Integrating proteomic and clinical data to discriminate major psychiatric disorders: Applications for major depressive disorder, bipolar disorder, and schizophrenia
Dongyoon Shin and Sang Jin Rhee contributed equally to this work as co-first authors.
Dear editor,
We report that integrating proteomic and clinical data enables objective differentiation between major depressive disorder (MDD), bipolar disorder (BD), and schizophrenia (SCZ). These major psychiatric disorders are associated with mortality and life-long disability.1 However, objective discrimination of these disorders remains a formidable challenge. Thus, this study aimed to distinguish MDD, BD, and SCZ by integrating targeted/untargeted proteomic data obtained from liquid chromatography-mass spectrometry (LC-MS) and clinical data.
The entire design of the current study is illustrated in Figure S1, and detailed information of the following methods is described in Supporting Information. The study included 675 subjects [171 SCZ, 170 BD, 174 MDD, and 160 healthy controls (HC)], aged 19 to 65 years, and proteomic analyses was performed from each plasma sample. After the final quantifiable 642 peptides for MDD, BD, SCZ, and HC were determined (Figure S2), LC-multiple reaction monitoring (MRM)-MS was performed on individual plasma samples, followed by LC-high resolution MS-based proteomic profiling on pooled plasma samples (Figure S3A,B). Logarithmic transformation was performed on the LC-MRM-MS data for the stable 588 peptides, followed by batch effect correction (Figure S4A,B). The 515 patients were divided into training, validation, and independent test sets (6:2:2). There were significant differences in demographics, medication use, and clinical features between groups (Tables S1–S4). Therefore, peptides that were significant with demographics, medication use, and chronicity of disease/medication, and not with disease types were excluded by ANCOVA, for each pairwise comparisons between groups, in the training sets. Furthermore, peptides with multicollinearity were excluded, resulting in 23, 29, and 30 proteomic candidate features (proteins) for differentiating MDD versus BD, MDD versus SCZ, and BD versus SCZ, respectively (Table S5). These proteins showed consistent expression level patterns across disease types, low inter-correlation with covariates (Figure S5A–C), and low interdependence between each other (Figure S6A–C).
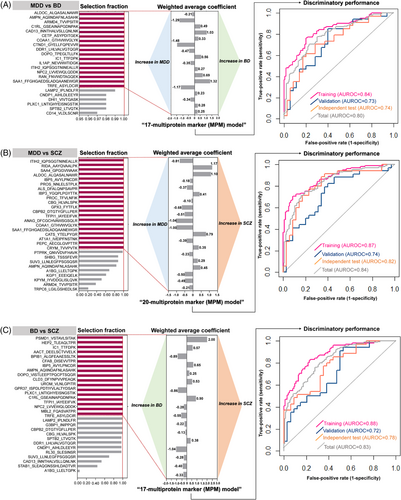
Multiprotein-marker (MPM) models were constructed by LASSO (least absolute shrinkage and selection operator) with 100-repeated 5-fold cross-validations, additionally with feature extraction and weighted model averaging,2 in the training sets (Table S6 and Figure S7A–C). After evaluating model performances in the validation sets based on selection fractions, the simplest models (selection fraction = 1) were selected, as the performances only mildly increased with selection fraction ≥.8 (Figure 1A–C; Figure S8A–C). The final MPM models for differentiating MDD versus BD, MDD versus SCZ, and BD versus SCZ consisted of 17, 20, and 17 proteins, and the AUROC values were .74, .82, and .78, respectively in the independent test sets (Figure 1A–C). Due to different analytical methods, the corresponding proteins differed with our previous study for discriminating MDD versus BD except for ITIH2.2 However, the current models were constructed with larger samples and expanded targets, and validated in an independent set; implying greater reproducibility. For each MPM model, the direction of each average coefficient corresponded to the alteration in expression (fold-change) (Figure 1A–C). The MPM models had similar performances in differentiating MDD, BD, and SCZ with different subgroups (Figure S9A–F), all of the proteins were less influenced by psychotropic medication (Figure S10), and only few proteins showed associations with specific symptoms (Table S7). Particularly for BD, the proteins were unrelated to depressive or manic symptoms. The mass spectral information of proteins in the MPM models is presented in Table S8, and the alterations in the expression of the proteins is presented in Table S9 and Figure S11. There was no protein that overlapped in all three MPM models.
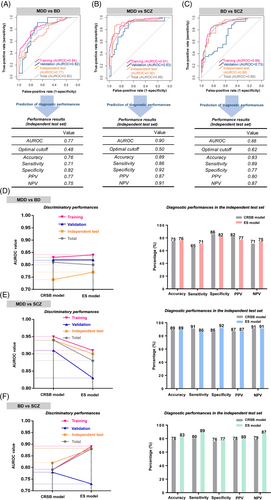
Symptom checklist-based (SCLB) models were constructed by generalized linear models (GLMs). The models with the highest discriminatory power considering all combinations of the Symptom Checklist-90-Revised (SCL-90-R)3 dimensions, were selected (Table S10 and Figure S12A–C). Then, ensemble (ES) models were constructed by combining MPM and SCLB models through the stacking ensemble strategy.4 At last, clinician rater score-based (CRSB) models were constructed by GLMs, combining the total scores of the Brief Psychiatric Rating Scale (BPRS),5 Hamilton Anxiety Scale (HAM-A),6 Montgomery–Asberg Depression Rating Scale (MADRS),7 and Young Mania Rating Scale (YMRS)8 (Table S10). The discriminatory and diagnostic performances of the ES and CRSB models were overall comparable (Figure 2A–F and Figure S13A–C).
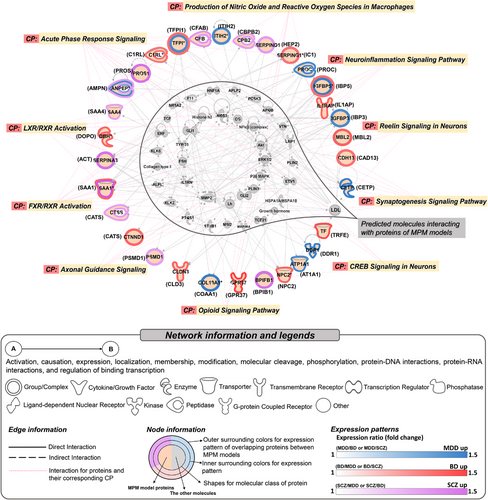
For 43 proteins from all MPM models, an integrated network comprising up to two networks was predicted (Table S11 and Figure 3). Diseases/functions associated with the network included cellular movement (p = 7.87 × 10-21–1.61 × 10-7), cell-to-cell signalling and interaction (p = 9.14 × 10-10–1.61 × 10-7), immune cell trafficking (p = 2.3 × 10-12–1.3 × 10-7), neurological disease (p = 7.47 × 10-12–8.17 × 10-8), and psychological disorder (p = 6.09 × 10-12–3.89 × 10-2). Furthermore, the network was related to significant canonical pathways including complement and coagulation cascade dysregulation, neural signalling, and oxidative and inflammatory pathways, which has been replicated in previous studies (Figure 3).2, 9 Especially, reelin signalling was a significant canonical pathway, which is known to regulate neuronal migration and synaptogenesis in the brain, and has been linked to MDD, BD, and SCZ.10
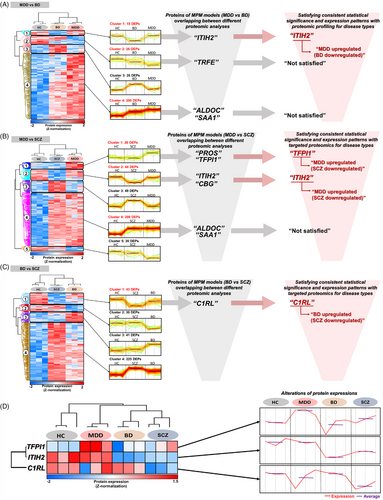
Through proteomic profiling, analytically stable plasma proteome (902 quantified proteins) were constructed in each pooled sample for the four groups (Table S12 and Figure S14A–D). Subsequently, 267 differentially expressed proteins (DEPs) with 4 clusters, 347 DEPs with 5 clusters, and 339 DEPs with 4 clusters were determined between MDD versus BD versus HC, MDD versus SCZ versus HC, and BD versus SCZ versus HC, respectively (Table S13). The DEPs that had consistent significance and expression patterns in both targeted proteomics and proteomic profiling were as follows; ITIH2 for the MPM model of MDD versus BD, TFPI1 and ITIH2 for MDD versus SCZ, and C1RL for BD versus SCZ. (Table S14; Figure 4A–C). The overall alterations in abundance of these 3 DEPs in each group is presented in Figure 4D. Further discussion of these key proteins is described in Supporting Information.
Our study has its limitations regarding sample size, the possibility of other potential confounders and proteomic targets including duration of the current episode, and medication dosage/duration, the cross-sectional study design, biological interpretations of proteins in peripheral blood, and limited practicalness to clinical practice as a diagnostic tool (Supporting Information). Nevertheless, we demonstrated the viability of integrating proteomic and clinical data in discriminating MDD, BD, and SCZ. We developed MPM and ES models for each pairwise comparison of groups, reporting their potential in differentiating and diagnosing these disorders.
ACKNOWLEDGEMENTS
This work was supported by the Industrial Strategic Technology Development Program, funded by the Ministry of Trade, Industry, and Energy (MOTIE, Korea) (No.20000134), and the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (No.HL19C0020 and HI19C1132). This study was also supported by a grant from Seoul National University Hospital (2022). Injoon Yeo was supported by BK21 FOUR, funded by the National Research Foundation of Korea. Dongyoon Shin and Jihyeon Lee received a scholarship from the BK21-Plus Education Program, provided by the National Research Foundation of Korea.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.




