A novel recombinant Salmonella vaccine enhances the innate immunity of NK cells against acute myeloid leukaemia cells Kasumi-1 in vitro
Abstract
Minor histocompatibility antigen HA-1-specific cytotoxic lymphocyte (CTL) clones have apparent anti-leukaemic efficacy, and the AML/ETO gene is a special fusion gene in leukaemic cells. Thus, we hypothesised that a vaccine targeting HA-1 and AML/ETO could stimulate NK cells to target leukaemia cells. Furthermore, we packaged the vaccine using attenuated Salmonella to enhance its immuno-activity. Expression of the NK cell-activating ligand ULBP2 was notably elevated upon packaging in a co-recombinant group. An AML/ETO single plasmid gave the weakest vaccine. The level of miR-182, which targets ULBP2, significantly decreased with increasing IFN-γ and granzyme B in a co-recombinant group. In summary, DNA vaccines including AML/ETO and HA-1 fragments significantly enhance the innate immunity of NK cells in vitro.
Introduction
Activation of the immune system as an oncolytic agent is of interest because of its ability to induce death in a broad range of human malignancies without affecting normal cells (Thomas and Badini, 2011). The innate immune system is primordially distinguished from the adaptive immune system because it is not subject to clonal selection and may develop immunological memory. However, new evidence suggests that a specific subset of mouse NK (natural killer) cells can develop long-lived and highly specific memory towards a variety of antigens (Paust and von Andrian, 2011). NK cells derived from bone marrow granular lymphocytes are effector lymphocytes of the innate immune system and are endowed with a constitutive cytolytic function (Meazza et al., 2011). Unlike T cells, NK cells immediately lyse target cells possessing appropriate activation signals without having to undergo effector differentiation (Gayoso et al., 2011). In vitro studies have shown that tumour cells are NK cell targets. In vivo analyses have relied on the antibody-mediated depletion of mouse NK cells including antibodies targeting either NK1.1 or the glycolipid asialo-GM1. However, due to the lack of Ab specificity, Ab-based depletion also hampers NK cell populations. Recent advances in NK cell biology have stimulated interest in manipulating these cells for anti-tumour responses (Gillgrass and Ashkar, 2011; Levy et al., 2011). Cells that express high levels of activating ligands and low levels of inhibitory ligands are candidate targets for NK cell-mediated attack. Several viruses, including natural viruses and those that are artificially generated via immune-activating gene insertion, stimulate anti-tumour immune responses and lead to direct oncolysis, suggesting that exogenous DNA may potentially be used as an immunotherapeutic and cytotoxic agent (Prestwich et al., 2008; Vidal et al., 2011). In human and mouse models, reovirus can either activate dendritic cells (DCs) to stimulate innate NK/T cell cytotoxicity against tumour cells or directly induce tumour cell apoptosis (Errington et al., 2008). It is not known whether the preprocessing of tumour-derived cells with exogenous viral DNA facilitates the priming of innate and adaptive anti-tumour responses to enable the autonomous behaviour of transformed cells. HA-1 is exclusively expressed in haematopoietic cells, and HA-1-specific CTL clones have apparent anti-leukaemic efficacy (Sellami et al., 2010). AML/ETO is a special leukaemic fusion gene (Dunne et al., 2011). We have inserted recombinant plasmids containing the HA-1 and AML/ETO immunogenic genes into Salmonella, and that infecting AML/ETO-positive Kasumi-1 cells with the resulting Salmonella triggers cytotoxicity along with the secretion of cytokines and chemokines. The underlying signalling pathways involved in the elevation of an NK cell-activating receptor and changes in miR expression have also been examined.
Materials and methods
Vector construction
The AML/ETO plasmid was purchased from Addgene (Cambridge, MA, USA), and full and mini HA-1 plasmids were generously provided by Prof. Dr. E.A.J.M. Goulmy of the Department of Immunohematology and Blood Transfusion and the Department of Hematology, Leiden University Medical Center, Leiden, the Netherlands and the Institute for Transplantation Diagnostics and Cell Therapeutics, Heinrich Heine University, Dusseldorf, Germany. The AML/ETO gene was cloned into the full and mini HA-1 plasmids, meaning HA-1F and HA-1M (the sequence shown in the Supporting Information, Appendix S1). A 207 bp DNA fragment was PCR amplified from the Addgene plasmid template with primers. Because the mini HA-1 plasmid does not contain BamHI or SacI restriction sites, we designed the following primers: 5′-AGGGATCCATGGAAGTGGAAGAGGGAAAAGC-3′ (forward) and 5′-ATGAGCTCGGGGGAGGTGGCATTGTT-3′ (reverse). For the full HA-1 plasmid, the reverse primer was altered to 5′-ATGGATCCGGGGGAGGTGGCATTGTT-3′ because the SacI restriction site was contained in the vector. The 207 bp PCR product was digested with BamHI with or without the SacI enzyme and cloned into the restriction enzyme site. Salmonella (SL7207) was kindly provided by Prof. Jiangdong Huang (Li Ka Shing Faculty of Medicine, The University of Hong Kong). The transformation of competent Salmonella cells was done by electroporation (Bio-Rad, Hercules, CA). Transformants containing single or double gene+ plasmids were selected on LB agar plates containing ampicillin. All plasmids were verified by sequencing.
Cell culture and Kasumi-1 cell infection
Kasumi-1, a leukaemic cell line containing a t(8; 21) chromosomal translocation, produces the chimeric AML1–ETO protein. The Kasumi-1 cell line was a generous gift from Prof. Jie Jin of The First Affiliated Hospital of Zhejiang University. Cells were cultured in RPMI 1640 medium supplemented with 10% foetal bovine serum. Prior to infection with Salmonella, Kasumi-1 cells were washed and resuspended in fresh culture media lacking antibiotics. Kasumi-1 cells were seeded for 1 h at 2 × 105 cells/mL in cell culture bottles. Bacterial cells were added to the Kasumi-1 culture in a 15:1 ratio (bacteria: Kasumi-1 cells), and the infection was allowed to proceed for 2 h at 37°C. Extracellular bacteria cells were killed by the addition of 100 mg/mL gentamicin (Sigma, St. Louis, MO, USA). Cells were harvested 24 h after infection to detect viability with trypan blue, the apoptosis ratio by FCM and ultrastructure by TEM.
Immature NK cell cultures derived from peripheral blood monocytes
Human immature NK cells were cultured from peripheral blood monocytes. Briefly, human PBMCs from healthy donors were isolated by processing the buffy coat with density gradient centrifugation using Ficoll–Hypaque (Pharmacia Biotech, Uppsala, Sweden). After a 1 h incubation to allow adherence, the culture dishes were rinsed with HBSS, and adherent cells were incubated overnight. The base medium for NK92 cells was alpha-minimum essential medium containing 2 mM L-glutamine and 1.5 g/L sodium bicarbonate without ribonucleosides and deoxyribonucleosides. To make complete growth medium, the following components were added to the base medium: 0.2 mM inositol, 0.1 mM 2-mercaptoethanol, 0.02 mM folic acid, 100–200 U/mL recombinant IL-2, 12.5% horse serum and 12.5% foetal bovine serum. After 72 h of culture, cells were collected and isolated with an NK cell isolation kit (Miltenyi Biotec, Germany).
Transmission electron microscopy cell processing
Cells were centrifuged at 2,000g for 5 min, prefixed in a solution containing 2% glutaraldehyde at room temperature for 48 h, washed three times in 0.1 M phosphate-buffered saline (pH 7.2), postfixed in 1% aqueous OsO4 for 2 h and washed again three times in 0.1 M phosphate-buffered saline (pH 7.2). The sample was placed in isoamyl acetate for 20 min before being dehydrated in a series of ethanol-water washes from 25 to 100%. Finally, the samples were dried using a critical-point drying apparatus, firmly mounted and sputter coated with a thin layer of gold before examination in a Hitachi S-570 electron microscope.
Cytotoxicity assay
NK cell cytotoxicity was determined after co-culturing with Kasumi-1 cells at effector-to-target (E:T) ratios ranging from 32:1 to 8:1. Effector cells were incubated with 5,000 target cells in 200 µL fresh NK cell culture medium in each well of a 96-well plate. After 4 h of co-culture, supernatants were prepared by centrifugation at 3,000 rpm and divided into two groups. One group was used for the ELISA assay, while the other was used for a CytoTox 96® non-radioactive cytotoxicity assay. The following equation was used to calculate the cytotoxicity percentage: (experimental − effector spontaneous − target spontaneous)/(target maximum − target spontaneous) × 100.
Real-time RT-PCR
After 4 h of infection with recombinant bacteria and co-culture with NK cells, total RNA was isolated from Kasumi-1 cells using TRIzol reagent (Invitrogen, USA). cDNA was synthesised from 1 µg total RNA using SuperScript III Reverse Transcriptase (Invitrogen) and miR-182 primers (Jima Com, Shanghai, China) in a 20 µL reaction. Samples were resuspended in 20 µL of the SYBR Green SuperMix (Invitrogen) containing the appropriate primers (Jima Com). Primers for U6 and miRs were purchased from Jima. The following NKG2D ligand primers were generated: ULBP1, 5′-TGCAGGCCAGGATGTCTTGT-3′ (forward) and 5′-CATCCCTGTTCTTCTCCCACTTC-3′ (reverse); ULBP2, 5′-CCCTGGGGAAGAAACTAAATGTC-3′ (forward) and 5′-ACTGAACTGCCAAGATCCACTGCT-3′ (reverse); ULBP3, 5′-AGATGCCTGGGGAAAACAACTG-3′ (forward) and 5′-GTATCCATCGGCTTCACACTCACA-3′ (reverse); ULBP4, 5′-TATGTCGACCTCCACAGTATGCGAAGAATATCCCTG-3′ (forward) and 5′-ATAGGCGGCCGCAGACTAAGACGTCCTCAA-3′ (reverse); GAPDH, 5′-GCCATCAATGACCCCTTCATT-3′ (forward) and 5′-TTGACGGTGCCATGGAATTT-3′ (reverse). The U6 and GAPDH expression levels were individually evaluated for each sample as a total RNA control. Each sample was assessed in triplicate to ensure the reproducibility of the quantitative measurements. Quantitative RT-PCR involved a LightCycler® 480 (Roche, Basel, Switzerland). Each gene was assessed on two identical array plates, which were loaded with equal amounts of cDNA in each well. The relative expression levels were calculated by the 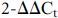 method. The mean ± SEM values were calculated from three independent experiments.
method. The mean ± SEM values were calculated from three independent experiments.
Apoptosis assay and the phenotypic characterisation of Kasumi-1 cells
Apoptotic cell counting followed Annexin V-FITC and PI staining (BioVision, USA). After a 24 h infection with Salmonella, cells were gently vortexed and resuspended in binding buffer at 3 × 106 cells/mL. Cell suspension (100 µL) was mixed with 5 µL Annexin V-FITC and 10 µL PI for 15 min in the dark at room temperature. Then 400 µL of PBS was added to the solution for analysis by FACScan (Becton Dickinson, San Jose, CA, USA) using CellQuest (BD Bioscience). To follow the Kasumi-1 cell phenotype, fluorescein isothiocyanate-, phycoerythrin- and allophycocyanin-labelled monoclonal antibodies specific for the following antigens were employed: ULBP1 (Clone: 170818), ULBP2 (Clone: 165903), ULBP3 (Clone:166510) and ULBP4 (FAB6285C) (all from R&D Systems, Inc., Minneapolis, MN, USA), and appropriate isotype-matched controls were included (BD Biosciences).
Measurement of IFN-γ and granzyme B secretion by ELISA
Supernatants from all groups were harvested, and IFN-γ (Human IFN-γ ELISA Kit II, BD Pharmingen, USA) and granzyme B (Granzyme B Human ELISA Kit, Diaclone, Stamford, CT, USA) secretion quantified. Briefly, supernatants and standards (100 µL) were each placed in precoated 96-well enzyme-linked immunosorbent assay (ELISA) plates and incubated for 2 h at 24°C. After washing, the plates were incubated with an equal volume of detection antibody for 1 h at 24°C. The plates were washed and incubated for 1 h with streptavidin–horseradish peroxidase at 24°C. After substrate addition, the plates were analysed in an ELISA plate reader (model 680; BioRad, Hercules, CA, USA).
Chip detection of miRs altered in Kasumi-1 cells
Based on the results from the above tests, we chose the AML/ETO/HA-1M and control groups to analyse changes in miR expression. Total miRNA from 1 × 108 cells was isolated using the mirVana™ miRNA isolation kit. A total of 186 miR genes were analysed and contained 153 miRs identified in the miR registry at http://rfam.sanger.ac.uk/, and 36 additional miRs that were either manually curated from published papers or extracted from GenBank at www.ncbi.nlm.nih.gov. A total of 19 human miRs (10%) were found by homology with cloned miRs from other species (mainly mouse). For all of these miRs, we determined the sequence of the precursor using the M Zucker RNA folding programme at http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form and selecting the precursor sequence that gave the best hairpin structure score.
Target in vitro and reverse test
Based on the miR array results and QRT-PCR, we predicted that the UUGCCAA sequence located at the 181–187 bp position was the seed sequence for miR-182, which regulates ULBP2. We cloned the ULBP2 3′-UTR sequence from Kasumi-1 cell total DNA using the following primers: 5′-CTCCTGTGAGCACGGTCTT-3′ (forward) and 5′-TTGTTTAGTCAGCCAGAA-3′ (reverse). The ULBP2 sequence was subsequently inserted into the pMIR-Check2 vector (Promega, USA). We measured the target gene change at the mRNA and protein levels by QRT-PCR and WB, respectively. All of the seed sequences were mutated from A to T using the Quick Change Kit (Stratagene, USA). Empty vector (pMIR-Check2) was used as a negative control. HEK-293 cells were transfected with 0.2 µg of the reporter plasmids, 0.01 µg of a pMIR-Check2 β-gal control plasmid, 200 nM of miR-182 mimics, a mimic control, called NC (Shanghai GenePharma Company, Shanghai, China), which has the same amount of bases with different sequence, per well in 96-well plates. Another control is the MO group, treated only with lipofectamine 2000. After 24 h of incubation, cells were assayed by the Luciferase Assay System (Promega, USA), normalised to β-galactosidase activity. All experiments were done in triplicate. To verify whether miR-182 and ULBP2 play any important role, we detected the immuno-sensitivity of Kausimi-1 cell pre-infected with recombinant plasmids, when we overexpressed miR-182 by its mimic or downregulated the ULBP2 by siRNA and its neutralising antibody respectively (Human ULBP-2 MAb 500 µg, MAB1298, R&D Systems, Inc.). As for ULBP2 siRNA sequence, sense is 5′-GGA CAU ACU UAC AGA GCA A-dTdT-3′ and antisense is 5′-UUG CUC UGU AAG UAU GUC C-dTdT-3′.
Western blot analysis
HEK-293 cells were transfected with a miR-182 mimic (100 nM) or an AMO inhibitor (200 nM). The cells were lysed in RIPA buffer in the presence of proteinase AMO (Shanghai Shenergy Biocolor BioScience & Technology Company, Shanghai, China). Protein concentration was determined by BCA (Bios, Beijing, China). Aliquots (25 mg) were separated in a 10% SDS–PAGE gel and transferred to a nitrocellulose membrane. The membranes were probed with primary antibodies directed against ULBP2 (sc-80419 mouse monoclonal; Santa Cruz, USA) at room temperature for 2 h, washed extensively with 0.1% Tween-20 in phosphate-buffered saline and incubated with secondary antibodies conjugated with horseradish peroxidase at a 1:1,000 dilution. The signals were detected using a fluorescently labelled antibody.
Results
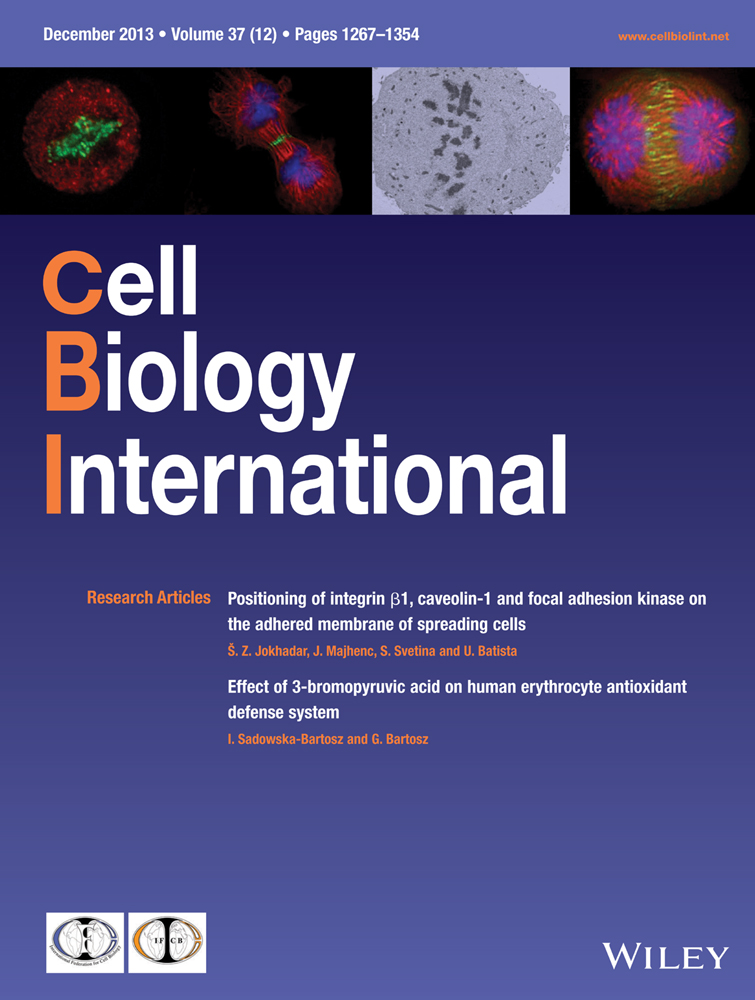
Effect of intracellular parasite infection on tumour cell viability and apoptosis
We constructed recombinant plasmids that were subsequently transfected into competent cells derived from the SL7207 Salmonella infection. The transfections were verified by sequencing. Salmonella is a facultative intracellular parasite. Kasumi-1 cells were incubated with a vaccine containing SL7207 derivatives for 1 h in medium lacking antibiotics and gentamicin – supplemented medium to block extracellular bacterial growth. TEM was used to confirm that SL7207 had efficiently infected tumour cells (Figure 1). Trypan blue and apoptosis data suggested that the infection was too weak to affect Kasumi-1 cell viability (Figure 1).
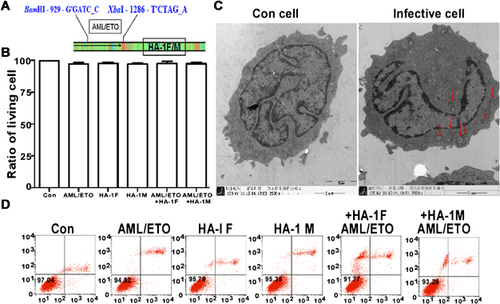
Infected Kasumi-1 cells have a high NKG2D ligand expression level and are sensitised to NK cell-mediated lysis
Bacteria or bacterial products are seen as promising applications for therapeutic cancer treatment. NK cells are effector lymphocytes of the innate immune system, which is the first line of defence against infection and tumours. The upregulation of NKG2D ligands can render tumour cells susceptible to NK cell-mediated lysis. How bacterial vaccination affects the regulation of NKG2D ligand expression remains unclear. To elucidate this point, the ULBP1–4 mRNA level was assessed in Kasumi-1 cells infected with various vaccines that were previously confirmed by QRT-PCR. ULBP1 and 2 were positive two- to threefold changes in double gene plasmid groups (P < 0.05; Figure 2A). In contrast, ULBP3 and 4 showed no obvious change. Based on the QRT-PCR results, we also assessed the change in the ULBP1–4 protein level by FCM (Figure 2B). Furthermore, the SL7207-based vaccines improved NK cell tumour surveillance and cytotoxicity (Figure 2C).
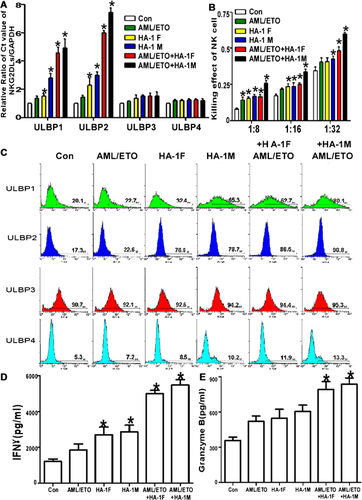
miR-182 regulated ULBP2 expression
Based on the miR array results (Figure 3A), we measured the miR-182 level in all groups. We observed a significant difference in miR-182 expression between the control and HA-1M-, AML/ETO + HA-1F- and AML/ETO + HA-M-infected cells (Figure 3B). We predicted the genes targeted by this miR using TargetScan software (http://www.targetscan.org/) and found a miR-182 ‘seed sequence’ in the 3′-UTR of ULBP2 (Figure 3C). We next examined whether miR-182 regulated ULBP2 transcription in HEK293 cells after transfection with miR-182 mimics or an NC using the luciferase method (Figure 3D). Furthermore, this regulatory effect was verified by assessing the expression of ULBP2 at the mRNA and protein levels by QRT-PCR and Western blot, respectively, after transfection with miR-182 mimics or an AMO in Kasumi-1 cells. The results show that miR-182 significantly regulates ULBP2 transcription (Figures 3E and 3F).
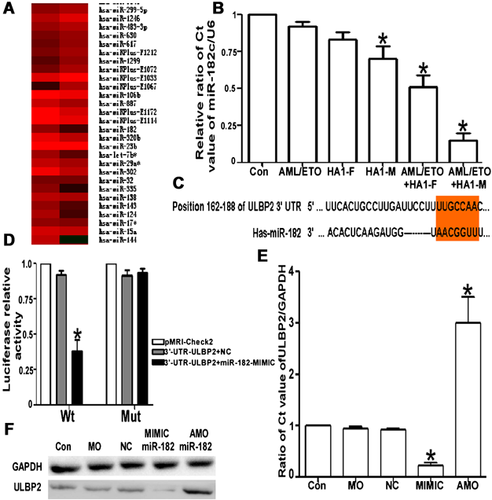
Interfering with miR expression reverses the vaccine effect
We previously showed that miR-182 plays an important role in the Kasumi-1 cell activation process induced by an SL7207-based DNA vaccine, but reintroduction experiments are more powerful. Kasumi-1 cells, which were infected with AML/ETO and HA-1M recombinant plasmids, were treated with miR-182 mimic, siRNA-ULBP2 and neutralising antibodies to inhibit the effect of ULBP2. The sensitivity of Kasumi-1 cells to NK cell-induced death was repressed and comparable with the ULBP2 expression level, which was significantly decreased at the mRNA and protein levels (Figures 4A and 4B). Thus, both the upregulation of miR-182 expression and suppression of ULBP2 counteracted the effects of an SL7202-based DNA vaccine (Figures 4D and 4E; P < 0.05).

Discussion
Minor histocompatibility antigens (mHags) are immunogenic peptides derived from polymorphic cellular proteins. From a clinical point of view, HA-1 appears to be one of the most interesting mHags (Mutis et al., 2010). HA-1-specific CTL clones efficiently display cytotoxic specificity against mHag-positive leukaemic cells from patients with AML and ALL but are not effective against non-haematopoietic cells (Rice et al., 2006; Hambach et al., 2010; Kim et al., 2011). As for immunologic reconstitution after allogeneic bone marrow transplantation, NK cells are in the first recovered lymphocyte subsets, which identify NK cells may are critical regulators in early stage of GVL. Whether NK cell has memory ability, how it is constituted and activated requires more investigation. Here we studied the effect of HA-1–AML/ETO recombinant double genes vaccine on NK cells. The t(8;21)(q22;q22) translocation, which occurs in 40% of patients with acute myeloid leukaemia that have an FAB-M2 subtype, results in the expression of AML1–ETO (RUNX1–CBF2T1) (Tonks et al., 2004), which may contribute to leukaemogenesis because of its interaction with nuclear corepressor complexes, including histone deacetylases. To optimise the effectiveness of such immunotherapy, we assembled HA-1 and AML/ETO vaccines, which both have a unique hematopoietic-restricted tissue distribution, to induce strong immune responses and enhance GVL. We found, as for NK activating effect, there were difference among HA-1M, HA-1F and AML/ETO single vaccines. The HA-1M plasmid has the most prominent stimulation effect on Kasumi-1 cells, almost double that of the AML/ETO plasmid. Correspondingly, the double genes plasmid, HA-1M combined with AML/ETO, is more powerful than the other. In any case, the double gene plasmids undoubtedly have more effect in promoting of ULBP2 expression and immuno-sensitivity of Kasumi-1 cell.
One major obstacle to achieving a tumour-specific response with a vaccine is immunological recognition, making it necessary to use an immunostimulatory messenger. Bacteria have been previously proposed as anticancer agents (Cronin et al., 2012). Live bacteria have the natural ability to sense the external environment and penetrate target tissues. Salmonella (Gram-negative bacterium) (Momiyama et al., 2012) infection is the most common non-sporulating and non-capsular infection in humans that occurs primarily via flagella and fimbriae. Salmonella has been widely used as a delivery vector for cytokines, chemokines, tumour antigens and DNA-based vaccines (Chorobik and Marcinkiewicz, 2011; Hegazy and Hensel, 2012; Zeng et al., 2012), largely because of its ability to preferentially bind to tumour sites. Avogadri et al. (2005) intra-tumourly injected Salmonella typhimurium into a neoplasm in a melanoma mouse model and eliminated the treated mass. Saccheri et al. (2010) demonstrated that connexin 43 (Cx43) was upregulated in human and murine melanoma cells that were administered certain molecules and effectively activated immune cells after Salmonella infection. To improve the therapeutic effects of a recombinant anticancer plasmid, we chose the SL7207-based DNA vaccine as a novel alternative to produce and locally release tumour antigens. NK cells are ideal for this purpose because they are a natural immunologic barrier for tumours (Curigliano, 2011; Stojanovic and Cerwenka, 2011) and bacteria (Souza-Fonseca-Guimaraes et al., 2011). SL7207 efficiently infected Kasumi-1 cells without impairing their viability. The evidence suggests that tumour cells become more susceptible to NK cell-mediated lysis, and NK cells produced a considerable amount of IFNγ and granzyme B. Furthermore, expression of MHC class I chain-related molecules in tumour cells became significantly elevated.
The recognition of ULBPs on tumour cells by NKG2D, the NK cell-activating receptor, induces NK cell cytotoxic activity and the subsequent lysis of their tumour targets. Many studies have reported tumour evasion via internalised or secreted soluble ligands (Hedlund et al., 2011; Morisaki et al., 2011; Sun et al., 2011) that downregulate the NKG2D expression level in NK and T cells, and lead to immune ignorance or tolerance. In our study, ULBP2, but not ULBP3 and ULBP4, increased on the surface of Kasumi-1 cells and contributed in promoting immuno-cytotoxicity. The molecular mechanisms underlying NKG2D ligand expression are ambiguous. MicroRNAs (miRNAs) are small noncoding RNAs that post-transcriptionally regulate the translation of their mRNA targets and are candidates for mediating this process (Lovat et al., 2011; Luo et al., 2011). Thus we analysed the difference in miRs between the control and AML/ETO + HA-1M groups. miR-182, an oncogenic miRNA, was significantly upregulated in melanoma (Segura et al., 2009), lung cancer (Zhu et al., 2011), medulloblastoma (Weeraratne et al., 2012) and ovarian carcinoma (Liu et al., 2012). The overexpression of miR-182 in non-sonic hedgehog-mediated medulloblastoma metastasis and miR-182 knockdown decreased cell migration in vitro. miRs 183, 96 and 182 are more highly expressed in prostate cancer tissue individually or as a cluster compared with normal prostate, and can diminish labile zinc pools and reduce zinc uptake (Mihelich et al., 2011). Our findings are novel concerning the effects of miR-182 on immunoregulation, since itis the most significantly reduced miR. Furthermore, miR-182 regulated ULBP2 via a 7-mer ‘seed sequence’. We provide evidence that ULBP2 is a direct miR-182 target in a luciferase assay. After transfecting Kasumi-1 cells with miR-182 mimics or an AMO, the ULBP2 expression level was altered at the mRNA and protein levels. To investigate the functional significance of miR-182, Kasumi-1 cells were inoculated with an SL7202-packaged vaccine and co-cultured in the presence or absence of a miR-182 mimic. Decrease in miR-182 in Kasumi-1 cells infected with SL7207-containing recombinant plasmids triggered the upregulation of ULBP2 (Figure 4). This effect was reversed by the miR-182 mimic. This result, at least in part, explains why infected Kasumi-1 cells are sensitive to NK cell-mediated cell lysis.
Conclusions
Many experiments have indicated that the control of the ‘dormant’ tumour state is largely dependent on the adaptive immune system. The lack of MHC class I expression or the upregulation of NKG2D ligands could render tumour cells susceptible to NK cell-mediated lysis. We propose two novel aspects that may be used to render tumour cells susceptible to NK cell-mediated lysis. First, we generated a recombinant HA-1 and AML/ETO cDNA vaccine in which the two proteins were exclusively expressed in leukaemic cells and their progenitors because NK cells play a key role in host defence against microbes and bacteria-mediated tumour therapy, which is an emerging treatment paradigm. The vaccine was packaged using the SL7207 Salmonella strain. Encouragingly, the SL7207-based DNA vaccine containing HA-1M and AML/ETO upregulated ULBP2 expression in Kasumi-1 cells, which subsequently modulated NK cell responsiveness. The mechanism for this phenomenon has been explained and evidence provided that miR-182 participates in this process by directly targeting ULBP2.
Conflict of interest
The authors declare that they do not have any conflicts of interest.