluorescence of the labeled peptic nucleic acid (PNA) probe (Alder et al., 2018; Aubert et al., 2012; Baerlocher et al., 2006). PNA probes differ from conventional DNA FISH probes, by having different underlying chemistry, allowing them to bind more readily to complementary nucleic acid sequence and with higher affinity (Genet et al., 2013). Telomere flow FISH also generates an intensity value which represents relative but not absolute telomere length. Using reference samples of known telomere length and calibration controls allows this data to be used to calculate approximate telomere length. (Rufer et al., 1998; Wand et al., 2016). Telomere flow FISH has been applied clinically to detect shortened telomeres. These can be due to inherited mutations resulting in decreased telomere length and causing accelerated aging (Armando et al., 2019; Mangaonkar & Patnaik, 2018; Stanley & Armanios, 2015).
2.3 Chromosome sorting
Fluorescence-activated cell sorting, introduced in the mid-1970s, can be used to isolate chromosomes with high yield and purity (Gray, Carrano, Moore, et al., 1975; Gray, Carrano, Steinmetz, et al., 1975). The ability to generate a large number of chromosomes has led to the widespread use of this technique in understanding DNA. The principle relies on the ability to generate droplets containing the chromosomes of interest. The instrument adds a charge to the droplets containing the desired chromosomes, which are then deflected in a static electric field into a collection vessel. The collected chromosomes can then be processed with other downstream techniques for further investigation (Doležel et al., 2011; Ibrahim & Van Den Engh, 2004). Flow sorting of chromosomes has been utilized in the preparation of genomic libraries, chromosome paints, mapping of genetic markers and was crucial in the early stages of the “Human Genome Project” (Ibrahim & Van Den Engh, 2004; Monard, 2016; Van Dilla et al., 1986; Van Dilla & Deaven, 1990). In addition to application in human disease research, flow karyotyping and sorting has been crucial to isolate and understand the chromosomes of plants and animal (Doležel et al., 2021; Yao et al., 2020). Yao et al., 2020 have shown the effectiveness of flow karyotyping and sorting in isolating complex chromosomes of sheep prior to sequencing. This approach has then been applied to study chromatin conformation, BAC library construction, optical mapping and sequencing amplified chromosomal DNA of plants (Doležel et al., 2021).
3 IMAGING FLOW CYTOMETRY: PRINCIPLES
- Camera based imaging device which utilizes either charged couple device (CCD) or complementary metal-oxide semiconductor, for example, ImageStreamX (Basiji et al., 2007) and virtual freezing imaging flow cytometry (Mikami et al., 2020).
- Photomultiplier (PMT) tube-based imaging device which utilizes PMT combined with a technique named spatial–temporal transformation, for example, 3D imaging flow cytometry (Han & Lo, 2015).
This review will focus on imaging flow cytometry utilizing CCD, specifically the Amnis ImageStreamX, as this has been most commonly reported for the analysis of human chromosomes.
Key components of the Amnis ImageStreamX are image magnification, extended depth of field and digital cameras. The microscope objectives in the device, commonly ×20, ×40, and ×60, give the operator the option to determine the degree of magnification required to identify the cell and chromosome of interest. Reports indicate for chromosome applications, both the ×40 and ×60 objectives enable high resolution analysis of the probe signal (Hui et al., 2019; Minderman et al., 2012; Sanderson & Simon, 2017). The “extended depth of field” (EDF) functionality is another key feature required for chromosome analysis. EDF incorporates a specialized element in the standard optical system that causes light from widely different focal positions in the object to be imaged and in focus on the detector plane simultaneously, a process called Wavefront Coding™ (Ortyn et al., 2007). Wavefront coding uses a special optical element near the aperture stop which makes the imaging system insensitive to a range of mis-focus related aberrations thereby extending the depth of field of the system. Images acquired using EDF have an effective depth of field of approximately 15 μm with all cellular components, even as small as 2 μm, in precise focus. This results in a high-resolution image of the cell as well as all cellular features simultaneously in focus (Basiji et al., 2007). This is especially important when analyzing large cells as the standard image focal plane is set to the center of the core stream however the part of the cell that is of interest (e.g., nucleus) may be at the front or back of the stream therefore appearing out of focus.
The digital cameras in the imaging flow cytometer capture images of all cells. There are different camera types, including CCD with time delay integration (TDI) and beam scanners with a scientific complementary metal-oxide semiconductor camera (Basiji & O'Gorman, 2015; Mikami et al., 2020). TDI works by accumulating multiple continuous exposures of a moving object at a rate that is synchronized with the motion of the object. By combining it with a CCD camera it produces an effect that is akin to panning the camera to track the object. The outcome is that CCD-TDI cameras allow cell images to be collected while in motion and without streaking (Basiji, 2016). The need to achieve synchrony and alignment between the motion of the object and the read-out rate of the camera does however hinder the capacity to integrate sorting capabilities (Han et al., 2016).
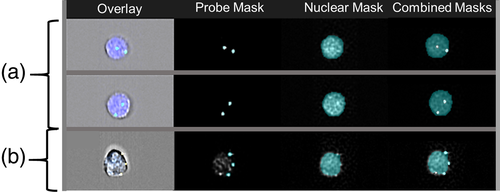
A number of software tools can be used to facilitate analysis of the digital images. First, “masks”, or specified regions within the cell of interest, can be selected and digitally “isolated” (Figure 2) (Dominical et al., 2017). These masks are akin to “gates” which are applied to interrogate a specific part of an object of interest on the digital images. For chromosomal analysis, masks have been used to target the cell nucleus or FISH probe signals and further evaluation is restricted to this selected cellular region. This makes the creation of accurate masks the cornerstone for image-based analysis; suboptimal masks can lead to other parts of the cells, that are not of interest, being evaluated (Dominical et al., 2017). Once a mask is determined, parameters within that area of interest can be assessed. For example, the localization, enumeration, size, shape, and fluorescence intensity of an object such as a chromosomal signal can be interrogated. It is this combination of in-focus digital images and ability to isolate a specific cell region of interest, that makes chromosomal analysis by imaging flow cytometry possible (Figure 2) (Hui et al., 2018).
4 CHROMOSOME ANALYSIS BY IMAGING FLOW CYTOMETRY
The application of imaging flow cytometry to study chromosomes on cells in suspension was pioneered by Minderman et al. (2012). Termed “FISH in solution” (FISH-IS), they showed that the number of copies of chromosomes X, Y and 8 in normal leucocytes and leukemic cells could be determined using centromeric FISH probes. The method they used required initial cell fixation, exposure to high temperature to denature double stranded DNA, followed by hybridization of the FISH probes to the complementary DNA sequence. DAPI stain was applied to highlight cell nuclei and a minimum of 20,000 cells acquired in the imaging flow cytometer. IDEAS software was used for evaluation of single cells in focus with bound FISH probe based on brightfield image analysis and probe fluorescence intensity. Masking of FISH signals (“spots”) was achieved by utilizing the software image analysis features to identify the centromeric FISH probe fluorescence signals. Accuracy of the masks applied was confirmed by manually visualizing the generated images from the digital cameras. The software was then able to enumerate the number of probes in each cell providing the overall spot count profiles (Figure 3). The capacity to interrogate FISH probe spots based on combined morphology and fluorescence features on tens of thousands of cells improved the sensitivity and utility of this technique.
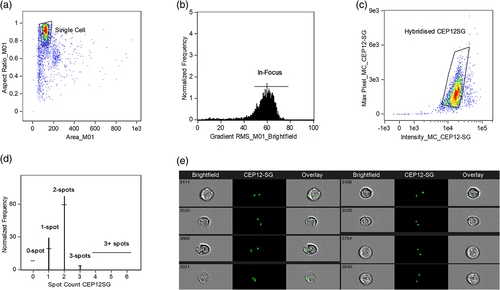
Source: Adapted from Minderman et al., 2012
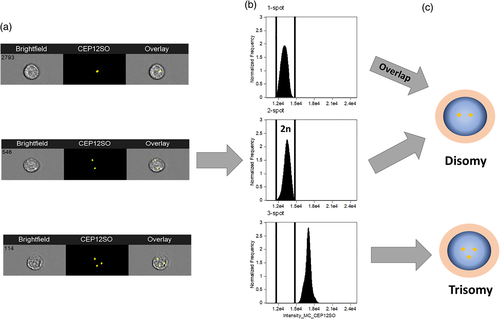
Minderman et al. then used fluorescence intensity measurements to take into account heterogeneity of FISH hybridization (e.g., due to incomplete saturation or asynchronous replication of sequences) and the possibility of overlapping FISH spots. This was to distinguish between the possibility of two aligned FISH signals being misinterpreted as monosomy due to the presence of a single spot on the imagery. This verification method was shown to be important as it was estimated there is at least 12.4% chance of overlapping signals for each FISH probe; further, this would increase with additional probes used (Minderman et al., 2012). Two overlapping spots gave twice the mean fluorescence intensity of a “real” single FISH signal (Figure 4). Cases of trisomy with a true third FISH signal would have 1.5× the fluorescence of true disomy (2n). By assessing the mean fluorescence intensity of FISH “spots”, the authors were able to detect monosomy, disomy, and trisomy for the chromosomes studied. This was accurate in detecting genetic aberrations with a sensitivity of 1% based on 20,000 cells analyzed and had high correlation (R2 = 0.99) with standard FISH (Minderman et al., 2012). However, at levels below 1% there was an increase in false positive rates requiring for the manual inspection of the selected population.
5 CHROMOSOME ANALYSIS IN IMMUNOPHENOTYPED CELLS
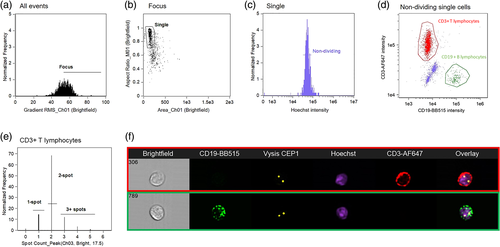
The next development was the incorporation of immunophenotyping to identify the cell of interest. This technique, developed by Fuller et al., was shown to increase precision by ensuring chromosomes were only assessed in cells with a specific antigenic profile (Fuller et al., 2016). Termed “Immuno-S-FISH” this method required no prior cell separation or isolation and could detect FISH probe signals in specific cell types identified by the phenotypic profile. Evaluated on normal lymphocytes, this method could detect chromosome 1, using a centromeric probe, in >90% of normal T and B lymphocytes identified with CD3 and CD19 antibodies (Figure 5).
Source: Data was based on 20,000 cells analyzed. Adapted from Fuller et al., 2016
In this seminal work, Fuller et al. demonstrated the importance of preserving cellular integrity, including the surface membrane antigens, while ensuring FISH probes could access nuclear material and hybridize to denatured DNA. This posed several challenges as optimal FISH conditions are damaging to cell structure, antigens and the antibody fluorescent signal. They highlighted the importance of cell phenotyping with fluorescent-conjugated antibodies being performed prior to FISH; reversing the order resulted in increased nonspecific antibody binding to the cells. To preserve the antibody-fluorochrome conjugate, an amine cross-linker was required to stabilize the antibody–antigen complex on the cell membrane prior to commencing the FISH process. The amine cross-linker bis(sulfosuccinimidyl)suberate (BS3) is a noncleavable, membrane-impermeable and water-soluble cross-linking agent. BS3 is commonly used to cross-link cell surface proteins in order to identify near-neighbor protein relationship and ligand-receptor interaction by stabilizing the protein-to-protein complexes (Suchanek et al., 2005). Fuller et al. found that when used after antibody staining BS3 can preserve immunophenotyping throughout acid incubation and the high temperatures required for FISH (Fuller et al., 2016). A post-phenotyping fixation step (4% formaldehyde) was then used to preserve cellular integrity while retaining sufficient immunophenotyping fluorescent signal for detection. Alternate fixatives, such as Carnoy's solution, were found to be less suitable as they caused shedding of some antibodies and/or quenching of the fluorophore signal.
FISH probe hybridization was performed after antibody labeling, cross-linking and fixation; the method used bore similarities to those developed by others. However, Fuller et al. found that an additional acid incubation step was necessary to promote denaturation of double-stranded DNA and allow probe hybridization to the complementary DNA sequence. Their protocol also included a cell membrane permeabilisation step with a nonionic detergent to allow probes to enter the DNA with minimal cellular structure disruption. FISH probes were then added and after overnight hybridization, samples were stained with a nuclear marker prior to analysis.
To evaluate the chromosomes, Fuller et al. gated in focus, single and nondividing cells (based on nuclear marker intensity) and used the “Spot Count” feature to count the number of probe “spots” per cell using the FISH probe fluorescence mask. This calculation analyses the pixel intensity of the probe spots to cell background ratio, the radius of the spots and the intensity of the probe fluorescence (Figure 5). The spot count profile was then verified and, by fluorescence intensity adjustment accounting for overlapping spots (as described by Minderman) and nonspecifically hybridized probes, an adjusted spot count result was generated. The data was presented as the number and percent of all cells, and those with the phenotype of interest, with the specific chromosomal change.
This principle of simultaneous assessment of chromosomes and phenotype had already been established in tissue pathology. “FICTION” (“fluorescence immunophenotyping and interphase cytogenetics as a tool for the investigation of neoplasms”) and immunoFISH are both methods used on sections of fixed tissue to assess FISH in immunophenotyped cells (Mattsson et al., 2007; Weber-Matthiesen et al., 1992, 1993, 1995). Immuno-S-FISH also assesses chromosomes by FISH in immunophenotyped cells. The differences are the starting material is fresh (i.e., blood samples), many thousand intact whole cells are evaluated, the analysis is automated on an imaging flow cytometer and numerical data as well as imagery is generated.
5.1 Applications to blood cancers
Source: Data was based on 20,000 cells analyzed. Adapted from Hui et al., 2019
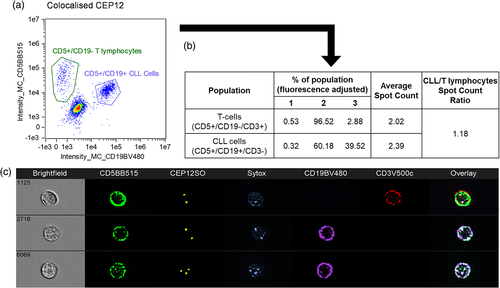
The 17p locus, a critical region of study because of TP53, has also been assessed by immuno-flowFISH in both CLL (blood) and plasma cell myeloma (bone marrow) (Erber et al., 2020; Hui et al., 2019). Deletion of 17p is predictive of poor prognosis in both malignancies, and treatment decisions are based on the presence of this chromosomal defects (Avet-Loiseau et al., 2007; Döhner et al., 2000). For CLL, analysis was with a 3-antibody panel and 2 FISH probes (17p12 and chromosome 17 centromeric probe [CEP17]) (Figure 7). The FISH probe fluorophores used were SpectrumGreen and OrangeRed, with emission spectra of 524 and 565 nm, respectively. Careful choice of antibody fluorophores was required as the standard channels were utilized by the FISH probes, leaving only channels suitable to tandem dyes available. Most protein-based tandem dyes (e.g., PECy5, PECy7, and APCCy7) were found to be unsuitable for Immuno-S-FISH and Immuno-flowFISH analysis. In contrast, synthetic polymer-based tandem dyes (e.g., BV480, BV605) were able to withstand the low pH and high temperature conditions required for optimal FISH hybridization (Fuller et al., 2016; Hui et al., 2018). For plasma cell myeloma, two antibodies (CD38 and CD138) were used to identify plasma cells (Figure 7). CEP17 was included as an internal numeric control to ensure diploid chromosome 17 signals. This would mean that if there was only one spot for the “test” probe (17p12), that it was due to loss of this region of the chromosome and not monosomy.
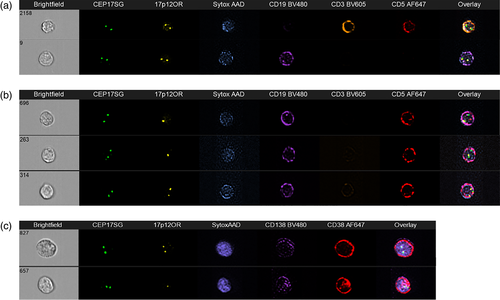
Del(17p) could be detected in both CLL and myeloma plasma cells, as identified by their phenotype (Figure 7). The number and percent CLL cells with del(17p) ranged from 270–35,441 cells (2–35% of CLL cells) with a limit of detection of 0.4% of all cells in the sample. For myeloma, del(17p) was detected in 2340–2964 plasma cells (13–19%), giving a limit of detection of 1% of cells (Erber et al., 2020). These impressive data suggest that with further validation, immuno-flowFISH may prove to be more sensitive than traditional FISH-on-a-slide which has a reportable positive cut-off of 5–7%. If confirmed, immuno-flowFISH could potentially be used to monitor residual disease following therapy (e.g., autologous transplantation for myeloma).
5.2 Application to telomeres
Telomere length has also been assessed by imaging flow cytometry by integrating FISH with antibody-based immunophenotyping for a range of other cell types such as HeLa cells and normal blood mononuclear cells (Figure 8) (Sanderson & Simon, 2017). These authors used a number of fluorophore-conjugated antibodies (including CD27, CD19, CD45RA, IgM, CD38, and CD8). Telomere detection was carried out by hybridizing unfixed cells with PNA-FITC probe and a total of 30,000 events were collected on ImageStream 100 or ImageStreamX markII flow cytometers. Similar to the previously described analyses, specific cell types identified by their phenotype were analyzed for telomere expression. The study by Sanderson et al. emphasized that image-based analysis of telomere length can provide a better correlation of telomeric length with age than fluorescence intensity measurements alone. The addition of immunophenotyping now also enables telomere length to be determined by using a spot count per cell rather than traditional intensity only.

Source: Clynick et al., unpublished data
6 ATTRIBUTES OF IMAGING FLOW CYTOMETRY FOR CHROMOSOMAL ANALYSIS
- the high number of cells that can be analyzed increasing sensitivity
- there is no need for any pre-analytical cell isolation or sorting
- high-throughput and high quantity analysis of whole cells in suspension
- the power of digital images which allows simultaneous visualization of cells, their nucleus, phenotype and chromosomal signal at high throughput with single cell resolution
- depth of field enabling the cell and FISH spot count images to be in focus
- improved specificity through the positive identification of cells of interest based on immunophenotype
FISH analysis by imaging flow cytometry does not require pre-analytical cell isolation as cell identification comes from standard immunophenotypic gating. This increases the specificity over traditional FISH which only assesses cell nuclei, and not specific cell populations. This has limitations when the cell of interest (e.g., neoplastic cell) makes up only a sub-population of the sample. Hence the chromosomal analysis could be of any cell type in the sample. This is addressed with some FISH applications, such as plasma cell myeloma where CD138-positive cell isolation is commonly performed.
Test sensitivity is also greatly increased as tens of thousands of cells are analyzed in a single test; this is in comparison with standard FISH which typically only assesses nuclei of 200 cells. To date, the limit of detection of immuno-flowFISH has been reported as being able to detect a chromosomal defect when present in less than 1% of cells in the sample (Erber et al., 2020; Hui et al., 2019). Acquiring data for thousands of cells and restricting analysis of cytogenetic features to cells with a specific phenotype, shows potential of this approach for chromosomal analysis for detecting rare events.
7 THE FUTURE OF IMAGING FLOW CYTOMETRY FOR CHROMOSOMAL ANALYSIS
Imaging flow cytometry for chromosomal analysis is still in its infancy and its precise place in the repertoire of cytogenomic testing for human disease requires further work. There are technical and analytical issues that are still to be assessed prior to the method being ready for clinical applications. These include the need for beta testing in multiple sites and clearing regulatory requirement for in vitro diagnostic products from the relevant governing body such as the U.S Food and Drug Administration. Testing to date has been restricted to blood and bone marrow which require minimal treatment for single cell analysis. Further work is required to evaluate the applicability of the technique to other cytological specimens (e.g., fine needle aspirate) and cells extracted from fresh solid tumor samples. Other aspects in relation to the sample that remain to be validated include the optimal anticoagulant and time from collection to analysis. Published literature has suggested that different anticoagulants and the length of time blood was in the anticoagulant may have a detrimental effect on the expression of some surface markers (Diks et al., 2019; Dorwal et al., 2014; Stetler-Stevenson et al., 2016). Lastly evaluation of the technique on larger patient cohorts and across multiple laboratories is required to confirm the clinical utility of this approach (Hui et al., 2019; Minderman et al., 2012).
Other antibodies and fluorophores need to be studied, specifically to determine whether they can withstand the low pH environment and high heat necessary for an immunophenotyping FISH technique. The effect of permeabilisation of cells for intracellular staining (e.g., TdT or myeloperoxidase, or cell cycle Ki67) prior to stabilization and fixation is unknown and will require additional investigation. Commonly used bright immunophenotyping fluorophores such as PE, APC and their tandem conjugates lose their fluorescence after incubation in acid and hybridization at high temperature (Fuller et al., 2016). FISH probes have been designed for fluorescent microscopy and are generally produced with a limited rage of fluorophores such as FITC and TexasRed. Use of these fluorophores for the FISH component has placed restrictions on the fluorophores that can be used for the immunophenotyping. New synthetic-based polymer fluorophores provide a promising avenue to expand the number of parameters with this technique providing a more stable base structure and tandem conjugation chemistry in low pH and high temperature protocols (Panchuk-Voloshina et al., 1999; Roura et al., 2016; Skotheim & Reynolds, 2007).
Imaging flow cytometry devices have significantly lower throughput than standard flow cytometers. The Amnis ImageStreamX markII, for example, has a throughput speed of up to 6000 cells/s, but with chromosomal analysis requires acquisition speeds of <1000 cells/s for highest sensitivity image analysis. This is ten times slower than that of standard flow cytometers such as the BD FACSCantoII or Beckman Coulter Aquios CL. However for an imaging flow cytometer which utilizes a CCD camera, there is a limitation in its maximum speed due to the manner in which fluorescence photons are collected and transferred without electron multiplication (Han et al., 2016). Although acknowledged, to date, this lower acquisition speed has not been recognized as a problem.
Advances in instrumentation are likely to occur over the coming years as the technology becomes more widely adopted. This could include auto-sampling and image-based sorting of specific chromosomal populations for further analysis. Prototype instruments such as the ENMA by CYBO, enable real-time cell sorting based on morphological features at a throughput of 100 cells/s (Isozaki et al., 2020; Nitta et al., 2018).
Advances in data analysis software are ongoing with the release of IDEAS v6.3 and Amnis Artificial Intelligence (AI) Image Analysis Software packages (Probst et al., 2020; Pugsley & Kong, 2020). These should enhance the sensitivity for detection of dim FISH spots, as seen with small probes, and reduce operator input into the analysis. The future could see machine learning software providing automated analysis of cytogenetic aberrations. This would increase automation, provide objective analysis and standardize interpretation. Machine learning could also add a new dimension to data handling and reveal FISH images as well as morphological profiles currently undetectable by eye or the software (Doan et al., 2018). This is exemplified in telomeric FISH where image-based analysis is already capable of achieving better separation of cells with different telomere lengths compared to operator selected fluorescence intensity measurement.
In summary, chromosomal analysis by imaging flow cytometry is showing enormous promise and has already been described as a “new frontier” in cytogenomics of cytological specimen (Weissleder & Lee, 2020). The ability to identify chromosomes and abnormalities in specific cell types in human disease has been a significant breakthrough. The techniques developed using imaging flow cytometry are likely to become a valuable addition to chromosome analysis in a range of disorders at diagnosis, for assessing low level circulating cells with a chromosomal defect and detecting residual disease following therapy. The technology may also open avenues for the detection of emergent and drug-resistant new clones following therapy.