LncRNA FGD5-AS1 potentiates autophagy-associated doxorubicin resistance by regulating the miR-154-5p/WNT5A axis in osteosarcoma
Abstract
Osteosarcoma is prevalent in children and adolescent. The oncogenic function of long-chain noncoding RNA (lncRNA) FGD5 antisense RNA 1 (FGD5-AS1) has been reported. However, the function of FGD5-AS1 in doxorubicin-resistance in osteosarcoma remains to be illucidated. Quantitative real-time PCR (qRT-PCR) and western blot analysis (WB) were used to measure the expression of FGD5-AS1, miR-154-5p, WNT5A and autophagy proteins. MTT assay was used to assess cell viability and transwell assay was performed to evaluate migration. A nude mouse xenograft model was developed to verify the function of FGD5-AS1 in vivo. FGD5-AS1 was upregulated in doxorubicin-resistant (DXR) osteosarcoma cells. Knockdown of FGD5-AS1 suppressed osteosarcoma cell proliferation, migration, and autophagy. FGD5-AS1 upregulated WNT5A expression via sponging miR-154-5p. Furthermore, FGD5-AS1 enhanced osteosarcoma cell chemotherapy resistance through upregulation of WNT5A by inhibiting miR-154-5p. Suppression of FGD5-AS1 significantly suppressed tumor growth in nude mice. FGD5-AS1 may promote chemoresistance through WNT5A-induced autophagy by sponging miR-154-5p in osteosarcoma cells.
1 INTRODUCTION
Osteosarcoma is a prevalent malignant bone disease originating in long bones and is characterized by a high tendency of metastasis. It is common in children and adolescents (Arndt et al., 2012; Ottaviani & Jaffe, 2009). Due to the rapid development of chemotherapy, the survival of osteosarcoma patients has been improved and the recurrence rate of local osteosarcoma has been significantly reduced (Kempf-Bielack et al., 2005). However, the resistance of osteosarcoma cells to chemotherapy drugs leading to tumor metastasis and recurrence is the main reason for the failure of osteosarcoma treatment (Hattinger et al., 2015; Kim & Lee, 2017; Shah et al., 2016). Therefore, the mechanism of osteosarcoma resistance to chemotherapy drugs should be better investigated. Additionally, previous study revealed that autophagy could resist doxorubicin-induced apoptosis in osteosarcoma (Zhao et al., 2014). Therefore, enhanced autophagy promotes doxorubicin resistance in osteosarcoma cells.
Recent publications have shown that lncRNA has a crucial role in regulating the occurrence, invasion, and metastasis of osteosarcoma due to its extensive mechanism of action (Huynh et al., 2017; Z. Li et al., 2016; Wang et al., 2020). LncRNA is a group of transcribed products with a length greater than 200 nucleotides (Charles Richard & Eichhorn, 2018). The behavior pattern of lncRNA is very complicated in osteosarcoma and its function was usually mediated by downstream miRNA and mRNA (Chen et al., 2017). The function of lncRNA FGD5-AS1 has been investigated in multiple cancer types (Fan et al., 2020; Gao et al., 2020; Ge et al., 2020). The expression of FGD5-AS1 is elevated in nonsmall cell lung cancer (NSCLC) cells which are resistant to cisplatin. Moreover, FGD5-AS1 inhibition suppresses cell malignancy and stimulates apoptosis of cisplatin-resistant NSCLC cells (Fu et al., 2020). The function of FGD5-AS1 in osteosarcoma has not been well illustrated. Study reported that FGD5-AS1 was enriched in osteosarcoma, and its upregulation predicted poor prognosis of the patient. The study in osteosarcoma cells revealed FGD5-AS1 inhibition interrupted the viability and invasion ability of osteosarcoma cells (Song et al., 2020). Further studies are required to characterize the function of FGD5-AS1 in osteosarcoma chemoresistance.
MicroRNA (miRNA) is a kind of noncoding RNA, which is widely expressed and has important biological functions. In recent years, many studies reported the levels of miRNAs in various cancers are abnormal (Kurozumi et al., 2017; Shah et al., 2016). In addition, various studies have shown that miRNAs are involved in tumor cell proliferation, metastasis, apoptosis, and autophagy (Yamamoto & Mori, 2016; Yu et al., 2015). The latest literature links miRNAs with the resistance of cancer chemotherapy drugs (Xie et al., 2016). Therefore, targeting miRNAs to reverse drug resistance has a great potential against osteosarcoma. Previous study revealed miR-154-5p was downregulated in osteosarcoma tissues. MiR-154-5p significantly inhibited cell malignancy of MG63 cells (Tian et al., 2020). However, studies of miR-154-5p in osteosarcoma are limited.
Wnt/β-catenin signaling is critical in cancer development, migration, and treatment resistance (X. Zhang et al., 2015). However, it has recently been discovered that noncanonical Wnt signaling pathways regulated by WNT5A also play a carcinogenic or tumor suppressor role in cancer cell types. Many studies have shown that WNT5A was highly expressed in osteosarcoma and can promote the progression of osteosarcoma by promoting the proliferation, migration, and invasion of osteosarcoma cells (Yong et al., 2017; A. Zhang et al., 2017). At the same time, there are literatures showing that the Wnt pathway is related to autophagy. Wnt signaling and upregulation of β-catenin repress the level of autophagy (Jati et al., 2018). Bioinformatics found that miR-154-5p has targeted binding sites with FGD5-AS1 and WNT5A.
Based on the above research, this study hypothesizes that FGD5-AS1 may have an oncogenic function in the regulation of doxorubicin-resistance in osteosarcoma through miR-154-5p and WNT5A.
2 MATERIALS AND METHODS
2.1 Cell culture
hFOB1.19 cells (human osteoblast), HOS and MG-63 cells (osteosarcoma cells) were used here. All these cell lines were from the Shanghai Cell Collection. Cells were cultivated in Dulbecco's Modified Eagle's Medium (DMEM), containing 10% fetal bovine serum (FBS) and under 5% CO2, 37°C. Doxorubicin-resistant (DXR) cell lines were established according to previous studies (Oda et al., 2000). In brief, osteosarcoma cells were exposed to elevating dose of doxorubicin (Sigma-Aldrich). The surviving cells were selected and cultured in medium supplemented with 1 μg/ml doxorubicin.
2.2 Cell transfection
Short hairpin RNA (shRNA) for FGD5-AS1 and negative controls (sh-NC) were from GenePharma. miR-154-5p mimics, inhibitor, and respective NC were also purchased from GenePharma. PcDNA3.1-WNT5A (p-WNT5A) and lentivirus encapsulating sh-FGD5-AS1 were also purchased from GenePharma. Lipofectamine 3000 reagent (Life Technologies Corporation) was utilized for direct transfection.
2.3 RNA extraction and qRT-PCR
Total RNA extraction was performed with RNAzol RT reagent kit (Sigma-Aldrich, R4533). miRNeasy Mini Kit (Qiagen 217004) was used for miRNA extraction. MicroRNA cDNA Synthesis was carried out with MystiCq™ microRNA cDNA Synthesis Mix Kit (Sigma-Aldrich, MIRRT). SuperScript kit (Invitrogen) was used for reverse transcription for 1 μg total RNA from each sample, and after that PCR amplification was carried out with Power SYBR Green PCR Master Mix (Thermo Fisher). qPCR analysis was performed according to manufacturer protocol. SYBR Green qPCR assay was used for qPCR. The levels of RNAs were measured via the method of 2-ΔΔCt. Primers were listed below:
FGD5-AS1: forward 5′-TCCTTCCCTGTTTCAGCACT-3′,
FGD5-AS1: reverse 5′-TCTCACTCACTGGGCACTTG-3′;
miR-154-5p: forward 5′-GGCGAATCATACACGGTTGA-3′,
miR-154-5p: reverse 5′- GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAATAGG-3′;
WNT5A: forward 5′-CCAACTGGCAGGACTTTCTC-3′,
WNT5A: reverse 5′-CCTGCCAAAAACAGAGGTGT-3′;
GAPDH: forward 5′-CCAGGTGGTCTCCTCTGA-3′,
GAPDH: reverse 5′-GCTGTAGCCAAATCGTTGT-3′;
U6: forward 5′-CTCGCTTCGGCAGCACA-3′,
U6: reverse 5′-AACGCTTCACGAATTTGCGT-3′.
2.4 Dual-luciferase reporter assays
The fragment from wild-type 3′-untranslated regions (3′-UTR) of FGD5-AS1 and WNT5A containing wide type (WT) miR-154-5p binding sites (FGD5-AS1-WT, WNT5A-WT) or mutant (MUT) 3′-UTR region of FGD5-AS1 and WNT5A without miR-154-5p binding sites (FGD5-AS1-MUT, WNT5A-MUT) were inserted into Plasmid pmirGLO vectors (Promega). For determining the interaction between miR-154-5p and FGD5-AS1 or WNT5A, cells were treated with miR-154-5p mimics, mimics NC, miR-154-4p inhibitor or inhibitor NC along with WT/MUT-FGD5-AS1 or WT/MUT- WNT5A. The luciferase activity was assessed with kits (Promega) after 48 h.
2.5 RIP assay
RNA immunoprecipitation(RIP) assays were conducted with Magna RIP Kit according to instruction. Briefly, cells were harvested and lysed. Then the products were treated with magnetic beads of anti-Ago2 antibody or normal mouse IgG. The immunoprecipitated products were purified and the presence of FGD5-AS1 or miR-154-5p were evaluated by qRT-PCR.
2.6 3-(4,5-Dimethylthiazol-2-ylMTT) assay
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide(MTT) Assay Kit (Cell Proliferation) was sourced from Abcam to examine cells viability. Samples were treated and incubated at 37°C for 24 h. Grow media was discarded and serum-free media and MTT reagent were added. Samples were incubated at 37°C for 3 h. MTT Solvent was added and shaken on an orbital shaker for 15 min. The absorbance of samples was measured at OD490 nm with a MTT reader. Change as percentage of control was determined after background subtraction.
2.7 Transwell assay
Tumor cells (2 × 104) in serum-free DMEM were put onto the upper chambers. DMEM plus 10% FBS was used in the lower chamber. Remove the surface cells of the filter after 24 h of incubation. 100% methanol was used to fix and crystal violet (Sigma-Aldrich) was utilized for staining. The migration abilities were calculated with the average cell number.
2.8 Western blot
To extract protein from tissue, transfer the tissue into a mortar filled with liquid nitrogen, and use the grinding method to grind the bone tissue into powder. Then collect the powdered bone tissue into a 1.5 ml EP tube. Add protein lysis solution (200 μl lysis solution per 100 mg tissue) to the EP tube, and lyse on ice for 30 min. Transfer the lysate from the EP tube to another EP tube, 12,000 rpm, and centrifuge at 4°C for 15 min. The supernatant is the extract containing the total protein. Ice-cold protein extraction buffer with 1% protease inhibitor (Solarbio) was used to isolate total protein. NanoDrop Spectrophotometer was used to quantified the protein and the concentration was adjusted to 3 μg/μl. Ten microliters of protein samples were loaded on each lane and underwent sodium dodecyl sulfate-polyacrylamide gel electrophoresis after denaturized in boiling water bath and transferred to nitrocellulose membranes (Solarbio). Then, membranes were blocked with a 5% fat-free milk. Primary antibodies were added and incubated for overnight and secondary antibody for 2 h after that. The antibodies and the dilution used in this study were following: LC3I (1:1000; Sigma-Aldrich), LC3II (1:1000; Sigma-Aldrich), Beclin-1 (1:1000; Cell Signaling Technology), P62 (1:2000, Abcam), WNT5A (1:1000; Cell Signaling Technology), GAPDH (1:1000, Abcam) and horseradish peroxidase-conjugated IgG (ab205718, 1:20000). Enhanced chemiluminescence (Thermo Fisher) were used to developed the signals and then exposed to X-ray films. QuantityOne software (Bio-Rad) were used to measure the values and the relative protein abundance was normalized to GAPDH expression.
2.9 In vivo tumor formation
All animal experiments were conducted in accordance to institutional guidelines for the care and use of laboratory animals of the China-Japan Union Hospital of Jilin University, and met the regulations published by the National Institutes of Health Guide for Care and Use of Laboratory Animals. BALB/c nude mice were from SJA. All the mice were assigned with 1 × 107 sh-NC or sh-FGD5-AS1-treated HOS/DXR cells, respectively. Tumor sizes were identified every 7 days. The tumor volume was calculated based on the equation volume = width2 × length × 0.5. When the tumor volume reached 50 mm3 (0 days), 5.0 mg/kg of doxorubicin was injected into nude mice by tail vein injection every other day, and the nude mice were killed four weeks post injection and tumor tissues were collected. The tumors were then excised and photographed.
2.10 Statistical analysis
Data were presented as mean ± SD of triplicate experiments unless specified otherwise. Statistical significance was evaluated with GraphPad Prism (version 8.0). The differences of the data were compared via Student's t-test or one-way analysis of variance with Tukey's post-hoc test. p < .05 was considered as statistically significant.
3 RESULTS
3.1 FGD5-AS1 suppression inhibited cell malignancy in DXR osteosarcoma cells
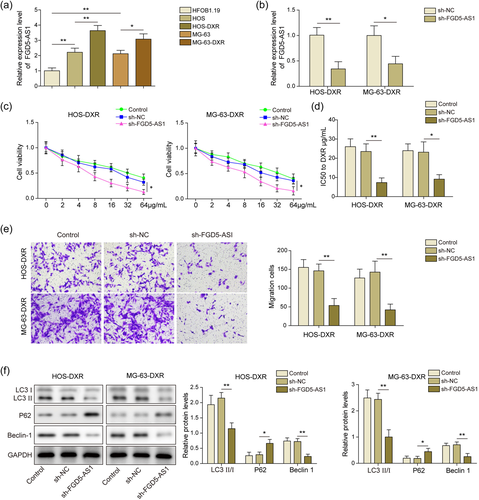
In our research, the expression of FGD5-AS1 in DXR chemoresistant and chemosensitive osteosarcoma cells was characterized. An increase of FGD5-AS1 in chemosensitive osteosarcoma cell lines (HOS and MG-63) was visualized compared to that of normal cells (HFOB1.19). Moreover, FGD5-AS1 was further upregulated in DXR chemoresistant cells (HOS-DXR and MG-63-DXR) (Figure 1a). Then the efficiency of sh-FGD5-AS1 was validated and it reduced FGD5-AS1 obviously in DXR chemoresistant cells (Figure 1b). Compared with control and sh-NC groups, cell viability was impaired after FGD5-AS1 knockdown (Figure 1c). Consistently, IC50 to doxorubicin in sh-FGD5-AS1 group reduced sharply (Figure 1d). Transwell assay indicated that the migration ability was suppressed when FGD5-AS1 was silenced (Figure 1e). Additionally, autophagy level was characterized by the expression of related proteins (LC3II/I ratio, P62, and Beclin-1). LC3II/I ratio and Beclin-1 were reduced and P62 was activated after sh-FGD5-AS1 treatment (Figure 1f). Therefore, FGD5-AS1 was enriched in chemoresistant cells, and its knockdown alleviated cell malignancy.
3.2 FGD5-AS1 targeted miR-154-5p in the osteosarcoma doxorubicin resistance
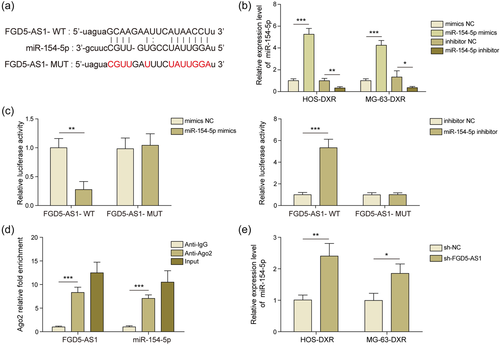
Bioinformatics predicted that FGD5-AS1 and miR-154-5p may interact directly (Figure 2a). Then we examined the efficiency of miR-154-5p mimics and miR-154-5p inhibitor in osteosarcoma cells and they can manipulate miR-154-5p efficiently (Figure 2b). In dual luciferase reported assay, the administration of miR-154-5p mimics significantly reduced the luciferase activity of FGD5-AS1-WT group, miR-154-5p inhibitor significantly increased the luciferase activity of FGD5-AS1-WT group, while there was no obvious difference in the MUT group (Figure 2c). This interaction was further validated by RIP assay. Compared with anti-IgG-treated group, FGD5-AS1 and miR-154-5p were enriched in anti-Ago2 group (Figure 2d). Moreover, miR-154-5p increased in osteosarcoma cell lines after FGD5-AS1 knockdown (Figure 2e). Collectively, FGD5-AS1 could directly bind to miR-154-5p and suppress its expression.
3.3 miR-154-5p reversed the effect of FGD5-AS1 in osteosarcoma DXR cells
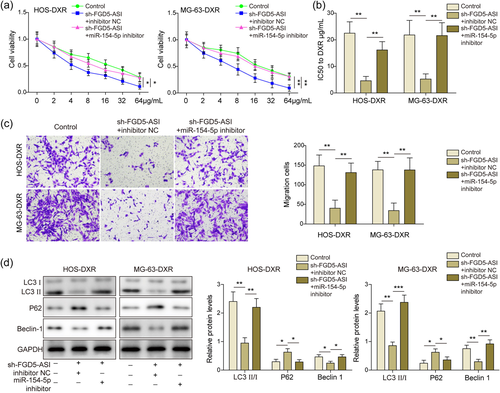
To identify the hierarchy relationship of FGD5-AS1 and miR-154-5p, cells were treated with sh-FGD5-AS1 and miR-154 inhibitor. After miR-154-5p inhibitor administration, cell viability was improved compared with sh-FGD5-AS1 group (Figure 3a) in MTT assay. IC50 of doxorubicin was elevated again in sh-FGD5-AS1 plus miR-154-5p inhibitor group, indicating an enhanced resistance to doxorubicin (Figure 3b). Similarly, migration ability was restored when miR-154-5p was suppressed (Figure 3c). The effect of FGD5-AS1 knockdown on autophagy was also reversed by miR-154-5p suppression, showed by the changes of autophagy-related proteins (Figure 3d). Taken together, miR-154-5p inhibition reversed the effect of FGD5-AS1 knockdown on osteosarcoma cells, which meant miR-154-5p acted at the downstream of FGD5-AS1.
3.4 miR-154-5p targeted WNT5A
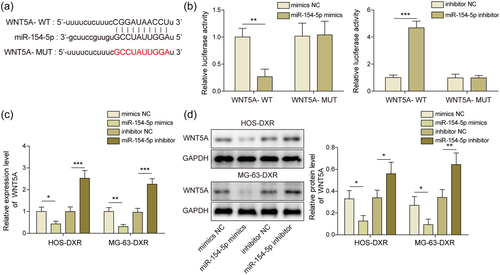
Bioinformatics suggested the existence of binding sites between miR-154-5p and WNT5A (Figure 4a). The administration of miR-154-5p mimics reduced the luciferase activity of WNT5A-WT group, miR-154-5p inhibitor enhanced luciferase activity of WNT5A-WT group, while there was no obvious difference in the WNT5A-MUT group (Figure 4b). The regulation relationship was confirmed by qRT-PCR and WB. Knockdown of miR-154-5p induced WNT5A while overexpression of miR-154-5p inactivated WNT5A (Figure 4c,d). Above results suggested miR-154-5p directly inhibited WNT5A.
3.5 MiR-154-5p inhibited osteosarcoma DXR cell malignancy by WNT5A
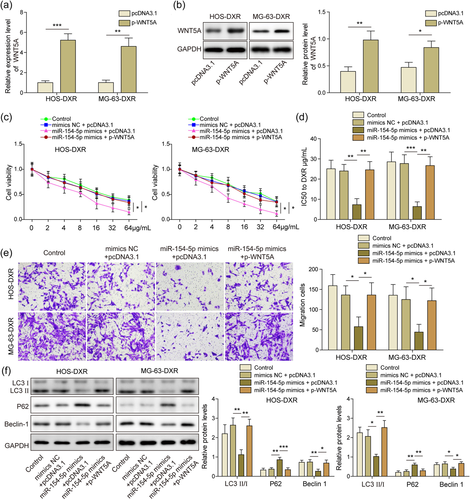
WNT5A was overexpressed by transfection of p-WNT5A plasmid. qRT-PCR and WB results demonstrated that transfection of this plasmid significantly elevated WNT5A expression in osteosarcoma cells (Figure 5a,b). Then osteosarcoma cells were cotransfected with miR-154-5p mimics and p-WNT5A plasmids. MiR-154-5p gain of function reduced cell viability, and overexpression of WNT5A attenuated the effect of miR-154-5p overexpression on cell viability (Figure 5c). IC50 of doxorubicin calculated based on MTT assay showed an accretion after p-WNT5A treatment (Figure 5d). Furthermore, overexpression of WNT5A reversed the effect of miR-154-5p, as evidenced by transwell assay (Figure 5e). LC3II/I ratio and Beclin-1 were reduced and P62 was activated after miR-154-5p mimics transfection, and reversed by WNT5A overexpression (Figure 5f). Overall, miR-154-5p inhibited WNT5A expression, thereby suppressing osteosarcoma DXR cell malignancy.
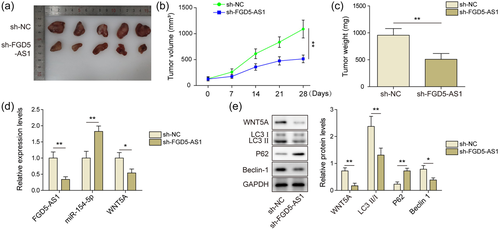
3.6 Knockdown of FGD5-AS1 suppresses DXR osteosarcoma cells tumorigenesis in vivo
HOS-DXR cells were used to establish tumor transplantation model in nude mice. As shown in Figure 6a–c, tumor volume and weight were smaller in sh-FGD5-AS1 group than that in sh-NC group. In sh-FGD5-AS1 group, miR-154-5p was activated and WNT5A was suppressed, which was consistent with in vitro results (Figure 6d,e). Moreover, WB results showed that autophagy was also limited in mouse tumor tissues after sh-FGD5-AS1 treatment (Figure 6e). Therefore, suppression of FGD5-AS1 also interrupted DXR osteosarcoma cells tumorigenesis in vivo.
4 DISCUSSION
Osteosarcoma is a malignant connective tissue tumor in which cancer cells can directly produce tumor bone and bone-like tissue. At present, the main treatment is surgical resection combined with preoperative and postoperative chemotherapy. Drug resistance in some patients leads to tumor recurrence and metastasis, which seriously hinders the efficacy and survival rate (Hattinger et al., 2015, S. Li et al., 2015). At present, the mechanism of resistance to chemotherapy drugs for osteosarcoma remains to be further explored. In this study, FGD5-AS1 was increased in osteosarcoma/DXR cell lines. Knockdown of FGD5-AS1 enhanced osteosarcoma cells' sensitivity to doxorubicin. Mechanistically, FGD5-AS1 could regulate proliferation, migration and autophagy in osteosarcoma/DXR cells via miR-154-5p/WNT5A pathway.
The role of lncRNA in tumors is complex. The same lncRNA can inhibit or promote tumor development in different cancer types. For example, TUG1 is a tumor suppressor in NSCLC and an oncogenic factor in hepatocellular carcinoma (W. Li et al., 2020; P. C. Lin et al., 2016). In osteosarcoma, a lncRNA may have contrary roles in different stage (Ulaner et al., 2003). Long intergenic nonprotein coding RNA 161 (LINC00161) was overexpressed in osteosarcoma cells treated with cisplatin. It activated cell apoptosis in osteosarcoma cells via miR-645/IFIT2 signaling (Reich, 2013). Osteosarcoma doxorubicin resistance-related upregulated lncRNA (ODRUL) is one of the most upregulated 1ncRNAs in osteosarcoma/DXR cells. Results demonstrate that the expression level of ODRUL in the MG63/DXR cell line is obviously higher than that of the MG63 cell line. In vitro experiments have shown that one of the possible mechanisms that ODRUL promotes resistance to osteosarcoma is through the induction of the classic multidrug resistance-related gene ABCB1. ODRUL can be used as an important molecular marker to predict the sensitivity of osteosarcoma to doxorubicin (C. L. Zhang et al., 2016). The role of FGD5-AS1 in doxorubicin-resistance in osteosarcoma was not investigated yet. Here, we reported that FGD5-AS1 was upregulated in osteosarcoma/DXR cell lines and promoted cell proliferation, migration, and autophagy.
In recent years, miRNAs were reported to play an important role in the pathogenesis of human cancers. In solid tumors, many miRNAs function as oncogenes and can be used as new targets for tumor therapy. MiR-154-5p participates in the pathogenesis of various diseases (Ren et al., 2019) and is investigated in multiple cancer types. Previous studies reported that miR-154-5p was suppressed in multiple cancer types, where it acted as a tumor suppressor (Bolandghamat Pour et al., 2019; X. Lin et al., 2015; Pang et al., 2015). Interestingly, Lin et al. reported miR-154-5p was activated in renal cell carcinoma and serves as an oncogene (C. Lin et al., 2018). These studies indicate miR-154-5p may have dual roles, depending on the tumor type. The function of miR-154-5p in chemotherapy resistance was rarely touched previously. In breast cancer, miR-154 suppresses nicotinamide phosphoribosyl transferase and enhanced tumor cells' susceptibility to doxorubicin (Bolandghamat Pour et al., 2019). In the current study, we further linked miR-154-5p to doxorubicin resistance in osteosarcoma. Knockdown of miR-154-5p reduced osteosarcoma/DXR cells' sensitivity to doxorubicin and enhanced proliferation, migration, and autophagy, which is consistent to the results in breast cancer.
As a component of noncanonical Wnt signaling pathways, WNT5A promotes chemotherapy resistance in various cancers, such as melanoma (Anastas et al., 2014) and colon cancer (Bordonaro et al., 2011). As described above, autophagy and Wnt signaling interacts through dual mechanisms. Wnt5a/β-catenin signaling modulated autophagy in glioma cells. However, whether WNT5A affects drug resistance in osteosarcoma is not clear yet. Our study revealed that WNT5A served as an oncogene and drove drug resistance in osteosarcoma, which was consistent with the roles other cancer types. WNT5A acted at the downstream of FGD5-AS1 and miR-154-5p. Manipulation of WNT5A abolished the effects of FGD5-AS1 knockdown or miR-154-5p overexpress on DXR chemoresistant Osteosarcoma cells.
In summary, FGD5-AS1 is enriched in DXR chemoresistant Osteosarcoma cells. FGD5-AS1 suppression inhibits cell proliferation, migration, and autophagy. Mechanistically, FGD5-AS1 directly interacted with miR-154-5p, thereby upregulating WNT5A. Therefore, application of FGD5-AS1 as biomarkers or therapeutic targets may be promising, which requires more data to validate.
ACKNOWLEDGMENTS
We would like to give our sincere gratitude to the reviewers for their constructive comments. This study was supported by the Natural Science Foundation of Jilin Provincial Department of Science and Technology (Jilin Provincial Key Laboratory Project and Theme Scientist Project) (No. 20190201282JC).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
All animal experiments were conducted in accordance to institutional guidelines for the care and use of laboratory animals of the China-Japan Union Hospital of Jilin University and met the regulations published by the National Institutes of Health Guide for Care and Use of Laboratory Animals.
Open Research
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.




