Genetics in liver diseases: From diagnostics to precise therapy
Potential conflict of interest: Nothing to report.
Abstract
Watch a video presentation of this article
Abbreviations
-
- ABC
-
- ATP-binding cassette
-
- AP
-
- alkaline phosphatase
-
- ALT
-
- alanine aminotransferase
-
- BRIC
-
- benign recurrent intrahepatic cholestasis
-
- BSEP
-
- bile salt export pump
-
- CYP
-
- cytochrome P450
-
- GWAS
-
- genome-wide association study
-
- HCC
-
- hepatocellular carcinoma
-
- HLA
-
- human leukocyte antigen
-
- ICP
-
- intrahepatic cholestasis of pregnancy
-
- LT
-
- orthotopic liver transplantation
-
- MDR
-
- multidrug resistance
-
- NASH
-
- nonalcoholic steatohepatitis
-
- OR
-
- odds ratio
-
- p
-
- protein
-
- PEBD
-
- partial external biliary diversion
-
- Pi
-
- protease inhibitor
-
- SBP
-
- spontaneous bacterial peritonitis
-
- UDCA
-
- ursodeoxycholic acid
Diagnosis of Monogenic Liver Diseases and Genetic Susceptibility
The past two decades have markedly broadened our understanding of the genetics of liver diseases, starting with the cloning of the Wilson's disease gene ATP7B in 1993.1 This discovery set the stage for diagnostic tests of monogenic diseases, where mutations in single genes cause liver diseases. Examples are hereditary hemochromatosis type 1 (HFE gene mutation p.C282Y), α1-antitrypsin deficiency (SERPINA1 mutations p.E264V and p.E342K), and Wilson's disease. Most patients with hemochromatosis and α1-antitrypsin deficiency carry the listed mutations, which can be easily genotyped (e.g., by PCR-based allelic discrimination assays), but the genetic diagnosis of Wilson's disease requires sequencing of the whole ATP7B gene (Table 1).
| Liver Disease/Phenotype | Gene/Locus | Genetic Tests |
|---|---|---|
| Diagnostic tests | ||
| Hemochromatosis type 1 | HFE | p.C282Y |
| Wilson's disease | ATP7B | Sequencing |
| α1-Antitrypsin deficiency | SERPINA1 |
p.E342K (PiZ) p.E264V (PiS) |
| Familial intrahepatic cholestasis | ATP8B1, ABCB11, ABCB4, TJP2 | Sequencing |
| Gilbert syndrome | UGT1A1 | Promoter variant A(TA)7TAA |
| Crigler-Najjar syndrome | UGT1A1 | Sequencing |
| Conjugated hyperbilirubinemia | ABCC2, SLCO1B1, SLCO1B3 | Sequencing |
| Mechanistic tests | ||
| Fatty liver disease |
PNPLA3 TM6SF2 MBOAT7 |
p.I148M (OR 3.5) p.E167K (OR 1.6) rs641738 (OR 1.2) |
| Therapeutic tests | ||
| Hepatitis C virus infection |
IL28B ITPA |
rs12979860 (OR 7.3 for response to therapy with interferon-α) p.P32T, g.8838 A>C |
| Irinotecan toxicity | UGT1A1 | Promoter variant A(TA)7TAA (OR 11 for diarrhea) |
| Statin toxicity | SLCO1B1 | p.V174A (OR 4.5 for myopathy) |
| Pharmacogenetic tests |
CYP2C19 CYP2D6 |
|
| Drug-induced liver injury | HLA | |
- Relative risk for mechanistic and therapeutic tests (where available) is given in brackets.
- Abbreviations: CYP, cytochrome P450; HLA, human leukocyte antigen; OR, odds ratio; p, protein; Pi, protease inhibitor.
In contrast with monogenic diseases, a large ensemble of gene variants affect polygenic diseases, such as gallstone disease or liver cirrhosis. Whereas mutations are defined as DNA variants occurring with a frequency <0.1% in a population, polymorphisms are more common but confer often only incrementally small genetic susceptibility.1 The first genome-wide association study (GWAS) that screened a large number of single nucleotide polymorphisms (500,000) identified a common variant (p.D19H) of the sterol transporter ABCG5/G8 as a susceptibility factor for gallstones.2 In GWAS, a single nucleotide polymorphism in the IL28B gene encoding interferon-λ3 was associated with the response to therapy with interferon-α in chronic hepatitis C virus infection.3 Although its clinical relevance has vanished with direct antiviral agents, it improved understanding of disease susceptibility, because genetic variation at this locus is also associated with spontaneous virus clearance.
Novel Mechanistic Insights and Disease Classifications
Progressive familial intrahepatic cholestasis represents a group of hereditary cholestasis syndromes (Table 2) that have been defined and classified by genetics.4 The diseases are caused by mutations of the genes encoding the hepatocanalicular lipid transporters for phosphatidylserine (ATP8B1), bile acids (ABCB11), or phosphatidylcholine (ABCB4), respectively. For diagnosis, sequence analysis of the complete genes is recommended (Table 1). Recently, ABCB4 variants have been classified as follows: (I) nonsense variants, (II) missense variants that affect maturation, (III) activity, or (IV) stability. Class II mutations might be corrected by chaperones such as 4-phenylbutyrate.5
| Type | ||||
|---|---|---|---|---|
| 1 (Byler syndrome) | 2 | 3 | 4 | |
| Gene | ATP8B1 (FIC1) | ABCB11 (BSEP) | ABCB4 (MDR3) | TJP2 |
| Transport | Phosphatidylserine | Bile salts | Phosphatidylcholine | Tight junction protein |
| Phenotypes | Biliary cirrhosis | Neonatal giant cell hepatitis Biliary cirrhosis | Biliary cirrhosis with ductular proliferation | Biliary cirrhosis in childhood |
| BRIC type 1 | BRIC type 2 | Gallstones (LPAC) | Familial hypercholanemia | |
| Extrahepatic manifestations: malabsorption, pancreatitis, deafness, pneumonia | ICP Gallstones Drug-induced cholestasis HCC | ICP Cholangiocarcinoma, HCC | Deafness HCC | |
| Hotspot mutations | p.I661T in 45% of BRIC patients | p.E297G and p.D482G in 60% of European patients | None | |
| Laboratory tests |
Low γ-GT AP/ALT ↑ |
Low γ-GT ALT/AP ↑ | γ-GT ↑ | Low γ-GT |
| Therapy | LT PEBD | PEBD LT | UDCA LT | LT |
- Abbreviations: ABC, ATP-binding cassette; AP, alkaline phosphatase; ALT, alanine aminotransferase; BRIC, benign recurrent intrahepatic cholestasis; BSEP, bile salt export pump; GT, glutamyl transpeptidase; HCC, hepatocellular carcinoma; ICP, intrahepatic cholestasis of pregnancy; LPAC, low phospholipid-associated cholelithiasis; LT, orthotopic liver transplantation; MDR, multidrug resistance; PEBD, partial external biliary diversion; UDCA, ursodeoxycholic acid.
The most common hereditary unconjugated hyperbilirubinemia is Gilbert syndrome, which is caused by a promoter variant of the UGT1A1 gene encoding uridine diphosphoglucuronate glucuronosyltransferase 1 (Table 1). This variant is also associated with gallstones and leads to decreased glucuronidation of an active irinotecan metabolite.4 Thus, a reduced starting dose of irinotecan should be considered for homozygous carriers. More severe mutations of the UGT1A1 gene cause Crigler-Najjar syndrome; type 1 in children is due to lack of enzyme activity, and type 2 is caused by partial inactivation, hence patients may also present in adulthood, and the enzyme can be induced with phenobarbital.4
Two rare types of hereditary conjugated hyperbilirubinemia are caused by mutations of hepatic transport proteins (Table 1). Deficiency of the hepatocanalicular bilirubin transporter ABCC2 causes Dubin-Johnson syndrome. Disruption of hepatic reuptake and “hepatocyte hopping” of bilirubin glucuronide (due to coexisting SLCO1B1 and SLCO1B3 mutations) underlies Rotor syndrome. Of note, the common SLCO1B1 mutation p.V174A also confers risk of statin-associated myopathy.
The development of fatty liver disease is associated with overnutrition and/or alcohol consumption (Fig. 1). In GWAS, a common variant of the PNPLA3 gene encoding the triglyceride hydrolase adiponutrin has been associated with both nonalcoholic6 and alcoholic fatty liver disease.7 The PNPLA3 variant p.I148M is carried by almost 50% of Europeans and leads to a 3-fold increased risk for steatosis, nonalcoholic steatohepatitis, cirrhosis, and hepatocellular cancer, pointing to shared critical mechanisms in nonalcoholic and alcoholic liver diseases.1 Two other common genetic risk factors affect hepatic lipid metabolism: Variants of the TM6SF28 and the MBOAT7 genes9 impair the lipidation of very low density lipoproteins and phosphatidylinositol acyl-chain remodeling, respectively (Table 1). Because carriers of the PNPLA3 p.I148M and the TM6SF2 p.E167K variants have consistently higher liver fat contents and increased risk for NASH, genotyping is recommended in selected patients,10 for example, lean and young individuals.11 Because the PNPLA3 variant has been associated with an increased HCC risk, it might also contribute to patient-risk stratification for tailored HCC surveillance in NAFLD,10 that is, at least very strict adherence to 6-month surveillance intervals.
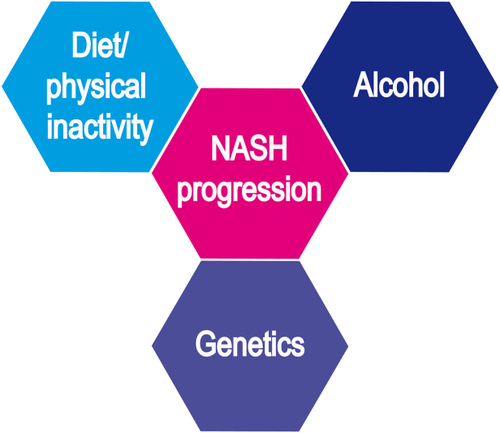
Schematic illustration of risk factors for fatty liver disease. Together with environmental factors such as diet, physical inactivity, and alcohol, genetic risk factors predispose to fatty liver phenotypes. NASH, nonalcoholic steatohepatitis.
Patients with liver cirrhosis are at increased risk for bacterial infections. Bacteria are recognized by membrane-bound Toll-like and intracellular Nod-like receptors, including NOD2, which senses the cell wall component muramyl dipeptide of gram-negative bacteria and leads to the activation of nuclear factor-κB and the release of antimicrobial peptides. In patients with cirrhosis and ascites, Appenrodt and colleagues12 demonstrated an association between spontaneous bacterial peritonitits and NOD2 variants that also predispose to Crohn's disease. In addition, SBP has been reported to be more frequent in patients with cirrhosis who carry risk variants of the Toll-like receptor 2 receptor, which recognizes triacylated lipoprotein from gram-positive bacteria.12 Common gene variants linked to pathological bacterial translocation might thus represent genetic risk factors for SBP and are currently being tested in clinical trials.
Genetic Tests in Clinical Practice
These examples illustrate how genetic testing may help clinicians and patients to diagnose and understand liver diseases (Table 1). After informed consent, the laboratory needs ethylene diamine tetraacetic acid–anticoagulated blood, which can be sent at ambient temperature by regular mail. Costs for sequence analyses are decreasing rapidly. Hence, genetic testing of specific cytochrome P450 (CYP) polymorphisms (e.g., CYP2D6, CYP2C19), which account for the most frequent variations in phase I metabolism of drugs, has also been included in drug labels (http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm). HLA testing can serve to assess causality in individual cases of drug-induced liver injury, and because of the high negative predictive value, HLA genotypes can exclude immune-mediated liver injury caused by specific drugs.5 For patients with unexplained liver diseases, exome sequencing of selected genes, whole-exome sequencing of all genes, or even whole-genome sequencing is now feasible, as demonstrated in children with acute liver failure of indeterminate etiology, atypical iron disorders, or peculiar cholestatic diseases.1, 5