Inhibition of CXCR4 regulates epithelial mesenchymal transition of NSCLC via the Hippo-YAP signaling pathway
Abstract
CXCR4 has been shown to play a key role in the metastasis of non-small cell lung cancer (NSCLC). And CXCR may be associated with the Hippo—Yes kinase-associated protein (YAP) pathway, thus involving in the occurrence and progression of NSCLC. This study aims to investigate the effect of CXCR4 inhibition on epithelial-mesenchymal transition (EMT), invasion and migration of NSCLC cells via the Hippo-YAP pathway. QRT-PCR and Western blot were employed to detect CXCR4 expression in NSCLC cell lines. A549 and H1299 cells were treated with WZ811 (0, 10, 30, and 50 µM), and A549 cells were also divided into the Control, WZ811, YAP siRNA, and WZ811 + YAP groups. Wound-healing, Transwell assay, immunofluorescent staining, and a luciferase reporter gene assay were performed in this experiment. Compared with human bronchial epithelial (HBE) cells, CXCR4 expression was up-regulated in NSCLC cell lines. WZ811 increased E-cadherin; decreased expression of Twist, vimentin, Snail, p-YAP, CTGF, and BIRC5; blocked GTIIC reporter activity; and reduced migration and invasion of A549 cells, all in a dose-dependent manner. YAP siRNA had a similar effect to WZ811 by inhibiting EMT, invasion and migration of A549 cells. However, compared with A549 cells in the YAP siRNA and WZ811 groups, cells in the WZ811 + YAP group showed a dramatically enhanced EMT phenotype as well as invasion and migration abilities. Inhibition of CXCR4 may reduce EMT, invasion and migration of NSCLC cells, thereby providing a new therapeutic target for NSCLC.
Abbreviations
-
- CXCR
-
- chemokine receptors
-
- EMT
-
- epithelial-mesenchymal transition
-
- NSCLC
-
- non-small cell lung cancer
-
- YAP
-
- Yes kinase-associated protein
-
- HBE
-
- human bronchial epithelial
-
- SDF-1
-
- stromal-derived-factor-1
-
- ECL
-
- chemiluminescence
Introduction
With the aggravation of air pollution, the incidence of lung cancer is increasing yearly, and lung cancer has become one of the most common tumors worldwide with the highest incidence (Tao et al., 2012). Non-small cell lung cancer (NSCLC), which accounts for approximately 75–85% of all lung cancer (Qi and Fan, 2010), can be classified into many histologic types, including adenocarcinoma (AD), squamous cell carcinoma (SCC), large cell carcinoma, and mixed histologic types (Ciborowski et al., 2017). Although tremendous progress has been made in diagnostic technology and surgical operations and many chemotherapy and targeted drugs have come onto the clinical market, the prognosis of lung cancer patients is still very poor with the 5-year survival rate lower than 20% (Xie et al., 2017). Therefore, early diagnosis and treatment are extremely important. In China, 70–80% of patients diagnosed with lung cancer cannot undergo radical surgery, and they have no choice but receive palliative chemotherapy, radiotherapy, targeted therapy, and biological therapy (Kan et al., 2015). However, whether receiving chemotherapy or targeted therapy, most patients eventually experience recurrence, metastasis, and further progression of the tumor, ultimately leading to treatment failure. Therefore, it is of great importance to explore the mechanism of tumor metastasis and to identify indicators that can predict prognosis as well as prevent and control tumor metastasis.
Chemokines, small soluble molecules known for their potent ability to induce cellular migration, are highly expressed in a variety of cancer cells, as are chemokine receptors (CXCR) (Zhang et al., 2013). Known as fusin or CD184, CXCR4 is an alpha-chemokine receptor specific for CXCL12, also known as stromal-derived-factor-1 (SDF-1) (Xu et al., 2015). In recent years, CXCR4 has been shown to play an important role in the metastasis of many cancers. For example, Zhang et al. (2013) found that mesenchymal stem cells can promote CXCR4-mediated osteosarcoma growth and pulmonary metastasis through the regulation of VEGF. Meanwhile, the study by Yang et al. (2014) also revealed that CXCR4 is a prognostic factor that influences the survival of breast cancer patients after visceral metastasis. More importantly, increased CXCR4 expression is correlated with many clinicopathological features of NSCLC patients, particularly the metastasis and prognosis of NSCLC (Wang et al., 2015). Therefore, researchers have begun to investigate the role of CXCR4 in the metastasis of NSCLC. A previous study revealed that a hyperactivated Hippo-YAP signaling pathway can induce CXCL5 expression in cancer cells via the modulation of the YAP-TEAD complex and the recruitment of myeloid-derived suppressor cells (MDSCs) (Wang et al., 2016a). In addition, Sharif et al. (2015) also found that cell growth density can modulate cancer cell vascular invasiveness by affecting the activity of the Hippo pathway and CXCR2 signaling pathway, suggesting that CXCR may be associated with the Hippo-YAP pathway, and thus involved in the occurrence and progression of the disease. Moreover, the Hippo-YAP pathway may greatly influence both the development and tumorigenesis of lung tissues (Yeung et al., 2016). The Hippo-YAP pathway also regulates the invasion and metastasis of cancer cells, thereby providing an alternative target for therapeutic intervention (Nallet-Staub et al., 2014). Nevertheless, whether CXCR4 can regulate the Hippo-YAP pathway to influence the metastasis of lung cancer and what the specific mechanism is have yet to be elucidated. Therefore, different concentrations of the CXCR4 antagonist WZ811 were administered to A549 cells, and the effects on cell invasion and migration were observed. Additionally, we treated A549 cells with WZ811 and YAP lentiviral activation particles simultaneously to explore whether CXCR4 can modulate the invasion and migration of A549 cells by mediating the Hippo-YAP signaling pathway.
Materials and methods
Cell culture
The normal HBE cell line and NSCLC cell lines (A549, H1299, H1975, SPCA1, and PC9) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin and 100 mg/mL streptomycin, in an incubator at 37°C with 5% CO2. When the cells covered 80% of the visual field under the microscope, they were digested by 0.25% trypsin for passaging, and the cells at logarithmic and stationary phases were used for subsequent experiments.
Cells treatment
Logarithmic growth phase of the 3rd passage A549 cells were cultured in RPMI-1640 medium with 10% fetal calf serum (FCS) in an incubator at 37°C with 5% CO2. When the cell confluence reached 80-90%, they were digested by 0.25% trypsin and then treated with different concentrations of WZ811 (0, 10, 30, and 50 µM) to investigate the effects of WZ811 on the Hippo-YAP signaling pathway, EMT process, invasion, and migration. Besides, the H1299 cell in logarithmic growth phases were treated with different concentrations of WZ811 (0, 10, 30, and 50 µM) to verify the above mentioned effect. Moreover, to further verify whether WZ811 influences the Hippo-YAP signaling pathway to regulate EMT, invasion and migration of A549 cells, we assigned cells into a Control group, WZ811 group (cells treated with 30 µM WZ811), YAP siRNA group (cells transfected with YAP siRNA), and WZ811 + YAP group (cells treated with 30 µM WZ811 and transfected with YAP lentiviral activation particles). YAP lentiviral activation particles are transcription activation system of synergistic activation mediator (SAM), which could specifically and efficiently upregulate the YAP expression through cell lentiviral transduction. Both the YAP siRNA (sc-38637) and the YAP lentiviral activation particles (sc-400040-LAC) were purchased from Santa Cruz Biotechnology, Inc., (CA, USA), and WZ811 was provided by Selleckchem (S2912, Houston, USA).
qRT-PCR
Cells were plated onto 60-mm culture dishes with 5 mL RPMI-1640 medium. When the cell confluence reached approximately 85%, total RNA was extracted using the Trizol reagent (Invitrogen, USA) and the OD260/280 value was determined using a UV spectrophotometer. The extracted RNA was preserved at −80°C for subsequent experiments. Additionally, the software Primer 5.0 was applied to design the primer sequences. The total RNA was reverse transcribed into cDNA on ice following the instructions of the cDNA transcription kit (Thermo Fisher Scientific, USA). qRT-PCR was performed according to the manual of the SYBR Green PCR Master Mix kit (Takara, Japan). With GADPH as the reference gene, the 2−△△Ct method was used to calculate the relative expression of target genes and the experiment was repeated three times independently.
Western blotting
Proteins were extracted from cells and their concentrations were determined using a bicinchoninic acid (BCA) protein assay. The protein samples were loaded with 45 μg per lane. The proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE), followed by transfer onto a polyvinylidene fluoride (PVDF) membrane. Next, the membrane was incubated with 5% bovine serum albumin (BSA) for 1 h of blocking at room temperature before the addition of primary antibodies overnight at 4°C, including anti-CXCR4 (ab124824, 1/1000), anti-E cadherin (ab1416, 1/50), anti-Twist (ab175430, at 1/500), anti-vimentin (ab45939, at 1 μg/mL), anti-Snail (ab53519, 1/500), anti-YAP1 (ab205270, at 1/1000), anti-CTGF (ab125943, at 1 μg/mL), and anti-survivin (ab469, at 1 μg/mL) (all purchased from Abcam (Cambridge, MA, USA). Then, the membrane was washed with TBST [50 mM Tris-HCl (pH 7.5), 500 μL of the wash was suspended in SDS-containing buffer, heated at 95°C, and 50 mM NaCl, 0.5% Tween-20] for 15 min three times and then exposed to the corresponding secondary antibody for 1 h at room temperature. After incubation, the membrane was washed for 15 min three times before detection and visualization using an enhanced chemiluminescence (ECL) substrate kit. With β-actin as the loading control, the gray value of the target bands was analyzed with Image J software (NIH, USA). The experiment was repeated three times independently.
Wound-healing assay
Cells were placed on six-well plates, and the sterilized tip (200 µL) of a pipette was used to scratch a line across the plates. Next, PBS buffer was used to wash the plates three times. Then, serum-free medium was added to the plates, which were observed and photographed under an inverted microscope at 0 and 48 h after incubation. The distance between scratches was measured using the software Image-Pro Plus 6.0. The experiment was repeated three times.
Transwell invasion assay
A transwell assay was performed in Boyden chambers coated with Matrigel according to the instructions of the manufacturer (BD Biosciences, San Jose, CA). A549 cells were placed on the upper side of the Matrigel in the upper chamber while culture medium containing EGF (10 ng/mL) was added into the bottom chamber as a chemoattractant. Later, the invasive cells on the underside of the Boyden chamber membrane were fixed with paraformaldehyde, stained with crystal violet and counted. All experiments were performed independently three times.
Immunofluorescent staining
Cells were cultured in 24-well plates at 5 × 104 cells/well. Later, the culture medium was removed, and the cells were washed with PBS buffer three times. After, the cells were fixed with 4% polyoxymethylene solution for 20 min and washed with PBS buffer three times. Next, the cells were incubated with primary antibodies and then with corresponding secondary antibodies. DAPI was used for nuclear staining. Images were acquired at 37°C using a fluorescence microscope (Zeiss Axiovert 135 TV; Zeiss) equipped with a 20× 0.50 NA air objective lens (Zeiss) and recorded on a Hamamatsu Retiga Exi camera. The experiment was repeated three times.
Dual-luciferase reporter gene assay
Cells were co-transfected with an 8× GTIIC-luciferase plasmid (Addgene, Cambridge, MA) and Renilla luciferase pRL-TK plasmid (Promega, Madison, WI). After 48 h, the cells were collected and transferred into a 96-well plate for analysis using the Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI). Subsequently, the luminescent signaling was detected with the GloMax-96 Microplate Luminometer (Promega, Madison, WI) according to the manufacturer's instructions. The experiment was repeated three times.
Statistical methods
All data were analyzed using the statistical software SPSS 21.0 (SPSS, Inc., Chicago, IL, USA). Measurement data were presented by mean ± standard deviation (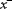 ± s). The comparison among multiple groups was conducted using one-way ANOVA while inter-group differences were analyzed using the least significant difference test (LSD-t). P < 0.05 indicated a statistically significant difference.
± s). The comparison among multiple groups was conducted using one-way ANOVA while inter-group differences were analyzed using the least significant difference test (LSD-t). P < 0.05 indicated a statistically significant difference.
Results
Expression of CXCR4 in different NSCLC cell lines
The expression of CXCR4 in HBE cells and different NSCLC cell lines (A549, H1299, H1975, SPC-A1, and PC9) was detected by qRT-PCR and Western blot. According to the results, CXCR4 expression was dramatically up-regulated in A549, H1299, H1975, SPC-A1, and PC9 cells when compared with that in HBE cells (all P < 0.05). Additionally, A549 cells had the highest expression of CXCR4, so the A549 cell line was chosen for subsequent experiments (Figure 1A–C).
The impact of different concentrations of WZ811 on the migration and invasion of A549 cells
A wound-healing assay was utilized to detect the migration ability of A549 cells. By observing the dynamic changes of cells in each group at the 0 h and 48 h after scratching, we found that WZ811 could effectively reduce the migration ability of cells (P < 0.05) (Figures 2D and 2E). Meanwhile, the results of the transwell invasion assay showed that compared with A549 cells in the Control group, the number of invasive cells treated with WZ811 was statistically decreased (P < 0.05) (Figures 3D and 3F). Of note, the migration and invasion abilities of cells decreased with increasing WZ811 concentration.
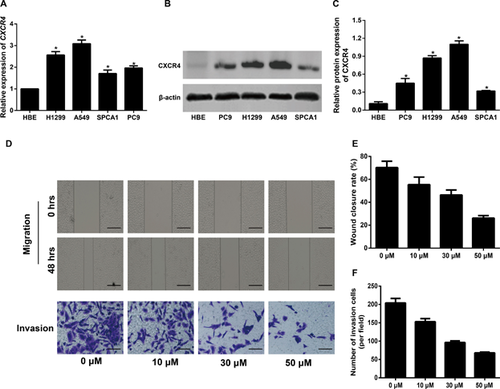
The impact of different concentrations of WZ811 on the EMT phenotype in NSCLC cells
The expressions of EMT-related proteins was detected by qRT-PCR and Western blot after A549 cells and H1299 cell were treated with different concentrations of WZ811. WZ811 remarkably up-regulated the expression of E-cadherin and reduced the expression of Twist, vimentin, and Snail, all in a concentration-dependent manner (all P < 0.05, Figure 2A–F). Additionally, the immunofluorescent staining showed results consistent with the qRT-PCR and Western blot analyses, regarding E-cadherin expression in A549 cells (Figure 2G). And the WZ811 decreased the expressions of N-cadherin and Vimentin in a concentration-dependent manner in A549 cells by using the immunofluorescent staining (Figure 2G).
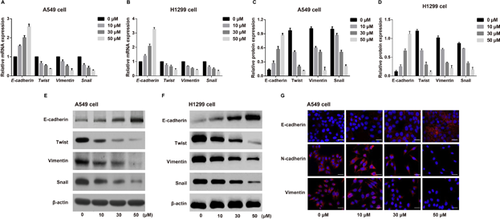
Effects of different concentrations of WZ811 on the Hippo-YAP pathway in A549 cells
After A549 cells were treated with WZ811, the phosphorylation of YAP decreased significantly, and the mRNA and protein expression of Hippo pathway downstream genes (CTGF and BIRC5) also decreased substantially. Additionally, WZ811 inhibited the GTIIC reporter activity of the Hippo pathway, suggesting that WZ811 can block the Hippo-YAP signaling pathway in A549 cells in a concentration-dependent manner (Figure 3).
WZ811 blocks the Hippo-YAP pathway to inhibit the EMT phenotype, invasion, and migration of A549 cells
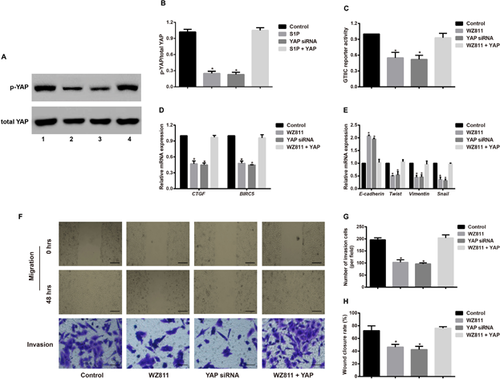
To further verify the impact of the WZ811-mediated Hippo-YAP signaling pathway on the EMT phenotype, invasion, and migration of A549 cells, we compared the YAP siRNA group with the Control group and found that YAP siRNA can suppress the p-YAP expression and GTIIC reporter activity, block the mRNA expression of downstream genes (CTGF and BIRC5), enhance E-cadherin expression, reduce the expression of Twist, vimentin and Snail, and inhibit cell invasion, and migration. The results from YAP siRNA were similar to WZ811 (30 µM) in A549 cells. However, when cells were simultaneously treated with WZ811 and YAP lentiviral activation particles, we observed an increase in the EMT phenotype, invasion, and migration when compared with the results from the YAP siRNA group and the WZ811 group; however, there was not an observable difference from the results in the Control group (Figure 4).
Discussion
CXCR4, consisting of 352 amino acids, is a highly reserved G-protein coupled 7-transmembrane chemokine receptor (Peng et al., 2018). In fact, abnormally high expression of CXCR4 has been observed in a variety of human tumors, including lung cancer, esophageal cancer, gastric cancer, endometrial cancer, and breast cancer (Hartmann et al., 2005; Kaifi et al., 2005; Yasumoto et al., 2006). In this study, we also found higher expression levels of CXCR4 in NSCLC cell lines than in HBE cells, suggesting that CXCR4 may act as an oncogene in the occurrence and development of NSCLC.
After treating A549 cells with different concentrations of a CXCR4 antagonist WZ811 (0, 10, 30, and 50 µM), we found that WZ811 can effectively reduce the invasion and migration ability of cells, indicating that decreased CXCR4 expression can block the metastasis of NSCLC cells. Similarly, the study by Lefort et al. (2017) revealed that two CXCR4 inhibitors, AMD3100 and TN14003, can significantly block the growth and metastasis of both Herceptin-sensitive and Herceptin-resistant HER2 breast cancer (Lefort et al., 2017). Consistent with our finding, Dragoj et al. (2017) also observed that WZ811 could dramatically reduce the migration and invasion of NSCLC cells in vitro. The possible explanation is that inhibiting CXCR4 expression can block the EMT phenotype and thereby suppress the invasion and migration of tumor cells, including NSCLC cells (Yin et al., 2016; Liu et al., 2017). EMT refers to the biological process during which epithelial cells transform into interstitial cells under specific physiological and pathological conditions (Hardin et al., 2014). The major manifestations of EMT include decreased expression of E-cadherin, the loss of cell polarity, and enhanced cell migration and invasion. Therefore, EMT plays a critical role in the metastasis of cancers (Hou et al., 2016; Kim et al., 2016; Zhou et al., 2016). EMT is often accompanied by decreased expression of epithelial markers, such as E-cadherin, and increased expression of interstitial phenotype markers, such as Twist, vimentin, and Snail (Lu et al., 2017; van Zijl et al., 2009). In this study, we observed that WZ811 can up-regulate E-cadherin expression and down-regulate the expression of Twist, vimentin, and Snail, in a concentration-dependent manner. Moreover, immunofluorescent staining was consistent with qRT-PCR and Western blot regarding the expression of E-cadherin, suggesting that cells with high CXCR4 expression may acquire mesenchymal characteristics to promote the invasion, migration and metastasis of tumor cells.
In recent years, accumulating studies have demonstrated that CXCR4 can play an essential role in the development and progression of tumors by mediating the transduction of signaling pathways (Shen et al., 2010; Wang et al., 2008). For example, the Hippo pathway (the Salvador-Warts-Hippo) is correlated with the renewal and differentiation of stem cells and it has been identified as a critical link of oncogenic transformation in NSCLC (You et al., 2015). In fact, YAP, as a Hippo signaling transcriptional coactivator, can greatly affect the proliferation of stem cells, the control of organ size, and the development of tumors through the regulation of downstream transcriptional target genes Ctgf and Birc5/surviving (Liu et al., 2015; Yeung et al., 2016). Additionally, a recent study reported that YAP transcriptional activity was associated with the metastasis of lung cancer (Huang et al., 2015). CXCR3 can effectively impair liver regeneration by reducing the expression of YAP1 (Ma et al., 2016). Moreover, long-term exposure toimatinib mesylate (IM) can lead to the up-regulation of the transcription coactivator YAP, thereby increasing CXCR4 and deregulating the Hippo signaling pathway (Chorzalska et al., 2017), suggesting that CXCR may influence the Hippo-YAP signaling pathway. After treating A549 cells with WZ811, we found that the phosphorylation of YAP was significantly reduced, the GTIIC reporter activity was significantly inhibited, and the expression of Hippo pathway downstream genes (CTGF and BIRC5) was dramatically down-regulated, all in a concentration-dependent manner, suggesting that inhibition of CXCR4 expression in A549 cells can block the Hippo-YAP signaling pathway. The Hippo-YAP signaling pathway is associated with the EMT phenotype, invasion, and migration of many tumors. For example, Wang et al. (2016b) found that inhibiting the Hippo pathway was critical in promoting the migration and invasion of breast cancer cells via Twist-mediated EMT. Additionally, the study by Zhang et al. (2017) revealed that furin may promote EMT phenotype formation in pancreatic cancer cells thorough the Hippo-YAP pathway, thereby constituting a potential therapeutic target to treat pancreatic cancer. Consistent with the aforementioned studies, we also found that YAP siRNA can significantly reduce EMT of A549 cells and inhibit their invasion and migration. Finally, we also found that when A549 cells were concurrently treated with WZ811 and YAP, their EMT phenotype, invasion, and migration was remarkably enhanced compared to cells in the YAP siRNA group and the WZ811 group but was not significantly different from cells of the Control group, further suggesting that WZ811 may influence the Hippo-YAP signaling pathway to affect EMT, invasion, and migration of A549 cells.
In summary, CXCR4 was highly expressed in different NSCLC cell lines. Additionally, the CXCR4 antagonist WZ811 significantly blocked the Hippo-YAP signaling pathway; up-regulated expression of E-cadherin; reduced expression of Twist, vimentin, and Snail; and inhibited migration and invasion of A549 cells, all in a concentration-dependent manner. After A549 cells were treated with WZ811 and YAP simultaneously, they significantly increased the EMT phenotype, invasion, and migration compared to those in the YAP siRNA group and WZ811 group.
Acknowledgments and Funding
We thank all subjects who participated in this study. We acknowledge the helpful comments on this paper received from our reviewers.
Conflicts of interest
The authors declare that they have no competing interests.




