An oleanolic acid derivative, K73-03, inhibits pancreatic cancer cells proliferation in vitro and in vivo via blocking EGFR/Akt pathway
Zheng Zhou, Yaokun Dong, and Na Li contributed equally to this study.
Abstract
Oleanolic acid (OA) and its derivatives show potent anticancer function. Pancreatic cancer (PC) is the fourth core motive of cancer-related deaths worldwide. Epidermal growth factor receptor (EGFR) has been implicated in PC and has been validated as a therapeutic target. Our study demonstrated that K73-03, an OA derivative, was identified as a potent inhibitor of EGFR by using reverse pharmacophore screening and molecular dynamics simulation assays. Moreover, Western blot analysis showed that K73-03 markedly suppressed the levels of phosphorylated-EGFR (p-EGFR) and phosphorylated-Akt (p-Akt). The inhibitory effect of K73-03 on PC cells was assessed in vitro and in vivo. Mechanistically, K73-03 effectively inhibited the cell proliferation of PC cells, and induced apoptosis and autophagy of ASPC-1 cells in a dose-dependent manner. Additionally, pretreatment with chloroquine, an autophagy inhibitor, significantly inhibited K73-03-induced autophagy and enhanced K73-03-induced apoptotic cell death. K73-03 also strongly repressed ASPC-1 cells xenograft growth in vivo. Thus, all these findings provided new clues about OA analog K73-03 as an effective anticancer agent targeted EGFR against ASPC-1 cells, it is worth further evaluation in the future.
1 INTRODUCTION
Pancreatic cancer (PC), one of the most aggressive and lethal solid malignancies, has become the fourth leading cause of cancer deaths worldwide (Cykowiak et al., 2021; Siegel et al., 2019). It is associated with lack of symptomatology and extremely dreary prognosis, less than 10% of patients would be alive at 5 years after diagnosis. Currently, chemotherapy is of vital importance therapy for sufferers with PC (Balsano et al., 2019). However, serious adverse side effects and the problem of multidrug resistance limit their clinical applicability (Cykowiak et al., 2021; Tuan Anh et al., 2018). Therefore, developing novel drugs with high efficiency and minimal adverse effects is urgently required.
Natural products and their derivatives have been shown considerable structural diversity and better drug-like properties compared to the synthetic compounds in the aspects of preventive and anticancer effects, and remain a major resource of new drugs discovery (Li et al., 2015; Y. Wang, Lia, et al., 2018). Oleanolic acid (OA) is a pentacyclic triterpenoid that is widely present in plant species. OA and its natural derivatives exerted a variety of biological activities, including hepatoprotective, anti-inflammatory, and especially anticancer effect in many cancer types (Baer-Dubowska et al., 2021; Tang et al., 2022). Therefore, OA derivatives remain essential fields regarding their synthesis and potential anticancer efficacy to PC.
The epidermal growth factor receptor (EGFR) family comprises four closely related receptors (EGFR/HER1/ErbB1, HER2/ErbB2, HER3/ErbB3, and HER4/ErbB4) and plays a major regulatory role in cell proliferation, differentiation, survival, and angiogenesis (Grapa et al., 2019; Mirgany et al., 2021). Structurally, EGFR is composed of an extracellular ligand-binding domain, a transmembrane region, and an intracellular tyrosine kinase domain. EGFR activation is involved in both the PC initiation and its progression to metastatic cancer (Navas et al., 2012). The inhibition of EGFR activity has been considered to be a prospective strategy for clinically treating pancreatic tumors. It has been reported that OA inhibited proliferation possibly through modulating EGFR activity in A375 cells which would qualify it as a potent anticancer agent (Ghosh et al., 2014). We previously synthesized several OA derivatives, named SZC009, 013, 014, 015, 017. Among them, SZC015 exhibited antitumor activity against PC cells (Chu et al., 2017; Song et al., 2018). Recently, we synthesized a novel OA derivative called K73-03. Our previous study has elucidated that K73-03 induced mitochondrial damage that led to apoptosis of PC through epigenetic SPINK1 suppression by miR-421 upregulation (Shopit et al., 2020). In the current study, we investigated the antitumor ability of K73-03 as a potent inhibitor of EGFR on PC, which induced apoptosis and autophagy through blocking EGFR/(Akt) pathway. In this respect, our data suggest that K73-03 may be a potential candidate for treating PC.
2 MATERIALS AND METHODS
2.1 Materials
K73-03 are provided in our previous study (Shopit et al., 2020). The primary antibodies against β-actin, EGFR, p-EGFR, Akt, p-Akt, LC3B, Beclin1 Caspase-3, Cleaved-Caspase-3, Caspase-9, Bax, and Bcl-2 and all the secondary antibodies were purchased from Proteintech. All other chemicals were purchased from Sigma Chemical Co. Human pancreatic ductal epithelial cells (HPDE6-C7), human PC cell lines ASPC-1, and PANC-1 were purchased from Shanghai Institute of Biochemistry and Cell Biology.
2.2 Targets predicted by PharmMapper
PharmMapper is an open-source online platform, which aims identify potential target candidates for the given probe small molecules via a pharmacophore mapping approach (Liu et al., 2010; X. Wang, Shen, et al., 2017). Molecular file of K73-03 was uploaded to the PharmMapper servers for in silico screening. The number of reserved matched targets was set to 300, whereas other parameters were considered as default.
2.3 Molecular docking
The high-resolution crystal structure of EGFR was obtained from the PDB bank (PDB code: 3W2S), Molecular docking analysis was performed using AutoDock 4.2 with MGL tools 1.5.6. The protein structures were cleaned by removing crystallographic water and prepared by the addition of polar hydrogen atoms, followed by the addition of Gasteiger charges. The residual interactions at the protein drug interface were evaluated using LigPlot (Laskowski & Swindells, 2011).
2.4 Molecular dynamics (MD) simulation
The conformation of the complexes formed between ligands and EGFR target protein were predicted using AutoDock 4.2 program, and all of the MD simulations were carried out by Gromacs 5.1.5 with Amber99sb force field (Y. Wang et al., 2019). After MD simulation, the molecular mechanics energies combined with the Molecular Mechanics/Poisson Boltzmann Surface Area (MM/PBSA) methods were employed to calculate the binding free energy of OA and K73-03 with EGFR target protein.
2.5 Cell culture and cell viability assay
HPDE6-C7, ASPC-1, and PANC-1 cells were cultured in high-glucose DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 µg/ml streptomycin and maintained in 5% CO2 at 37°C in saturated humidity. Cell viability was measured by Cell Counting Kit-8 (CCK-8; Dojindo) assay. Cells were seeded into 96-well plates with 5 × 103 cells/well and incubated for the cell attachment, and were treated with K73-03 with different concentrations for 24 h. Then, each well was treated with 10 µl CCK-8 solution. Two hours later, the absorption values were measured at 450 nm using a microplate reader (Varioskan™ LUX; Thermo Co).
2.6 Transient transfection of EGFR small interfering RNA (siRNA) in ASPC-1
Transfection was performed with Lipofectamine 3000 (Invitrogen) according to the manufacturer's protocol. Briefly, Cells were seeded in 6-well or 96-well plates at approximately 50% confluency for 24 h before siRNA treatment. Then, siRNA transfection was performed 24 h before K73-03 treatment. The siRNAs were synthesized by GenePharma (Shanghai, China) with following sequences: siNC: 5′-UUCUCCGAACGUGUCACGUdTdT-3′, siEGFR:5′-CCUAUGCCUUAGCAGUC UUTT -3′.
2.7 Western blot analysis
Equal amounts of cell lysate proteins were loaded onto a 8%−12% sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE), separated by gel electrophoresis and then electrically transferred onto a polyvinylidene difluoride membrane. The membrane was sealed with 5% skim milk for 2 h at room temperature and incubated with specific antibody. The blots were then detected using enhanced chemiluminescence reagent and photographed by Bio-Spectrum Gel Imaging System (Bio-Rad).
2.8 In vitro migration assay
Cell migration was observed by Scratch assay (wound healing assay). The cells were inoculated into six-well plates, incubated at 37°C and 5% CO2 overnight, and then wounded with a 200 µl sterile pipette tip after 6 h of starvation in incomplete medium. Cells were treated with indicated doses of K73-03 in full medium. After incubation at 37°C for 24 h, cells were washed three times with phosphate buffered saline (PBS), and images of the cells were obtained using microscope.
2.9 Cell cycle analysis
To evaluate the cell cycle distribution after exposure to K73-03, ASPC-1 cells were incubated with K73-03 for 24 h. After the treatment, the cells were collected following centrifugation and fixed in ice-cold 70% ethanol overnight at 4°C. The samples were then suspended with staining reagent (50 µg/ml of PI and 100 µg/ml of RNase A in PBS) in the dark at 37°C for 30 min. Finally, the DNA content of cells was detected using FACScan flow cytometry (BD FACSAria II; BD Co).
2.10 Apoptosis assay
To determine whether K73-03 can trigger apoptosis in ASPC-1 cell line. Cells (5 × 105) cultured in six-well plates were treated with different concentrations of K73-03 for 24 h. The cells were collected and stained with annexin-V–FITC and propidium iodide (PI) for 30 min in the dark at 37°C according to the instructions of the manufacturer. The samples were immediately analyzed using flow cytometry (BD FACSAria II; BD Co). Flow cytometry data were finally analyzed using FlowJo version 7.6.1 (FlowJo).
2.11 Transmission electron microscopy
ASPC-1 cells were plated in six-well plates, incubated at 37°C and treated with K73-03 for 24 h. Cells were collected and fixed with 2% glutaraldehyde in 0.1 M PBS overnight at 4°C. The samples were subsequently dehydrated, embedded, sectioned, and double stained with uranyl acetate and lead citrate. Electron micrographs were taken on a Transmission Electron Microscope (TEM) (JEM-2000EX; JEOL Co).
2.12 In vivo studies
The laboratory animal center of Dalian Medical University (SCXK: 2013-0006) provided BALB/c nude mice with a body weight of 15−20 g for 4−6 weeks. All animals were maintained under sterile conditions, and animal experiments are conducted in accordance with the guidelines of the Animal Care and Use Committee. To generate tumor xenografts, 5 × 105 ASPC-1 cells in 0.2 ml of PBS were injected into the subcutaneous area of the right flank of mice. When the tumor diameter reached 6−9 mm, these mice were randomly divided into three groups (n = 5 per group): Control group, low-dose (25 mg/kg) of K73-03-treated group, and high-dose (50 mg/kg) of K73-03-treated group. Mice from the experimental groups were administered (intraperitoneal injection, [ip]) with either 25 or 50 mg/kg K73-03 once a day for 3 weeks. Tumor volume was measured every 3 days with Vernier calipers and calculated using the following formula: V = A × B2/2, where A and B represent the maximum and minimum diameters (Peirce et al., 2011). Finally, mice were Euthanized, and tumors were weighed, measured and photographed.
2.13 Hematoxylin and eosin (H&E) staining
Before paraffin embedding, primary tumors were fixed in 4% paraformaldehyde overnight. 5-µm serial slicing, sections were dyed with H&E as previously described. For H&E, histopathological changes were observed using a light microscope (Olympus).
2.14 Statistical analysis
The results were expressed as mean ± SD. Statistical differences between groups were assessed using a One-way analysis of variance. Two groups were compared using two-tailed Student's t-test using Graph Pad Prism 8.0 (Graph Pad Software, Inc.). Bonferroni test was used to correct multiple comparison and the p < 0.05 were considered statistically significant.
3 RESULTS
3.1 Potential target prediction and screening using PharmMapper and molecular docking
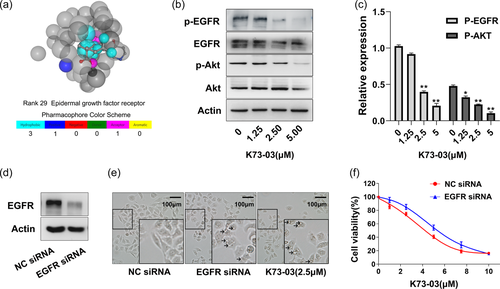
The chemical structure of OA and K7303 are shown in Supporting Information: Figure S1A. Potential protein target for K73-03 was identified by virtual screening procedures with PharmMapper. Significantly, EGFR ranked 29 among 2241 human protein targets (Figure 1a). Moreover, total 22 kinases from top 100 ranked potential protein targets were subjected to further molecular docking computation using Autodock 4.2. The results of this analysis are shown in Table 1, where the docking score in the EGFR complex (−11.34 Kcal/mol) was best among the top 100 of potential protein targets.
| PharmMapper rank | PDB ID | Name | Target gene | Docking score (kcal/mol) |
|---|---|---|---|---|
| 8 | 1UNH | Cyclin-dependent kinase 5 activator 1 | CDK5R1 | −9.39 |
| 13 | 3BGQ | Proto-oncogene serine/threonine-protein kinase Pim-1 | PIM1 | −6.15 |
| 14 | 2G01 | Mitogen-activated protein kinase 8 | MAPK8 | −8.97 |
| 19 | 2RKU | Serine/threonine-protein kinase PLK1 | PLK1 | −8.54 |
| 29 | 2ITZ | Epidermal growth factor receptor | EGFR | −11.34 |
| 41 | 1QPD | Proto-oncogene tyrosine-protein kinase LCK | LCK | −9.13 |
| 42 | 1SM2 | Tyrosine-protein kinase ITK/TSK | ITK | −3.14 |
| 44 | 2BU5 | Pyruvate dehydrogenase kinase 2 | PDK2 | −7.68 |
| 47 | 2UW9 | RAC-beta serine/threonine-protein kinase | AKT2 | −9.69 |
| 61 | 2VX0 | Ephrin type-B receptor 4 | EPHB4 | −5.73 |
| 63 | 2I6B | Adenosine kinase | ADK | −9.75 |
| 65 | 1RW8 | TGF-beta receptor type-1 | TGFBR1 | −9.46 |
| 69 | 1PMN | Mitogen-activated protein kinase 10 | MAPK10 | −9.15 |
| 72 | 2F57 | Serine/threonine-protein kinase PAK 7 | PAK5 | −9.55 |
| 82 | 1S9J | Dual specificity mitogen-activated protein kinase kinase 1 | MAP2K1 | −8.77 |
| 87 | 1NXK | MAP kinase-activated protein kinase 2 | MAPKAPK2 | −10.36 |
| 89 | 2E9V | Serine/threonine-protein kinase Chk1 | CHEK1 | −9.15 |
| 91 | 1OKY | 3-phosphoinositide-dependent protein kinase 1 | PDPK1 | −5.92 |
| 92 | 1P2A | Cell division protein kinase 2 | CDK2 | −9.51 |
| 94 | 1Q3D | Glycogen synthase kinase-3 beta | GSK3B | −7.19 |
| 96 | 1M51 | Phosphoenolpyruvate carboxy kinase | PCK1 | −5.10 |
| 97 | 1XJD | Protein kinase C theta type | PRKCQ | −9.66 |
- Note: The K73-03 potential targets with High Fit Score were screened by PharmMapper. Total 22 kinases from top 100 ranked potential protein targets were subjected to further molecular docking computation using Autodock4.2.
3.2 K73-03 decreases the levels of p-EGFR and downstream targets of EGFR signaling pathway in ASPC-1 cells
CCK8 assay were performed to examine the effect of OA and K73-03 on cell viability in ASPC-1, PANC-1, and HPDE6-C7 cell lines. The results showed that K73-03 decreased the cell viability of the ASPC-1 and PANC-1 cell lines in a concentration and time-dependent manner. However, the toxicity of k73-03 to HPDE6-C7 cells was less, suggesting that K73-03 possesses a promising anti-PC effect (Supporting Information: Figure S1B, C). Interestingly, as an OA derivative, K7303 exhibited a more significant reduction in the cell viability of ASPC-1 cells when compared with OA (Supporting Information: Figure S1D). The 50% inhibitory concentration (IC50) after 24 h of K73-03 treatment was 3.3 µM in ASPC-1 cells and 4.5 µM for PANC-1 cells respectively. These results demonstrated that the K73-03 exhibited potent antitumor activity and ASPC-1 cell line showed more sensitive to the agent. Therefore, ASPC-1 cell line was selected to make a further exploration due to its low IC50 value.
Western blots were used to detect the protein level of p-EGFR and its downstream PI3K/AKT signaling transduction. As shown in Figure 1b,c, K73-03 significantly suppressed the protein expression levels of both p-EGFR and p-AKT in a concentration dependent manner. Moreover, although both K73-03 and OA were able to decrease the expression of p-EGFR, 2.5 µM K73-03 treatment could almost completely inhibit the expression of p-EGFR in ASPC-1 cells (Supporting Information: Figure S2).
To better understand whether the effects of K73-03 were mediated directly through EGFR, we compared the effects of ASPC-1 cells transfected with NC siRNA or EGFR siRNA. Western blot analysis confirmed the downregulation of EGFR in ASPC-1 cells (Figure 1d). Transfected and NC ASPC-1 cells were cultured, and then cell viability was determined using CCK-8 assay. The results showed that the proliferation of NC siRNA cells was decreased with K73-03 treatment compared to knocked down cells (Figure 1f). These results suggested that EGFR could be a direct target for K73-03 to suppress cancer cell growth. Interestingly, treatment EGFR siRNA markedly caused cellular morphological changes, the vacuolation was observed as they were consistent with treatment with 2.5 µM K73-03 (Figure 1e).
3.3 Binding mechanism of K73-03 to kinase domain of EGFR
A large number of EGFR crystal structures have been reported in the literature. The availability of EGFR crystal structure allowed us to utilize molecular docking to identify the novel EGFR inhibitors (Ahmed et al., 2018; Haddad et al., 2021). In this study, high resolution crystal structures (PDB ID:3W2S) was further considered for the molecular docking and MD simulations studies. As shown in Figure 2a,b, the interactions showed by LigPlot are mediated by hydrogen bonds and by hydrophobic interactions. Specifically, In the adenosine-triphosphate (ATP) binding site of the EGFR kinase domain, K73-03 interacted with the active amino acid residues (Asp837 and Cys797) to formed two hydrogen bonds. Furthermore, K73-03 maintains hydrophobic interactions with the catalytic residue Asp837 and its surrounding residues Gly796, Leu844, Leu718, Val726, Lys745, Asp855, Phc723, Asn842, Ser720, Ala722, Gly721, Arg841, Gly719, and Ala743 in the active site. Collectively, these results suggested K73-03 inhibits EGFR activity by targeting the ATP binding site.
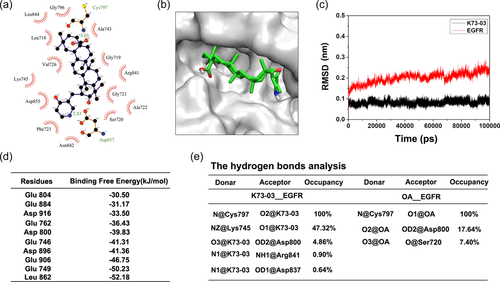
3.4 MD simulations
The K73-03 is subjected to MD simulation of 100 ns to make sure the ligand stays inside the active site pocket and to observe the interaction properties to ensures the ligand functionality. The RMSD trajectory of K73-03 was stable, indicating that the small molecule K73-03 was stable in combination with EGFR protein receptor (Figure 2c). The interaction between the K73-03 and each residue was analyzed by the MM/PBSA methods. Binding free energy calculation listed in Table 2 where the binding energy in the EGFR-K73-03 complex (−242.78 kJ/mol) was better than in the EGFR-OA complex (−164.95 kJ/mol). The results also showed Top 10 hot residues, which made a favorable contribution to the binding of the inhibitor and protein (Figure 2d).
| Complex | ΔEvdW | ΔEele | ΔGPB | ΔGSA | ΔGbinding |
|---|---|---|---|---|---|
| EGFR-OA | −191.78 | −109.38 | 158.35 | −21.89 | −164.95 |
| EGFR-K73-03 | −226.41 | −232.14 | 240.35 | −24.71 | −242.78 |
- Note: Snapshots extracted from the last 0.5 ns MD simulation were submitted to MmPbSaStat.py for the free energy calculation. All the energies are in kJ/mol.
- Abbreviations: MM/PBSA, Molecular Mechanics/Poisson Boltzmann Surface Area; OA, Oleanolic acid.
Further, hydrogen bond analysis showed that the hydrogen bond interaction between Cys797-K73-03 was stable during the last 5 ns simulation in the binding system, and the hydrogen bond occupancy was 100%. The hydrogen bond occupancies between Lys745-K73-03, Asp800-K73-03, and Arg841-K73-03 in the system were 47.32%, 4.86%, and 0.90%, respectively. Figure 2e summarized hydrogen-bond interactions and hydrogen bond occupancy between ligand atoms and residues during simulations.
3.5 Suppression migration of ASPC-1 cells by K73-03
As shown in Figure 3a, in the control group of the migrating ASPC-1 cells, the part of gap or wounding space between cell layers after making a scratch was occupied almost 24 h after making a scratch, while in the cells treated with K73-03, due to the damage of migration ability, the migration failed to occupy the scratched space.
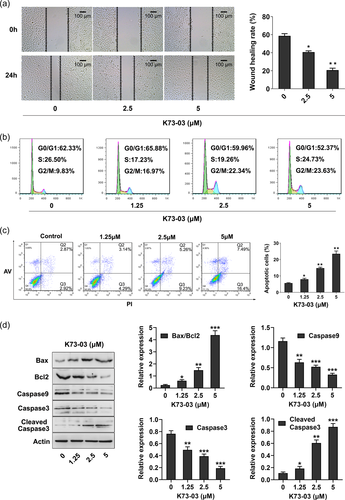
3.6 Effect of K73-03 on cell cycle distribution of ASPC-1 cells
Cell cycle arrest is an important mechanism to inhibits cancer cell growth (F. Wang, Tian, et al., 2018; Yao et al., 2019). We next study the effect of K73-03 on cell cycle progression in ASPC-1 cells. Our data showed that K73-03 treatment caused an accumulation of ASPC-1 cell lines in the G2/M-phase. As seen in Figure 3b, the percentage of ASPC-1 cells in the G2/M-phase was increased from 9.87% in the control group to 22.83% in the 5 µM K73-03 group. Our findings suggest that K73-03 induces G2/M-phase arrest of the ASPC-1 cells.
3.7 K73-03 induces apoptosis in the PC cells
To detect whether the antitumor activity of K73-03 was associated with apoptosis in ASPC-1 cells, flow cytometry analysis using Annexin V-FITC and PI double staining was performed. The cells apoptotic rates were noticeably increased from 5.79% (control group) to 7.43%, 14.49%, and 23.89% in ASPC-1 cells treated with 1.25, 2.5 and 5 µM of K73-03 for 24 h, respectively (Figure 3c). The ratio of Bax/Bcl-2 which determines the susceptibility to apoptosis by regulating mitochondrial functions was increased by K73-03. The levels of procaspase-9 and procaspase-3 were suppressed, while the level of cleaved caspase-3 was increased by K73-03 (Figure 3d). The results confirmed that K73-03 induced apoptosis in ASPC-1 cells in a concentration-dependent manner.
3.8 K73-03 induces autophagy in ASPC-1 cells
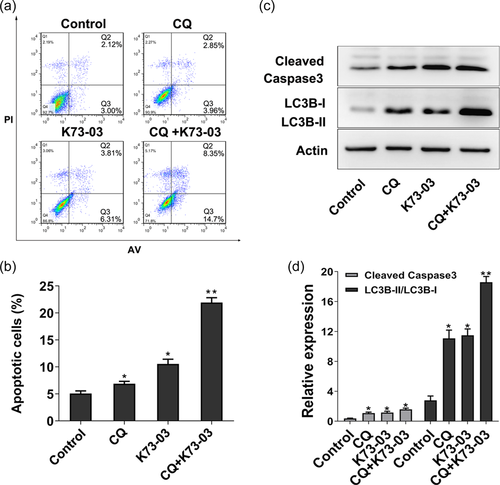
The micro-morphological change of K73-03 treated ASPC-1 cells were also observed by TEM, which is the most convincing standard method to verify autophagy (Gao et al., 2015; Song et al., 2017; F. Wang, Xu, et al., 2017). The intact cell and nuclear membrane, rough endoplasmic reticulum, distinct nucleus, and homogeneous cytoplasm maintained normal micromorphology in the untreated cells, while the results showed that 2.5 µM K73-03 treatment resulted in numerous vacuoles in the cytoplasm changes of autophagosomes in the ASPC-1 (Figure 4a). Moreover, microtubule-associated protein 1, light chain 3 (LC3), the specific marker of autophagy was detected by Western Blots. As shown in Figure 4b, compared with the control group, the ratio of LC3-II/LC3-I was increased after treatment with K73-03. Our data showed that K73-03 induced autophagy in ASPC-1 cells in a concentration-dependent manner. Furthermore, to determine the relationship between autophagy and apoptosis induced by K73-03, we evaluated level of cleaved caspase-3 after pretreatment with Chloroquine, a late-stage inhibitor of autophagy. Our study showed pretreatment with 10 µM Chloroquine dramatically increased expression of cleaved caspase-3, as compared with only K73-03 (2.5 µM) group (Figure 5). These data suggest that K73-03-induced autophagy not only prevent cells from K73-03-induced cell death, but also inhibit apoptosis triggered by K73-03 in ASPC-1 cells.

3.9 K73-03 suppresses ASPC-1 cell xenograft growth
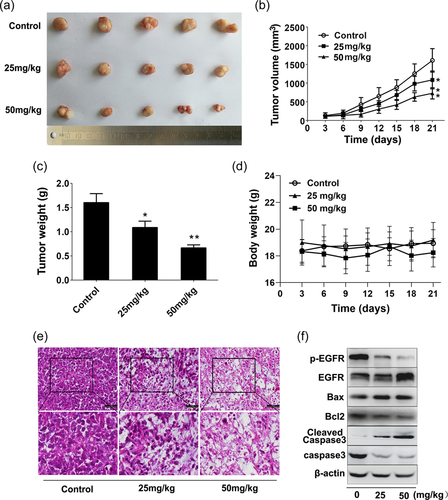
To investigate the effect of K73-03 on tumor formation of PC in vivo, ASPC-1 cell xenograft mouse models (BALB/c nude mice) were used to evaluate antitumor activity of K73-03. As illustrated in Figure 6a−c, K73-03 significantly decreased the growth of ASPC-1 cell xenograft tumors compared with those of control tumors. Moreover, 50 mg/kg K73-03 treatment exhibited a more significant inhibition of tumor volumes and weights compared with 25 mg/kg K73-03, whereas the body weight was comparable between K73-03-treated and control groups (Figure 6d). H&E staining further showed that K73-03 administration could lead to several morphological changes such as the decrease of tumor cell number, tumor tissue cavitation, tumor cell density and necrosis area in tumor tissue (Figure 6e). Meanwhile, results from immunoblotting analyses confirmed that p-EGFR expression was markedly reduced in xenograft tissues receiving K73-03 administration (Figure 6f). Therefore, our results indicate that K73-03 exerts potent antitumor effect tumors against PC in vivo mediated by EGFR.
4 DISCUSSION
In recent years, the technologies of target fishing fulfill an important role in investigating the mechanism of bioactive small molecules (Alvarez et al., 2006; Giordano et al., 2013). It has become a hot spot in drug research for using computer tools to identify novel interactors or novel targets for drugs. OA is a pentacyclic triterpenoid that is widely present in plant species. OA and its natural derivatives exerted a variety of biological activities, including hepatoprotective, anti-inflammatory, and especially anticancer effect in many cancer types. However, the molecular mechanism of action still be obscure. Recently, we synthesized a novel OA derivative called K73-03. In this study, the potential protein targets of K73-03 were screened via reverse pharmacophore screening and molecular docking study. Our primary investigation showed that EGFR was the best potential kinase protein target among 2241 human protein targets. Moreover, EGFR overexpression was one of the indicators for clinical outcome implicated in pancreatic carcinomas (Guo et al., 2016). Furthermore, our result using Western blot also confirmed that K73-03 suppressed the protein level of p-EGFR and its downstream PI3K/AKT signaling transduction in ASPC-1 cells.
To better understand whether the effects of K73-03 were mediated directly through EGFR, we compared the effects of ASPC-1 cells transfected with NC siRNA or EGFR siRNA. The proliferation of NC siRNA cells was decreased by the treatment with K73-03 comparing to knocked down cells (Figure 1f), suggesting that EGFR could be a direct target for K73-03 to suppress cancer cell growth. Interestingly, the treatment with EGFR siRNA markedly caused cellular morphological changes, the vacuolation was observed as they were consistent with treatment with 2.5 µM K73-03 (Figure 1e).
Molecular docking calculation was performed to gain insight into the binding mode of K73-03-EGFR complex and OA-EGFR complex. Subsequently, 100 ns MD simulation was subjected to further observe the interactions between K73-03 and EGFR. As shown in Table 2, the binding free energy in the EGFR-K73-03 complex (−242.78 kJ/mol) was better than in the EGFR-OA complex (−164.95 kcal/mol). Further, the hydrogen bond interaction between Cys797-K73-03 was stable during the last 5 ns simulation in the binding system, and the hydrogen bond occupancy was 100%. The results indicated the carboxy of K73-03 is a reactive group and Cys797 could be a vital residue instead of Thr790. Therefore, K73-03 has the potential to become an EGFR tyrosine kinase inhibitor to overcoming the mutation of T790M resistance.
PC is a highly lethal malignancy, and the treatment outcomes including chemotherapy at present are rather ineffective (Rabi & Catapano, 2016; Rossi et al., 2014). EGFR is a rational target for PC treatment because it is usually expressed at high levels in PC. Similarly, the present study demonstrated that K73-03 markedly inhibited cell viability in ASPC-1 and PANC-1 cells. Interestingly, as an OA derivative, K73-03 exhibited a more significant reduction in the cell viability of ASPC-1 cells when compared with OA, and exerted a less pronounced antiproliferative effect on HPDE6-C7 normal pancreatic cells. Moreover, K73-03 exerts potent antitumor effect tumors against PC in vivo mediated by EGFR, suggesting that K73-03 could be used as a protective agent in therapies targeted against PC.
In multicellular organisms, there exists a delicate balance between the cell-generating effects of mitosis and apoptosis-induced cell death (Cotter, 2009; Huang et al., 2016; Prakobwong et al., 2011). The induction of apoptosis and cell cycle arrest are currently two important processes in the development of anticancer drugs (Curtin, 2013; Gao et al., 2016). Consequently, flow cytometric analysis was performed to determine whether K73-03 could induce PC cells death, which was associated with apoptosis. As shown in Figure 3 and 6, K73-03 markedly increased in the Bax/Bcl-2 ratio and caspase 3 activity both in vitro and in vivo, suggesting that the action of K73-03 likely has been linked with intrinsic apoptosis, which is mediated via targeting of the mitochondria (Y. Wang et al., 2019; Yao et al., 2019). The cell cycle, featuring its various components and stages, and cell growth and division, forms a complex biological process (Martín-Renedo et al., 2008; Tao et al., 2014). In the present study, K73-03 induced antiproliferative effects on pancreatic cells by restricting cell cycle progression, which was verified by blocking ASPC-1 cells in the G2/M phase of cell cycle.
Several natural products have been demonstrated to possess anticancer activity, mainly through autophagy-dependent mechanisms (Bredholt et al., 2009; Nie et al., 2016; Zhang et al., 2013). Autophagy, as an evolutionarily conserved lysosomal degradation process, has an important role in regulating prosurvival and prodeath signaling pathways in numerous diseases, especially in cancer (Mizushima et al., 2010). Our result using TEM revealed that the autophagosomes of the ASPC-1 cells upon treatment with K73-03 exhibited the typical appearance of containing cytoplasmic organelles and an engulfed bulk cytoplasm. Furthermore, a significant transformation from LC3I to LC3II, which is a typical sign of autophagy, was also detected (Baehrecke, 2005; Mizushima et al., 2010). Therefore, to determine the association between autophagy and apoptosis induced by K73-03, the extent of apoptosis in the cells, and the level of cleaved caspase-3 following pretreatment with chloroquine, were evaluated. The findings supported that apoptosis triggered by K73-03 was significantly enhanced by blocking autophagy (Figure 5), suggesting that autophagy induced by K73-03 may serve as an inhibitor role in the process of apoptosis in ASPC-1 cells.
5 CONCLUSIONS
In summary, we report the OA derivative K73-03 as a novel type inhibitor of EGFR. Moreover, K73-03 inhibited the cell proliferation and cell migration of ASPC-1 cells, and subsequently induced autophagy and apoptosis of ASPC-1 cells, which were associated with the EGFR/AKT signaling pathway. Furthermore, K73-03 also strongly inhibited ASPC-1 xenograft growth in vivo. These findings provide a strong theoretical basis that K73-03 possesses a potential to be further developed in the treatment of PC.
ACKNOWLEDGMENTS
This study was supported by the Natural Science Foundation of China (no. 82073768) and the Dalian High-level Talent Innovation Support Program (no. 2019RD03).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The animal study was reviewed and approved by the Animal Experimental Ethics Committee of Dalian Medical University.
Open Research
DATA AVAILABILITY STATEMENT
The data or materials generated or used during the study are available from the corresponding author upon reasonable request.




