Reciprocal tumor-platelet interaction through the EPHB1-EFNB1 axis in the liver metastatic niche promotes metastatic tumor outgrowth in pancreatic ductal adenocarcinoma
Lin-Li Yao, Wei-Ting Qin, Li-Peng Hu, and Tie-Zhu Shi contributed equally to this work.
Abstract
Background
The interaction between the metastatic microenvironment and tumor cells plays an important role in metastatic tumor formation. Platelets play pivotal roles in hematogenous cancer metastasis through tumor cell-platelet interaction in blood vessels. Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal malignancy distinguished by its notable tendency to metastasize to the liver. However, the role of platelet in the liver metastatic niche of PDAC remains elusive. This study aimed to elucidate the role of platelets and their interactions with tumor cells in the liver metastatic niche of PDAC.
Methods
An mCherry niche-labeling system was established to identify cells in the liver metastatic niche of PDAC. Platelet depletion in a liver metastasis mouse model was used to observe the function of platelets in PDAC liver metastasis. Gain-of-function and loss-of-function of erythropoietin-producing hepatocellular receptor B1 (Ephb1), tumor cell-platelet adhesion, recombinant protein, and tryptophan hydroxylase 1 (Tph1)-knockout mice were used to study the crosstalk between platelets and tumor cells in the liver metastatic niche.
Results
The mCherry metastatic niche-labeling system revealed the presence of activated platelets in the liver metastatic niche of PDAC patients. Platelet depletion decreased liver metastatic tumor growth in mice. Mechanistically, tumor cell-expressed EPHB1 and platelet-expressed Ephrin B1 (EFNB1) mediated contact-dependent activation of platelets via reverse signaling-mediated AKT signaling activation, and in turn, activated platelet-released 5-HT, further enhancing tumor growth.
Conclusion
We revealed the crosstalk between platelets and tumor cells in the liver metastatic niche of PDAC. Reciprocal tumor-platelet interaction mediated by the EPHB1-EFNB1 reverse signaling promoted metastatic PDAC outgrowth via 5-HT in the liver. Interfering the tumor-platelet interaction by targeting the EPHB1-EFNB1 axis may represent a promising therapeutic intervention for PDAC liver metastasis.
Abbreviations
-
- ADAM9
-
- A distegrinin and metalloprotease 9
-
- ATP
-
- Adenosine triphosphate
-
- CCK-8
-
- Cell counting Kit-8
-
- CK19
-
- Cytokeratin 19
-
- CLEC-2
-
- C-type lectin-like-2 receptor
-
- DAB
-
- Diaminobenzidine
-
- DAPI
-
- 4ʹ,6-diamidino-2-phenylindole
-
- DCs
-
- Dendritic cells
-
- EFNB1
-
- EphrinB1
-
- EGF
-
- Epidermal growth factor
-
- ELISA
-
- Enzyme-linked immunosorbent assay
-
- EPHB1
-
- Erythropoietin-producing hepatocellular receptor B1
-
- EphB1-Fc
-
- Recombinant EphB1 Fc chimera protein
-
- ERK
-
- Extracellular signal-regulated kinase
-
- FBS
-
- Fetal bovine serum
-
- FDR
-
- False discovery rate
-
- FGF2
-
- Fibroblast growth factor 2
-
- FPKM
-
- Fragments Per Kilobase of transcript per Million mapped reads
-
- GFP
-
- Green fluorescent protein
-
- GO
-
- Gene Ontology
-
- GPVI
-
- Glycoprotein VI
-
- GSEA
-
- Gene set enrichment analysis
-
- GSVA
-
- Gene set variation analysis
-
- H&E
-
- Hematoxylin and eosin
-
- HEPES
-
- 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
-
- HexA
-
- Hexosaminidase A
-
- HRP
-
- Horseradish peroxidase
-
- IL1β
-
- Interleukin-1β
-
- Integrin α6β1
-
- Integrin alpha6beta1
-
- IVIS
-
- In vivo imaging system
-
- JNK
-
- c-Jun N-terminal kinase
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- KPC
-
- KrasG12D/+; Trp53R172H/+; Pdx-1-Cre
-
- LGALS3
-
- Galectin-3
-
- MAPK
-
- Mitogen-activated protein kinase
-
- NES
-
- Normalized enrichment score
-
- NK
-
- Natural killer cell
-
- One-way ANOVA
-
- One-way analysis of variance
-
- PBS
-
- Phosphate buffered saline
-
- PDAC
-
- Pancreatic ductal adenocarcinoma
-
- PDGFα
-
- Platelet-derived growth factor α
-
- PDPN
-
- Podoplanin
-
- PF4
-
- Platelet factor 4
-
- PI3K
-
- Phosphatidylinositol 3-kinase
-
- QPCR
-
- Quantitative real-time polymerase chain reaction
-
- SD
-
- Standard deviations
-
- SDF-1α
-
- Stromal cell-derived factor-1α
-
- SERT
-
- 5-HT transporter
-
- SNARE
-
- Soluble N -ethylmaleimide-sensitive factor attachment protein receptor
-
- SRA
-
- Sequence Read Archive
-
- TGF-β1
-
- Transforming growth factor β1
-
- TMA
-
- Tissue microarray
-
- Tph1−/−
-
- Tph1 knockout
-
- Tph1
-
- Tryptophan hydroxylase 1
-
- VEGF
-
- Vascular endothelial growth factor
-
- WB
-
- Western blotting
-
- WT
-
- Wild-type
1 BACKGROUND
Cancer cell dissemination may be initiated early in tumor progression, but the majority of disseminated cells do not successfully establish colonies in distant organs. The survival, quiescence, and outgrowth of disseminated cells depend on interactions between tumor cells and the metastatic microenvironment including stromal cells, extracellular matrix and other factors, which create a supportive environment within the host tissue, facilitating the progression of tumor cells [1-4]. Therefore, fully understanding the metastatic microenvironment and identifying the key genes in tumor cells involved in this crosstalk to promote metastatic tumor formation is important for exploring effective approaches for the treatment of cancer metastasis.
Pancreatic ductal adenocarcinoma (PDAC) is highly lethal with a mere 13% 5-year survival rate [5]. The main reason for the poor outcome is early distant metastasis, mostly to the liver [6]. The majority (∼80%) of PDAC patients already exhibit liver metastasis at the time of diagnosis, and more than 60% of patients experience liver recurrence following surgical resection of primary pancreatic tumors [7, 8]. RNA-sequencing (RNA-seq) data have revealed a transcriptomic difference between distant metastases and primary tumors that contain microenvironment tissues, suggesting a different microenvironment between primary and metastatic tumors [9-11]. The unique nonparenchymal cell populations in the liver microenvironment include liver sinusoidal endothelial cells [12], hepatic stellate cells, Kupffer cells and other immune cells [13]. However, the function of other cellular components in the liver metastatic niche remains unclear.
Platelets, which are the smallest anuclear cytoplasmic bodies released from megakaryocytes in the bone marrow, not only serve as key contributors to thrombosis and hemostasis but also play pivotal roles in various pathophysiological processes, such as atherogenesis, inflammation and cancer [14]. Recent evidence indicates that platelets play a role in metastasis and adversely impact the prognosis of cancer patients with abnormal platelet counts or dysfunctional platelet function [15]. Antiplatelet therapy has been reported to be associated with reduced cancer development, including liver cancer [16], breast cancer [17] and head and neck cancer [18]. Extensive studies have focused on their role within blood vessels, including safeguarding tumor cells from shear forces and assault by Natural killer cells, summoning myeloid cells through the secretion of chemokines, orchestrating the arrest of the tumor cell platelet embolus at the vascular wall, and facilitating extravasation in remote organs [15]. However, a recent study demonstrated that platelets could enhance CD8+ T cell-mediated antitumor immunity in liver cancer, suggesting the complex roles of platelets in tumor regulation [19]. Moreover, an elevated mean platelet volume has been shown to be correlated with poorer survival outcome in PDAC patients with synchronous liver metastases, suggesting the participation of platelets in patients with PDAC [20]; however, their role needs experimental validation.
Hundreds of different molecules are stored in the dense, α and lysosomal granules of platelets. Different agonist stimulants in platelets selectively release certain cargo proteins [21], which regulate angiogenesis [22, 23], cell proliferation, adhesion, and immune responses [24, 25]. The molecules secreted by activated platelets are likely the major drivers of PDAC outgrowth in the liver. Serotonin, also known as 5-HT, is a major molecule that is stored in dense granules of platelets. Although it is well known as a neurotransmitter with critical cognitive and behavioral functions, approximately 95% of 5-HT is produced by enterochromaffin cells in the human gut and is stored in platelets. There is growing evidence linking 5-HT signaling to tumorigenesis and tumor progression, including PDAC [26, 27].
In the current study, we aimed to establish a metastatic niche labeling system to detect platelets in the liver metastatic niche of patients with PDAC and to investigate the interaction between tumor cells and platelets in patients with PDAC liver metastasis.
2 MATERIALS AND METHODS
2.1 Clinical samples
Human PDAC tissues, adjacent normal pancreas tissues and liver tissues with PDAC liver metastasis were obtained at Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University (Shanghai, China). The patients were diagnosed with liver oligometastasis at the first diagnosis before surgery between January 2010 and December 2018. None of the patients had received radiotherapy, chemotherapy, hormone therapy or other related antitumor therapies before surgery. A tissue microarray (TMA) was constructed using cores with a diameter of 1.5 mm, including 35 matched liver metastasis, primary PDAC, and nontumor tissue samples, as previously reported [9]. The overall survival of patients was calculated as the time from surgery to death. The study was approved by the Research Ethics Committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University (2015037). All the patients enrolled in this study provided written informed consent.
2.2 Data mining and bioinformatics analysis
RNA-seq data including synchronous surgically resected adjacent normal tissue, primary tumors and liver metastases from liver oligometastatic PDACs at Ren Ji Hospital, deposited in the Gene Expression Omnibus (GEO) database GSE151580 were used to investigate the differential transcriptomic signatures in PDAC patients with liver metastasis [9]. Among them, 6 patients had adjacent normal tissue-primary PDAC-liver metastatic tumor trios and 7 patients had only primary PDAC- liver metastatic tumor pairs. Therefore, 13 primary PDAC and 6 adjacent normal tissues were used to investigate the differential transcriptomic signatures in primary PDAC tissues from patients with liver metastasis. RNA-seq data from the BioProject database (PRJNA664673), which contains transcriptional data of liver metastatic tumors and livers from mice, were used to analyze the transcriptional changes in liver metastatic tumors [28]. GSE71729 was used to analyze the transcriptional difference between liver metastatic tumors and primary tumors in patients with PDAC [29]. The RNA-seq data were normalized and log2-transformed, and differential analyses were subsequently performed via the R package “Linear Models for Microarray Data”. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed using the KEGG pathway database (https://www.genome.jp/kegg/pathway.html). Gene Ontology (GO) analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/). Gene set enrichment analysis (GSEA) was performed on the Broad Institute Platform (https://www.broadinstitute.org/) and the statistical significance (false discovery rate, FDR) was set at 0.25. Reactome or KEGG gene sets in the GSEA MSigDB resource were used to identify the difference between the liver metastatic tumor and primary tumor, or adjacent tissue. The platelet activation genes were selected from the GSEA gene sets (https://www.gsea-msigdb.org/gsea/index.jsp/). The platelet activation score was determined using gene set variation analysis (GSVA) based on the platelet activation signature genes [30].
2.3 Mice
Tryptophan hydroxylase 1 (Tph1) is the rate-limiting enzyme in the 5-HT biosynthetic pathway in the periphery; therefore, Tph1 knockout mice were used to investigate the role of 5-HT in liver metastasis of PDAC. Tph1 knockout mice (C57BL/6 N-Tph1em1cyagen) were purchased from Cyagen Co., Ltd (Suzhou, Jiangsu, China). Gt (ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo (Rosa26 mT/mG) mice donated by Professor Xing-Xu Huang from School of Life Science and Technology, ShanghaiTech University (Shanghai, China), were used for isolating platelets. All animal experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The mice were housed and handled according to protocols approved by the Institutional Animal Care and Use Committee of Shanghai JiaoTong University. C57BL/6J mice were purchased from GemPharmatech Co., Ltd. (Shanghai, China). All mice used in the animal experiments were male mice aged 6-8 weeks.
2.4 Cell culture
The murine PDAC cell line KPC1199, which was derived from KrasG12D/+;Trp53R172H/+;Pdx-1-Cre (KPC) mice, and the murine PDAC cell line Panc02 were both gifts from Prof. Jing Xue (State Key Laboratory of Oncogenes and Related Genes, Renji-Med X Clinical Stem Cell Research Center, Shanghai Cancer Institute, Renji Hospital, School of Medicine, Shanghai Jiao Tong University). The 293T cells were maintained at the Shanghai Cancer Institute. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) or Roswell Park Memorial Institute 1640 medium (RPMI 1640) supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) antibiotics (penicillin-streptomycin, #15140122, Gibco, Grand Island, NY, USA) at 37°C in a humidified incubator under 5% CO2 conditions as suggested by the American Type Culture Collection (ATCC, Manassas, VA, USA). Cell line authentication was performed before experiment using Short Tandem Repeat (STR) profiling in Panc02 cells and single nucleotide variant (SNV) detection in KPC1199 cells.
2.5 Labeling system
The labeling system was established as previously reported [31]. Briefly, a soluble peptide and a modified trans-acting activator of a transcription peptide were cloned upstream of the mCherry cDNA. The secreted lipid permeable-mCherry (sLP-mCherry) sequence was subsequently cloned and inserted into a pRRL lentiviral backbone (GenePharma, Shanghai, China). Green fluorescent protein (GFP)-labeled KPC1199 cells were stably infected with sLP-mCherry lentiviral particles and subsequently sorted to obtain GFP+mCherry+ KPC1199 cells.
2.6 Mouse models for liver metastasis of PDAC
C57BL/6J mice were used to establish a mouse liver metastasis model of PDAC. A total of 2 × 106 luciferase-transfected KPC1199 or Panc02 mouse PDAC cells were suspended in 25 µL phosphate buffered saline (PBS) and then inoculated into the spleen of mice anesthetized with isoflurane. To monitor the tumors in the liver after KPC1199 cell injection, luciferin emission imaging of isoflurane-anesthetized animals was performed 7 and 14 days after injection using the in vivo imaging system (IVIS) Spectrum (PerkinElmer, Waltham, MA, USA) after intraperitoneal injection of 150 mg of D-luciferin (#P1043, Promega, Madison, WI, USA) into the mice. The mice were humanely euthanized by cervical dislocation after deep anesthesia at 14 days after tumor cell injection, and their livers were collected, fixed and prepared for the histological examination.
For the intrapancreatic xenograft model, luciferase transfected KPC1199 cells at a series of dilution: 1 × 105, 1 × 104 and 1 × 103 were suspended in 25 µL of PBS and then inoculated into the pancreas of C57BL/6J mice. Tumor growth in the mice was monitored via IVIS Spectrum and tumors in the liver were confirmed via IVIS Spectrum after the livers were collected 6 weeks after tumor cell injection or at the time of natural mouse death before 6 weeks.
For the platelet depletion experiments, C57BL/6J mice, Tph1−/− mice or wild-type (WT) control mice were first intrasplenically injected with KPC1199 cells (2 × 106). The mice were then randomly divided into two groups, and injected with 100 µL of Ultra-LEAF™ purified anti-mouse CD41 antibody (clone MWReg30, 0.5 mg/mL, #133940, Biolegend, San Diego, CA, USA) or control IgG into the tail vein on Day 2. CD41 antibody or IgG was injected every 3 days until the mice were euthanized at the end of the experiments.
2.7 Flow cytometry
Liver tissues were collected at 7 days after intrasplenic tumor cell injection and 4 weeks after intrapancreatic tumor cell injection. The liver was perfused using a perfusion medium (#17701038, Gibco), dissociated with Liver Digest Medium (#17703034, Gibco) and filtered through a 30-µm strainer. Single-cell suspensions were centrifuged at 60 ×g at 22°C for 1 min, and the cells in the suspensions were collected. To detect the presence of immune cells and platelets in liver metastatic tumors, single-cell suspensions were incubated with a mixture of labeled antibodies containing immune cell and platelet markers for surface staining for 30 min at 4°C. After two washes with PBS, the cell suspensions were treated with BD Cytofix/Cytoperm™ solution (#554722, BD Biosciences, San Diego, CA, USA) and wash buffer (#554723, BD Biosciences) for fixation and permeabilization and stained with mCherry (#167453, Abcam, Cambridge, MA, USA) followed by an anti-rabbit Alexa Fluor 594 secondary antibody. The cells were then stained using a Zombie Aqua™ Fixable Viability Kit (#423101, Biolegend). Flow cytometry analyses were performed on a BD LSR-Fortessa cell analyzer (BD Biosciences) and FlowJo v.10.4.2 software (BD Biosciences) was used for further analysis. Information on the antibodies is shown in Supplementary Table S1.
2.8 Mouse platelet isolation
Whole blood was drawn via cardiac puncture from anesthetized C57BL/6J male mice using a syringe preloaded with 3.2% sodium citrate and collected in tubes supplemented with 3.2% sodium citrate. Platelet-rich plasma was obtained by centrifugation at 200 ×g for 10 min at 22°C. Platelet-rich plasma was then recentrifuged at 1250 ×g for 10 min after mixing 1:1 (v/v) with Tyrode's buffer with prostaglandin I2 sodium salt (PGI2, #P6188, Sigma-Aldrich, CA, USA). The platelets were washed again with Tyrode's buffer, resuspended in Tyrode's buffer and adjusted to a final platelet count of 1 × 1010/mL.
2.9 Identification of platelets
For identification of platelets isolated from Rosa26 mT/mG mice, washed platelets were stained with fluorescein isothiocyanate (FITC)-conjugated anti-mouse CD41 (1:100, #133903, Biolegend) for 30 min at 4°C. For detection of CD62P expression in a separate coculture system, platelets in the upper chamber were collected in tubes and stained with phycoerythrin (PE) -conjugated anti-mouse CD62P. The stained cells were analyzed via flow cytometry on a BD LSR-Fortessa cell analyzer, and FlowJo v.10.4.2 software was used for further analysis.
2.10 Contact and noncontact coculture of tumor cells and platelets
Tumor cells (5 × 104) and platelets (5 × 107) were cocultured via a cell culture insert (pore size: 0.4 µm; #MCHT24H48, Millipore, Burlington, MA, USA) in a 24-well plate. Six groups were prepared as follows: medium group: DMEM with 2% FBS and no cells; platelet group: platelets were added in the upper chamber; platelet + thrombin group: platelets were added in the upper chamber and stimulated with murine thrombin (1 µg/mL, #HY-114164, MedChemExpress, Monmouth Junction, NJ, USA); KPC1199 cell group: tumor cells were added in the lower chamber; contact coculture group: platelets and tumor cells were added in the lower chamber; and noncontact coculture group: tumor cells were added in the lower chamber, and platelets were added in the upper chamber. After coculture for 2 h, the culture medium was collected and centrifuged at 1500 ×g for 10 min at 4°C to remove platelets and cell debris. The content of platelet factor 4 (PF4), 5-HT and hexosaminidase A (HexA) in the culture medium were determined via enzyme-linked immunosorbent assay (ELISA) kits.
After coculture for 3 days, the lower chambers were washed with PBS for 3 times, and cell viability was measured using a cell counting kit (CCK-8, #40203ES60, Yeasen, Shanghai, China) following standard protocols. The cell nuclei were stained with 4ʹ,6-diamidino-2-phenylindole (DAPI) and observed via microcopy.
2.11 Immunofluorescence staining
For tissue immunofluorescence staining, paraffin sections (5 µm thick) of mouse or human tumors were dewaxed with gradient ethanol and subjected to antigen retrieval in citrate-based buffer. For cell immunofluorescence analysis, the cells were grown on chamber slides and fixed with 4% paraformaldehyde. Both tissue and cell samples were blocked with 10% bovine serum albumin (BSA) for 1 h at room temperature, followed by incubation with primary antibodies at 4°C overnight. The slides were then washed three times with PBS, and incubated with secondary antibodies for 1 h at room temperature. The cell nuclei were visualized using DAPI staining. Digital images were captured using a laser scanning confocal microscope. The specific antibodies used in this assay were rabbit anti-cytokeratin 19 (CK19) (1:300, #ab52625, Abcam), rabbit anti-EPHB1 (1:100, #AP7622a, Abgent, San Diego, CA, USA), PE anti-mouse/rat CD62P (P-selectin) (1:200), rat anti-mouse CD41 (1:100), mouse anti-CD42b (1:200, #14-0429-82, Invitrogen, Carlsbad, CA, USA), mouse anti-CK19 (1:200, #GB12197, Servicebio, Wuhan, Hubei, China), anti-rat Alexa Fluor 647 (1:400, #4418, Cell Signaling Technology, MA, USA), anti-rabbit Alexa Fluor 594 (1:400, #711-545-152, Jackson ImmunoResearch, PA, USA), and anti-mouse Alexa Fluor 488 (1:400, #705-585-147, Jackson ImmunoResearch). Frozen sections (10-µm thick) of mouse liver were obtained after intrasplenically injecting KPC1199-labeled cells. The slides were incubated with mouse anti-mCherry (#125096, Abcam), rabbit anti-GFP (#ab290, Abcam), and rat anti-mouse CD41 antibodies for 1 h at room temperature, followed by staining with anti-mouse Alexa Fluor 594, anti-rabbit 488 and anti-rat Alexa Fluor 647 secondary antibodies.
To label KPC1199 cells, live cells growing in a µ-Slide chamber (#80826, ibidi GmbH, Germany) were directly captured using a laser scanning confocal microscope.
2.12 Hematoxylin and eosin staining
The livers of mice were removed, fixed with 4% paraformaldehyde, and then made to paraffin-embedded tissue sections (5 µm). Eight sections spaced 100 µm apart at the largest coronal section were selected for Hematoxylin and eosin (H&E) staining in each liver followed standard protocols. The nodules with the largest diameter above and below 500 µm were counted separately. The average number of nodules on each section was taken as the final number of liver metastatic nodules. The liver metastatic tumor area was determined via ImageJ software [32].
2.13 Immunohistochemistry
Immunohistochemistry (IHC) staining on TMAs and paraffin sections was performed routinely as previously described [9]. Briefly, the formalin fixed and paraffin-embedded slices (5-µm thick) were deparaffinized and rehydrated. The sections were blocked in 10% BSA and then incubated with primary antibodies overnight at 4°C and with horseradish peroxidase (HRP) conjugated secondary antibodies for 1 h at room temperature. The sections were then treated with diaminobenzidine (DAB) substrate (#S21024-2, Thermo Fisher Scientific, Waltham, MA, USA) and counterstained with hematoxylin for contrast. All the sections were observed and photographed with a photo microscope (Carl Zeiss, USA). The evaluation of EPHB1 staining adhered to a previously established scoring system, taking into account both the percentage of positively stained cells and the staining intensity. Specifically, based on the percentage of positive-staining cells: 0-5% scored 0, 6%–35% scored 1, 36%–70% scored 2, and >70% scored 3; and staining intensity: no staining scored 0, weakly staining scored 1, moderately staining scored 2, and strongly staining scored 3. The final score was calculated by multiplying the percentage and intensity scores, leading to the following categorization: “-” (0-1), “+” (2-3), “++” (4-6), and “+++” (> 6). Notably, Low expression was defined as a total score less than 4, and high expression was defined as a total score of 4 or greater [33]. To ensure objectivity, these scores were independently determined by two senior pathologists in a blinded manner. Specifically, positive staining in the endocrine pancreas was excluded from scoring. Specifically, 16 dots in the TMA were lost during the dewaxing or staining process, and 124 dots left were used for analysis. The specific antibodies used in this assay were EPHB1 (1:100) and Ki67 (1:100, #GB111499, Servicebio).
2.14 Lentivirus production and cell transduction
Full-length cDNA encoding mouse Ephb1 and shRNA constructs against mouse Ephb1 and scrambled sequences were purchased from GeneCopoeia (Shanghai, China). Lentivirus particles were generated using the psPAX2 and pMD2.G packaging system in 293T cells with Lipofectamine 2000 (#11668027, Invitrogen) following standard protocols. The cells were infected with 1 × 106 recombinant lentivirus-transducing units in the presence of 6 mg/mL polybrene (#H9268, Sigma-Aldrich). Twenty-four hours after infection, the cells were treated with 5 µg/mL puromycin (#A1113802, Gibco). The puromycin-resistant cells were selected in the presence of 5 µg/mL puromycin for 5 days. The puromycin-resistant cells were passaged, and the knockdown or overexpression efficiency of Ephb1 was verified by Quantitative real-time polymerase chain reaction (qPCR) and Western blotting (WB).
2.15 Recombinant EphB1 expression, purification, and verification
Monomeric EphB1 was prepared as follows. The extracellular domain of Ephb1 was cloned and inserted into the episomal expression vector with a 6 × His-tag. EphB1 was recombinantly expressed in 293T cells after transfection with the reconstructed plasmid using X-tremeGENE 9 DNA Transfection Reagent (#6365779001, Roche, Basel, Switzerland). After 48 h, the 293T cells were screened with puromycin (#P7130, Sigma-Aldrich) at a dose of 2 µg/mL in DMEM supplemented with 10% FBS for 7 days. Then, the culture media were collected, and monomeric EphB1 was purified with IMAC nickel columns (Ni-IMAC).
2.16 Western blotting
Cells or tissues were lysed using radioimmunoprecipitation assay lysis and extraction buffer (RIPA) (#P0013B, Beyotime, Shanghai, China) with 1× cOmplete™ Protease Inhibitor Cocktail (#4693116001, Roche) and 1× phosphatase inhibitors (#04906845001, Roche). The protein concentration was measured via a bicinchoninic acid assay kit (#WB6501, NCM Biotech, Suzhou, Jiangsu, China). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membrane. The membranes were blocked with 5% skim milk for 1 h and then incubated overnight at 4°C with one of the following primary antibodies: EPHB1 (1:500, Abgent, or 1:500, #PA5-14604, Invitrogen), phospho-Akt (Ser473) (1:1,000, #4060, Cell Signaling Technology), Akt (1:1,000, #9272, Cell Signaling Technology), c-Jun N-terminal kinase (JNK) (1:1,000, #24164-1-AP, Proteintech, Wuhan, China), phospho-JNK (Tyr185) (1:1,000, #80024-1-RR, Proteintech), phospho-p38 mitogen-activated protein kinase (MAPK) (Thr180/Tyr182) (1:1,000, #28796-1-AP, Proteintech), p38 MAPK (1:1,000, #14064-1-AP, Proteintech), p44/42 MAPK (extracellular regulated protein kinases1/2, Erk1/2) (1:1,000, #9102, Proteintech), p44/42 MAPK (Erk1/2) (137F5) (1:1,000, #4695s, Cell Signaling Technology), Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (1:1,000, #9101, Cell Signaling Technology) and anti-beta-actin (1:3,000, #AB0035, Abways, Shanghai, China). Protein detection was achieved using Alexa-Fluor or HRP-conjugated secondary antibodies, and signal was imaged using the LI-COR Odyssey CLx system (LI-COR Biosciences, Lincoln, NE), or the chemiluminescence Bio-Rad system (Bio-Rad Laboratories, Hercules, CA, USA). All images were analyzed using Image Studio Lite v5.2 software (LI-COR Biosciences, Lincoln, NE, USA) or Image Lab Software version 3.0 (Bio-Rad Laboratories).
2.17 Quantitative real-time polymerase chain reaction
Total RNA was isolated using RNAiso Plus (#9108Q, Takara Bio, Shiga, Japan), and reverse transcription was performed using the PrimeScript RT-PCR kit (#RR600A, Takara Bio) according to the manufacturer's instructions. Subsequently, Real-time PCR analyses were conducted on a 7500 real-time qPCR system from Applied Biosystems, employing SYBR Premix Ex Taq at the recommended thermal cycling settings: an initial denaturation phase at 95°C for 15 min, followed by 40 cycles of denaturation at 94°C for 15 s and annealing/extension at 60°C for 30 s. Relative mRNA expression was calculated by normalizing with β-actin gene expression, applying the 2(−ΔΔCt) method. Sequences of specific primers are listed in Supplementary Table S2.
2.18 RNA-seq
For RNA-seq analysis of mouse PDAC metastatic liver and normal control livers, total RNA was isolated utilizing the TRIzol reagent following the manufacturer's protocol. Subsequently, the isolated RNA was dispatched to Oebiotech (Shanghai, China) for clustering and sequencing. To quantify gene expression, the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) value for each gene was meticulously calculated, taking into account both the length of the gene and the number of reads that were successfully mapped to the gene. Characteristic gene sets were analyzed according to the genes presenting the strongest enrichment scores for each gene set.
2.19 Tumor cells and platelets coculture
To compare the differences in platelet activation and tumor cell viability between cocultures of platelets with Ephb1-overexpressing (OV-Ephb1) and control (OV-Vector) KPC1199 cells, OV-Vector or OV-Ephb1 KPC1199 cells (1 × 106 in 2 mL medium) were seeded into 6-well plates and starved for 24 h after adherence of the cells. Fresh medium with 2% FBS was provided, and the platelets were supplemented with different platelet: tumor cell ratios ranging from 0 to 1,000 and incubated at 37°C. The cells were incubated for 72 h, after which the cell viability was measured. The medium was collected from the cells cocultured with 1 × 109 platelets for 2 h to determine the 5-HT content. To compare the tumor cell viability of OV-Vector or OV-Ephb1 KPC1199 cells cocultured with platelets from WT C57BL/6J mice or Tph1−/− mice, OV-Vector or OV-Ephb1 KPC1199 cells were seeded into 96-well plates at 2,000 cells per well. Immediately thereafter, platelets were isolated from the blood of WT C57BL/6J mice or Tph1−/− mice, and 2 × 106 platelets were added to the number of cancer cells per well. Platelets added to empty wells without cancer cells were used as control wells. The cells were incubated for 72 h to measure cell viability.
2.20 Static tumor cell-platelet adhesion
For quantification of adhesive interactions between tumor cells and platelets, OV-Vector or OV-Ephb1 KPC1199 cells (5 × 104) were cultivated in µ-Slide 8-well chamber to confluence. Platelets isolated from Rosa26 mT/mG mice (platelets were tdTomato-positive) were resuspended in DMEM to a concentration of 5 × 108 platelets/50 µL. Subsequently, 50 µL/well platelet buffer was added to the top of the cells. After 30 min of incubation, the unbound platelets were removed by gently washing with PBS three times. Then, the tumor cell-bound platelets were fixed and stained with DAPI. Images were captured with a confocal microscope, and the number of tdTomato-positive platelets was calculated. To quantify the adhesive interactions between shNC or shEphb1 tumor cells and platelets, the experiment was conducted following the same protocol.
2.21 Tumor cell and platelet adhesion assays in suspension
OV-Vector or OV-Ephb1 KPC1199 cells (1 × 106) were suspended in 500 µL of 1 × PBS. Freshly isolated platelets (1 × 109) from Rosa26 mT/mG mice were added to tumor cell suspensions (1,000:1) and incubated for 20 min at room temperature. tdTomato-positive tumor cells were analyzed by flow cytometry for platelet-bound tumor cells. For monomeric EphB1 or Ephrin B1 (EFNB1) antibody blocking in the coculture system, monomeric EphB1 (2 µg/mL, #HY-P75748, MedChemExpress) or EFNB1 antibody (20 µg/mL, #MAB4731, R&D, MN, USA) was added to platelets, incubated for 30 min before mixing with tumor cells and then analyzed by flow cytometry.
2.22 Platelet stimulation in vitro
Platelets were isolated from mice and suspended in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer at a concentration of 1 × 108/100 µL. They were subjected to the following treatments: control: no stimulation; thrombin: platelets stimulated with murine thrombin (1 µg/mL); Fc: platelets stimulated with an Fc fragment (#AG714, Merck, Darmstadt, Germany); EphB1-Fc: platelets stimulated with disulfidelinked homodimeric recombinant EphB1 Fc Chimera Protein (2 µg/mL; #1596-B1, R&D); thrombin + EphB1-Fc: platelets stimulated with thrombin and EphB1-Fc; mono EphB1: platelets stimulated with monomeric His6-tagged proteins corresponding to the exodomains of EphB1 (2 µg/mL); mono EphB1 + EphB1-Fc: platelets stimulated with monomeric EphB1 for 30 min and then treated with EphB1 Fc; and EFNB1 Ab + EphB1-Fc: platelets stimulated with an EFNB1 antibody for 30 min and then treated with EphB1 Fc. The platelets were treated for 20 min and then stained with PE-conjugated anti-CD62P antibody. To investigate the role of LY294002 in EphB1-Fc-induced platelet activation, platelets were pretreated with LY294002 (10 µmol/L) for 20 min and then treated with EphB1-Fc for another 20 min, followed by PE-CD62P antibody staining. Flow cytometric analysis was performed, and the results were analyzed using FlowJo software.
2.23 Determination of 5-HT levels in mouse tissue
For the preparation of tissue homogenates, 5 liver metastatic tumors and adjacent liver tissues were isolated from mice intrasplenically injected with tumor cells. The tissues were washed with PBS to eliminate any residual blood components. Following homogenization in standard ELISA buffer, the resulting mixture was centrifuged at 12,000 ×g for 30 min at 4°C. The supernatant was carefully collected for subsequent quantification of 5-HT levels. The amount of 5-HT in the coculture medium and tissue homogenates was measured using an serotonin/5-hydroxytryptamine (ST/5-HT) ELISA kit according to the manufacturers' instructions (#E-EL-0033c, Elabscience, Houston, TX, USA).
2.24 Determination of platelet release factors after EphB1-Fc stimulation by ELISA
To determine the content of factors released by platelets, 5 × 108 platelets isolated from mice were suspended in 500 µL of HEPES in every sample. After treatment with Fc, thrombin, or EphB1-Fc for 30 min at 37°C, the platelets were centrifuged at 1,500 ×g for 10 min at 4°C, and the supernatant was collected for ELISA. Platelets treated with only HEPES were presented as vehicle controls. The adenosine triphosphate (ATP) level in the supernatant was determined via a bioluminescent ATP assay kit (#S0027, Beyotime) according to the manufacturer's instructions. Luminescence was measured using a luminometer (M1000 PRO, TECAN). The following ELISA kits were purchased from Aimeng Youning (Shanghai, China): Mouse CXCL4 & PF4 ELISA Kit (#LV30145), Mouse bFGF ELISA Kit (#LV30045), Mouse IL-1β ELISA Kit (#LV30300), Mouse PDGF-AB ELISA Kit (#LV30426), Mouse SDF-1α ELISA Kit (#LV30467), Mouse TGF-β1 ELISA Kit (#LV30503), Mouse EGF ELISA Kit (#LV30164), Mouse VEGFA/VEGF ELISA Kit (#LV30582), Mouse Histamine ELISA Kit (#LV30665), Mouse 5-HT ELISA Kit (#LV30594), and Mouse β-hexosaminidase ELISA Kit (#LV30666). All experiments were performed in triplicate and repeated three times.
2.25 Statistical analysis
All statistical analyses were performed using GraphPad Prism 8 software (GraphPad Software Inc., San Diego, CA). Appropriate statistical analyses were performed, depending on the comparisons made and are referenced in the text and figure legends. Reproducibility was ensured by performing more than three independent experiments. Data were presented as the means ± standard deviations (SD). P < 0.05 was considered significant.
3 RESULTS
3.1 Cancer-associated platelets were identified in the liver metastatic niche of PDAC
To investigate the cellular components involved in the liver metastatic niche of PDAC, we employed an ingeniously designed mCherry niche-labeling system. This system involves the release of a cell-penetrating fluorescent protein by metastatic cancer cells, which is subsequently taken up by neighboring cells, allowing us to directly identify and visualize the local metastatic cellular environment in vivo as previously reported [31]. We genetically engineered a murine PDAC cell line, KPC1199 cells derived from KPC mice, to express both GFP and a secreted fluorescent mCherry protein that incorporated a modified lipid-permeable transactivator of transcription peptide (sLP-mCherry) and these cells were referred to as labeled KPC1199 cells (Figure 1A–B). The labeled KPC1199 cells (GFP+mCherry+) were intrasplenically or intrapancreatically injected into the mice to induce liver metastases. The labeled surrounding host tissue cells in the liver metastatic niche were GFP−mCherry+ cells, whereas the unlabeled cells were GFP−mCherry− cells (Figure 1A, Supplementary Figure S1). CD45+ immune cells and CD41+ platelets were identified in GFP−mCherry+ cells from the metastatic niche in both intrasplenic (Figure 1C) and intrapancreatic injection models (Figure 1D). Among all immune cells and platelets, the percentage of platelets was the highest (Figure 1E), which attracted our attention. The existence of mCherry+ platelets in the liver metastatic niche was further confirmed by immunofluorescence staining (Figure 1F). Furthermore, the activation status of platelets was investigated by examining CD62P (P-selectin) expression. We observed a significantly greater percentage of activated platelets among mCherry+ platelets than among mCherry− platelets (Figure 1G–J), indicating that more activated platelets were present in the liver metastatic niche.
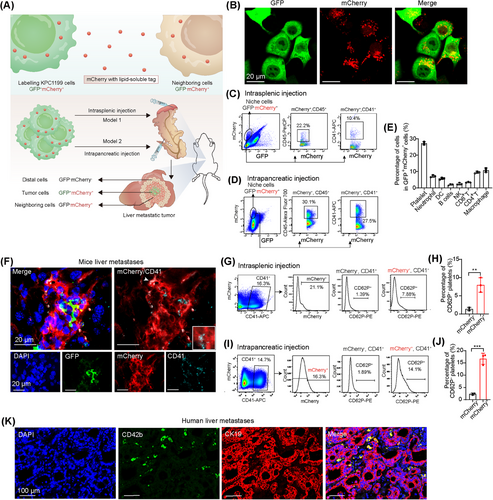
To further confirm the involvement of activated platelets in the liver metastatic niche of PDAC, we analyzed RNA-seq data of liver metastatic tumors from a mouse model of PDAC liver metastasis induced by intrasplenic injection of KPC1199 cells, as reported in our previous work [28]. GO analysis revealed that the genes whose expression was increased in liver metastatic tumors compared with normal liver tissues were enriched in platelet-related pathways (Supplementary Figure S2A). Furthermore, immunofluorescence staining of the platelet surface markers CD41 and CD62P revealed accumulation of platelets in the liver metastatic niche of the mouse model (Supplementary Figure S2B).
We next investigated whether activated platelets were also involved in liver metastatic PDAC in human patients. To address this issue, we collected tumor tissues that were simultaneously surgically resected from 13 patients with liver metastatic PDAC at Ren Ji Hospital. RNA-seq data, including those from 6 adjacent normal tissues and 13 paired primary PDAC-liver metastatic tumors, were used to examine the distinct transcriptomic signatures associated with PDAC-liver metastatic tumors (GSE151580). KEGG pathway analysis revealed that the genes whose expression was increased in liver metastatic PDAC tissue compared with adjacent normal pancreatic tissue were enriched in platelet activation in liver metastatic PDAC (Supplementary Figure S2C). We then analyzed the RNA-seq data from the GSE71729 dataset, which included 145 primary PDAC tumors and 25 PDAC liver metastatic tumors. GSEA of the transcriptome data revealed striking alterations in platelet activation and aggregation in liver metastatic tumors compared with their primary tumors (Supplementary Figure S2D). To directly observe the distribution of platelets in the liver metastatic niche, consecutive tissue sections from PDAC patients with liver metastasis were stained with H&E or an antibody against CD42b, a cell marker for human platelets. Interestingly, massive accumulations of tissue-infiltrating platelets were observed in the liver metastatic niche (Supplementary Figure S2E), which was further confirmed by coimmunostaining for CD42b and the PDAC cell marker CK19 in tumor samples from PDAC patients (Figure 1K). These results indicated the existence of activated platelets in liver metastases from both mouse and human PDAC samples.
3.2 Platelets promote PDAC cell growth via direct physical interactions with tumor cells in the liver metastatic niche
To further address the functional involvement of platelets in the PDAC liver metastatic niche, we depleted platelets in a PDAC liver metastasis mouse model with an anti-CD41 antibody, which specifically depletes platelets in mice, as previously reported [34]. The mice were intrasplenically injected with luciferase-labeled KPC1199 cells at Day 0 and treated with anti-CD41 antibody or control IgG every 3 days from Day 2 to sacrifice (Figure 2A). Treatment with anti-CD41 antibody was sufficient to decrease the number of platelets both in the circulation and in the liver metastatic niche (Supplementary Figure S3A–B). Furthermore, in vivo imaging of the mice, gross morphology of the liver and H&E staining of liver metastasis sections revealed a decreased tumor burden after platelet depletion (Figure 2B–D). Immunohistochemical staining for the cell proliferation marker Ki67 confirmed a decrease in tumor cell proliferation in the metastatic tumors after platelet depletion (Figure 2E).
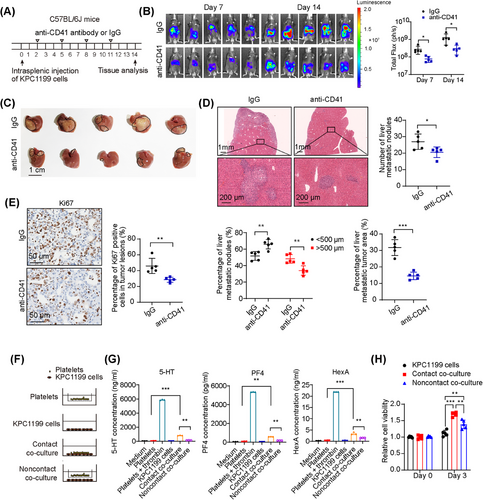
Although the tumor burden was reduced in the liver metastatic niche after the depletion of platelets, it is still unclear whether the influence of platelets on PDAC growth in the liver metastatic niche is caused by direct interactions with tumor cells or indirect effects through the modulation of the microenvironment. To investigate the possibility of direct interaction between platelets and tumor cells, we used two kinds of coculture systems: a direct contact coculture system and a coculture system with a permeable membrane with 0.4 µm pores in vitro (Figure 2F). The contents of PF4, 5-HT and HexA, which are molecules released by α, dense and lysosomal degranulation, respectively, were tested. The results revealed higher contents of PF4, 5-HT and HexA released by activated platelets when they were cocultured with tumor cells in direct contact conditions than in noncontact conditions (Figure 2G), indicating that more platelets were activated by tumor cells in a contact-dependent manner. CCK8 analysis and DAPI staining revealed that the increase in both cell viability and the number of tumor cells was more significant when tumor cells were cocultured with platelets in conditions of direct contact than when they were cultured in noncontact conditions (Figure 2H, Supplementary Figure S3C–D). Together, these data indicate that contact-dependent interactions between tumor cells and platelets enhance the activation of platelets and further promote the growth of PDAC cells.
3.3 EPHB1-expressing tumor cells mediate contact-dependent activation of platelets
To identify the molecules involved in the contact-dependent activation of platelets by tumor cells, we analyzed RNA-seq data from liver metastatic tumor of 25 patients with PDAC liver metastasis (GSE71729). The samples were divided into the high-score group and the low-score group on the basis of the platelet activation signature genes via GSVA. The median of the signature scores was selected as the cutoff for the high- and low-score groups [30]. By comparing the differentially expressed genes between those two groups, we found that axon guidance molecules, particularly the EPH/Ephrin family, were enriched in the high-score group (Figure 3A–B, Supplementary Figure S4). The molecule that was most positively associated with platelet activation was EPH receptor B1 (EPHB1) at the mRNA level (Figure 3B–C), which was further confirmed at the protein level by coimmunostaining with EPHB1 and CD42b antibodies in liver metastatic tumor samples from PDAC patients (Figure 3D).
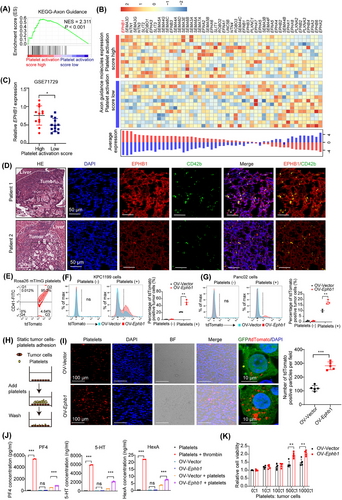
Immunohistochemical staining of a TMA containing 35 matched liver metastatic tumor, adjacent liver, primary PDAC tumor and corresponding adjacent normal pancreatic tissue samples from liver metastatic PDAC patients revealed that, the protein levels of EPHB1 in the tumor cells of primary PDAC and liver metastases were notably elevated compared with adjacent normal pancreas (Supplementary Figure S5A). Patients with high EPHB1 expression in liver metastatic tumors tended to have shorter overall survival (Supplementary Figure S5B).
OV-Ephb1 and OV-Vector cells were constructed from luciferase-labeled murine KPC1199 and Panc02 cells to further explore the function of EPHB1 in tumor cell-platelet interactions (Supplementary Figure S5C–D). With isolated tdTomato-positive platelets from Rosa26 mT/mG mice (Figure 3E), we first conducted a tumor cell-platelet adhesion assay in suspension after Ephb1 was overexpressed in both KPC1199 and Panc02 cells. Compared with OV-vector cells, OV-Ephb1 cells presented an increased percentage of tumor cells that aggregated with tdTomato-positive platelets (Figure 3F–G). Second, platelets were allowed to adhere to tumor cells in vitro under static conditions (Figure 3H). The results revealed that platelet adhesion to OV-Ephb1 tumor cells was much greater than that to OV-Vector tumor cells (Figure 3I). In addition, a loss-of-function study of Ephb1 also revealed that the adhesion of platelets to shEphb1 KPC1199 cells was significantly lower than that to shNC cells under both suspension and static conditions (Supplementary Figure S6).
Furthermore, in KPC1199 cells and platelet contact coculture under static conditions, the contents of PF4, 5-HT and HexA were measured, and the results revealed that the contents of all three molecules, particularly 5-HT, were significantly greater in the media of OV-Ephb1 tumor cells than in those of OV-Vector cells in contact with platelets (Figure 3J). In addition, OV-Ephb1 or OV-Vector tumor cells were cocultured with platelets at different ratios. The CCK8 assay revealed notably greater viability in OV-Ephb1 cells than in OV-Vector cells when they were cocultured with platelets (Figure 3K). Taken together, these findings indicate that Ephb1-overexpressing tumor cells facilitate direct contact between tumor cells and platelets, thereby increasing platelet activation.
3.4 The promoting effect of EPHB1 on PDAC outgrowth in the liver metastatic niche was dependent on platelet activation
To assess the functional involvement of EPHB1 in PDAC liver metastasis in vivo, we established both intrasplenic and intrapancreatic xenograft models in C57BL/6J mice using OV-Ephb1 and OV-Vector luciferase-labeled KPC1199 cells. In the intrasplenic injection model, bioluminescence imaging revealed an increased tumor burden, and histological observation via H&E staining and CK19 immunofluorescence revealed larger metastatic nodules in the livers of the OV-Ephb1 group than in those of the OV-Vector group (Figure 4A, Supplementary Figure S7). In addition, the C57BL/6J mice in the OV-Ephb1 group had much shorter survival than those in the OV-Vector group did (Figure 4B). Consistently, liver metastasis was also increased after Ephb1 was overexpressed in tumor cells in the intrapancreatic xenograft model (Supplementary Figure S8A–B). Next, we intrasplenically injected C57BL/6J mice with shNC or shEphb1 KPC1199 cells. In vivo imaging revealed a substantial reduction in the liver metastatic tumor burden in the shEphb1 group compared with the shNC group (Supplementary Figure S8C). Collectively, these in vivo data suggest that Ephb1 promotes liver outgrowth in liver metastatic PDAC.
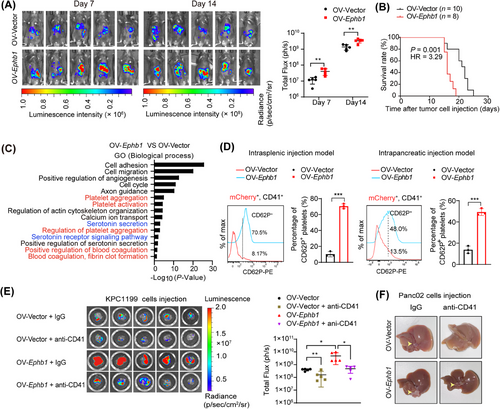
To decipher the molecular changes in liver metastatic tumors after EPHB1 overexpression, comparative transcriptomic analysis of liver metastatic tissues from the mice in the OV-Ephb1, OV-Vector and control (normal liver tissue) groups was performed. Both GO term and KEGG pathway analyses of the 1,980 upregulated genes in the OV-Ephb1 group compared with those in the OV-Vector group revealed several pathways related to platelet function, including platelet aggregation, the regulation of platelet aggregation, blood coagulation, fibrin clot formation and platelet activation (Figure 4C, Supplementary Figure S9A–B). In addition, GO term and KEGG pathway analyses of upregulated genes between the OV-Vector and control groups revealed pathways related to platelet function (Supplementary Figure S9C–E).
Next, we established an mCherry labeling system in OV-Vector and OV-Ephb1 KPC1199 cells, and then those cells were transferred into the mice via intrasplenic or intrapancreatic injection. Flow cytometric analysis revealed a significant increase in CD62P+ activated platelets in CD41+mCherry+ platelets from the OV-Ephb1 group compared with those from the OV-Vector group in both models (Figure 4D). Taken together, these results suggest that increased EPHB1 expression in tumor cells results in increased platelet activation in the liver metastatic niche of patients with PDAC.
We next demonstrated whether the promoting effect of EPHB1 on PDAC outgrowth in the liver metastatic niche was platelet-dependent in vivo by depleting platelets with an anti-CD41 antibody (Supplementary Figure S10A–B). Both bioluminescence imaging and gross morphology of the resected livers revealed that the increased tumor burden in the liver induced by Ephb1 overexpression was largely attenuated by treatment with the anti-CD41 antibody (Figure 4E–F).
3.5 EPHB1 expression in tumor cells promoted platelet activation by phosphorylation of AKT in platelets through EPHB1-EFNB1 reverse signaling
EFNB1 is the only reported ligand of EPHB1 in platelets [35]. The binding of EPHB1 with EFNB1 can mediate bidirectional signaling through either EPHB1- or EFNB1-expressing cells interdependently. Signaling toward EPHB1-expressing cells is called forward signaling, whereas signaling toward EFNB1-expressing cells is reverse signaling [36]. It has been reported that dimeric or multimeric forms of the extracellular domain of EPHB1 can replicate the effect of EPHB1 stimulation and lead to EFNB1 activation, whereas monomeric EPHB1 extracellular domain protein cannot induce signaling activation but interferes concurrently with signaling pathways, thereby serving as an antagonist of EPHB1/EFNB1 [37-39]. We hypothesized that EPHB1/EFNB1 interactions may mediate contact-dependent signaling between tumor cells and platelets. To test this hypothesis, we blocked the EPHB1/EFNB1 interaction by preincubation of platelets with EphB1 monomers (monomeric proteins corresponding to the extracellular domain of EPHB1, which act as antagonists by binding to EFNB1 [35, 40]) or antibodies raised against EFNB1. The results revealed that OV-Ephb1 tumor cells almost failed to further enhance the adhesion of platelets preincubated with EphB1 monomers or EFNB1 antibodies (Figure 5A–B). Furthermore, we investigated the effect of EPHB1/EFNB1 on platelet activation. Platelets were treated with recombinant EphB1-Fc (disulfidelinked homodimer of EphB1, which is capable of activating EFNB1 on platelets), an Fc fragment, EphB1 monomers, an EFNB1 antibody or thrombin (as a positive control). Flow cytometry analysis of CD62P expression revealed that EphB1-Fc activated platelets whereas the Fc fragment or EphB1 monomers did not; When platelets were pretreated with EphB1 monomers or an EFNB1 antibody to block the binding of EphB1-Fc to the Ephrin ligand, the platelets were no longer activated; EphB1-Fc further enhanced thrombin-induced platelet activation (Figure 5C). To identify the signaling pathway induced by EphB1-Fc, we investigated AKT signaling, which is activated in response to a number of platelet activation agonists [41-43]. Phosphatidylinositol 3-kinase (PI3K) and Akt signaling was reported to mediate granule secretion primarily through the activation of MAPKs and phosphorylation of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins [44-46]. Western blotting analysis revealed that the expression of p-Akt (Ser473), p-JNK, p-P38 and p-ERK was increased after EphB1-Fc stimulation (Figure 5D). The PI3K/AKT pathway inhibitor LY294002 suppressed platelet activation induced by EphB1-Fc (Figure 5E–F), indicating that the activation of AKT signaling was induced by EphB1-Fc in platelets. Taken together, these data indicate that the EPHB1/EFNB1 axis mediates the interaction between platelets and tumor cells in a contact-dependent manner.
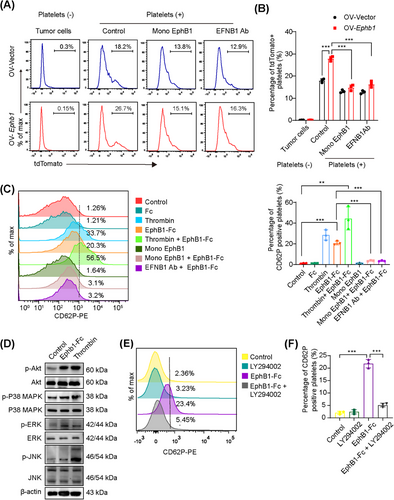
3.6 5-HT released by activated platelets supported PDAC outgrowth in the liver metastatic niche
Upon activation, platelets can secrete a plethora of molecules, which are likely the major drivers of PDAC outgrowth in the liver. Among the molecules released by activated platelets, PF4, fibroblast growth factor 2 (FGF2), interleukin-1β (IL-1β), platelet-derived growth factor α (PDGFα), stromal cell-derived factor-1α (SDF-1α), transforming growth factor-β1 (TGF-β1), epidermal growth factor (EGF), and vascular endothelial growth factor (VEGF) in α-granules, histamine, 5-HT and ATP in dense granules, and HexA in lysosomal granules have been reported to promote cell growth [21]. We therefore compared the levels of these factors released by EphB1-Fc-treated platelets with those released by resting platelets or thrombin-treated platelets. We found that the contents of most factors were greater in EphB1-Fc-treated platelets than in resting platelets. Notably, the levels of 5-HT were significantly increased in the supernatant of EphB1-Fc-treated platelets and comparable to those of thrombin-treated platelets (Figure 6A). Accordingly, via GO term analysis of the genes whose expression was upregulated in the OV-Ephb1 group compared with the OV-Vector group, several 5-HT-related biological processes, including the 5-HT receptor signaling pathway and 5-HT secretion, were enriched (Figure 4C). 5-HT is mainly stored in platelets in the periphery and is released after platelet activation. Previously, we demonstrated that endogenous 5-HT expressed by tumor cells promoted PDAC cell survival and glycolysis in an autocrine manner [27]. Therefore, 5-HT may be a putative candidate bioactive molecule from activated platelets involved in mediating metastatic PDAC outgrowth in the liver.
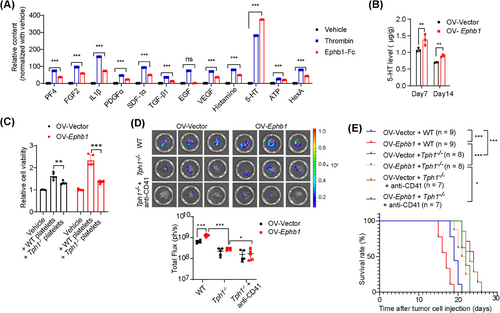
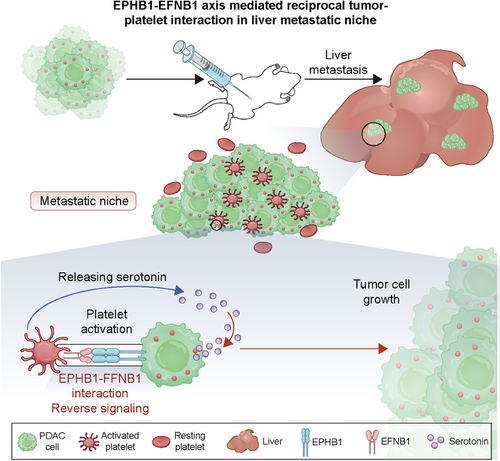
To validate the role of 5-HT, we first measured the concentration of 5-HT in mouse PDAC liver metastatic samples. As expected, the 5-HT concentration was significantly greater in the OV-Ephb1 group than in the OV-Vector group on both Days 7 and 14 after intrasplenic injection of tumor cells (Figure 6B). As a second line of evidence, we used a Tph1-knockout mouse, which lacks the rate-limiting enzyme in the 5-HT biosynthetic pathway in the periphery. In vitro, platelets derived from WT or Tph1−/− mice were cocultured with tumor cells. The results revealed that the viability of both OV-Vector and OV-Ephb1 cells cocultured with platelets from WT mice was much greater than that of those cocultured with platelets from Tph1−/− mice (Figure 6C). We then generated a mouse liver metastasis model by intrasplenic injection of OV-Vector or OV-Ephb1 KPC1199 cells into Tph1−/− mice or WT mice. The 5-HT content was significantly lower in PDAC liver metastatic tissues from Tph1−/− mice than in those from WT mice in both the OV-Ephb1 group and the OV-Vector group (Supplementary Figure S10C). Furthermore, the increased liver metastatic tumor burden induced by Ephb1 overexpression was notably attenuated in Tph1−/− mice. In parallel, platelet depletion in Tph1−/− mice further suppressed liver metastasis, albeit moderately, compared with that in Tph1−/− mice only (Figure 6D). In addition, the overall survival duration obviously increased in the Tph1−/− and platelet depletion groups (Figure 6E). Both in vivo and in vitro data suggest that the promotive effect of EPHB1 on metastatic PDAC outgrowth in the liver metastatic niche is platelet-dependent and relies mainly on 5-HT (Figure 7).
4 DISCUSSION
Platelets serve as legitimate factors in the progression of malignant diseases, affecting a variety of aspects of cancer development, particularly in the area of tumor cell metastasis [47]. In the present study, using the mCherry metastatic niche-labeling system, we identified cancer-associated platelets in the liver metastatic niche of PDAC patients and then further revealed direct platelet-tumor cell interaction through the EPHB1-EFNB1 axis.
The EPH/ephrin system is reported to play a pivotal role in developmental tissue patterning. This is due to its unique ability to perform interdependent forward and reverse signaling, which effectively modulates contact among interacting cell types [48]. Consistent with its capacity to regulate contact-dependent interactions between cells, the binding of Ephrins and Eph to adjacent platelets has been found to promote stable platelet aggregation [35, 49-52]. In the present study, we identified the functions of the EPHB1-EFNB1 axis in facilitating the interaction between platelets and tumor cells in a contact-dependent manner. Indeed, platelet-tumor cell interactions can be mediated by other receptors on platelets and molecules expressed on tumor cells, such as C-type lectin-like-2 receptor (CLEC-2)-podoplanin (PDPN) [53], glycoprotein VI (GPVI)-Galectin-3 (LGALS3) [54], integrin alpha6beta1 (integrin α6β1)-A distegrinin and metalloprotease 9 (ADAM9) [55], and so on [56]. However, whether these receptors mediate tumor-platelet interactions in the liver metastatic niche of patients with PDAC needs further experimental investigation. The increased expression of EPHB1 in liver metastatic tumor cells highlights the important role of EPHB1-EFNB1 in enhancing platelet activation in liver metastases. In addition, we found that EPHB1-Fc treatment notably promoted 5-HT secretion from platelets. Platelet secretion is contextually thematic, demonstrating the ability to discharge specific cargo sets in response to particular agonists [57]. Several studies suggest that alternative routes to fuse with the open canalicular system and the plasma membrane of the intragranular protein cargo and the kinetic differences in release are responsible for the selectivity [21, 58, 59]. However, the exact mechanism by which EPHB1 affects 5-HT secretion needs further investigation.
Eph/ephrin is a promising therapeutic target that is critical in tumor biology and in a variety of nonmalignant diseases [60, 61]. Notably, the research group led by Prof. Tognolini is dedicated to identifying and developing small molecules that target the Eph/ephrin system and develop multiple bioavailable small molecules that effectively inhibit Eph-ephrin interactions [37, 62-65]. The promising clinical performance of an EPHA3-agonist antibody in phase I trials provides a foundation for expanded research and development in the field of EPH-targeted cancer therapeutics [66]. A recent phase II trial in patients with metastatic urothelial carcinoma suggested that a soluble form of Ephb4 (sEphb4-HSA) disrupts the bidirectional signaling between Ephb4 and its ligand Ephb2, which significantly enhances the efficacy of PD-1 blockade through synergistic effects [67, 68]. Therefore, therapies targeting EPHB1 might be clinically effective in preventing liver metastasis in patients with PDAC.
Multiple 5-HT receptors are found expressed in PDAC cells [27], and 5-HT signaling has been associated with tumorigenesis and tumor progression [26]. Our previous studies revealed that 5-HT promoted the survival of primary pancreatic tumor cells through HTR2B mediated Warburg effect [27]. Here, we have not confirmed whether the HTR2B mediated 5-HT signaling activation also contributed to metastatic tumor formation in liver. Besides the classical receptor-dependent activities, 5-HT also takes a role via a post-translational modification-serotonylation after entry to cells through 5-HT transporter (SERT) [69]. In PDAC, 5-HT uptake via SERT promoted activation of Rac1 and was required for the early transdifferentiation of acinar cells into acinar-to-ductal metaplasia [70]. Inhibition of serotonylation via targeting SERT are found suppressed mTOR serotonylation, leading to mTOR inactivation and suppressed colon cancers development [71]. The potential signaling pathways and cellular processes activated by 5-HT in liver metastatic PDAC is of great interest and needs further investigation.
The targeting of 5-HT receptors has been shown to be effective in suppressing tumor growth in various cancers. Previous studies by our group revealed that PDAC progression in mice was obviously suppressed by the administration of 5-HT receptor inhibitors [27]. Skolnik et al. [72, 73] reported that blockade of 5-HT receptors or calcium channels hindered fibrosarcoma cell lodging in the liver and suppressed tumor liver metastasis [72, 73]. In addition to targeting receptors, targeting the 5-HT transporter to inhibit the cellular uptake of 5-HT has shown significant antineoplastic effects in diverse tumor types [71]. Schneider et al. [74] reported that the use of clinically approved 5-HT-targeting agents, either as monotherapies or in combination with programmed cell death protein 1 (PD-1) blockade, achieved long-term control of pancreatic and colorectal cancers in murine models [74]. Here, we provide evidence that 5-HT promotes metalized pancreatic tumor growth in liver, however, whether targeting 5-HT suppressed PDAC liver metastasis remain further investigation.
Despite the above, there were still several limitations in this study. First, a mouse model that can mimic the entire process of PDAC liver metastasis in human patients was lacking. In the absence of this information, we were unable to determine with certainty when platelets first accumulate and settle in liver metastases, i.e., before the arrival of tumor cells or thereafter. Second, the mechanism by which 5-HT promotes tumor growth remained undescribed in this work and needs further investigation in subsequent studies.
5 CONCLUSIONS
To summarize, we identified the existence of tissue-infiltrating platelets in liver metastatic PDAC and revealed that reciprocal tumor-platelet interaction through the EPHB1-EFNB1 axis promotes the outgrowth of metastatic PDAC. Importantly, our work highlights concern regarding the potential implications of targeting EPHB1 or platelets as therapeutic interventions in the liver metastasis of PDAC.
DECLARATIONS
AUTHOR CONTRIBUTIONS
X.-L. Zhang, X.-M. Yang, Z.-G. Zhang and S.-H. Jiang conceived the project, L.-L. Yao, D.-X. Li, X.-L. Zhang and X.-M. Yang designed experiments and interpreted the data in the manuscript.; L.-L. Yao performed research and wrote the manuscript; Z.-G. Zhang, X.-L. Zhang, X.-M. Yang, D.-X. Li and S.-H. Jiang edited the manuscript; T.-Z. Shi performed bioinformatics analyses; J.-Y. Yang and D.-J. Liu provided clinical specimens, made clinical pathology evaluations and contributed to interpretation of clinical data; D.-X. Li performed western blotting analysis and IHC analysis, analyzed and interpreted the data; L.-P. Hu, Q. Li, J. Li and X. Wang contributed to animal breeding and mice model establishment; W.-T. Qin performed platelet assay; X.-M. Yang, H.-Z. Nie, L. Zhu, Z.-G. Zhang and Y.-L. Zhang contributed to mouse model establishment and provided critical review. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
Thanks to Professor Jing Xue (State Key Laboratory of Oncogenes and Related Genes, Renji-Med X Clinical Stem Cell Research Center, Shanghai Cancer Institute, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China) for providing KPC1199 and Panc02 cells and Professor Xing-Xu Huang (School of Life Science and Technology, ShanghaiTech University, Shanghai, China) for sharing Rosa26 mT/mG mice. Thanks to Professor Eng-Ang Ling (Department of Anatomy, Yong Loo Lin School of Medicine, National University of Singapore, Singapore) and Enci Mary Kan (Agency for Science, Technology and Research, Duke-NUS Medical School, Singapore) for critically reading the manuscript. Thanks for FreeScience for drawing schematic diagram images. We thank the staff members of the Integrated Laser Microscopy System at the National Facility for Protein Science in Shanghai, Shanghai Advanced Research institute, Chinese Academy of Sciences, China, for sample preparation, data collection and analysis. This study was supported by the National Natural Science Foundation of China (No. 82230087, and No.82350123 and No. 92168111 to Z.-G. Zhang; No. 82173153 to S.-H. Jiang; No. 82073023 to J. Li; No. 82273228 to H.-Z. Nie; No. 82303278 to W.-T. Qin; No. 82103357, and No. 82372821 to L.-P. Hu); Shanghai Municipal Health Commission (No. 202340202 to L.-L. Yao); Natural Science Foundation of Shanghai (No. 22ZR1460000 to X.-L. Zhang); Innovative research team of high-level local universities in Shanghai (SHSMU-ZDCX20210802).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Research Ethics Committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University (ID: 2015037). The experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai JiaoTong University (A2020108). All patients enrolled in this study provided written informed consent.
Open Research
DATA AVAILABILITY STATEMENT
The PDAC liver metastases transcriptome data used in this study are available in the GEO database under accession code GSE71729, GSE151580, and BioProject database (PRJNA664673). The RNA-seq data generated in this study have been deposited in the Sequence Read Archive (SRA) repository under accession code PRJNA833240. The remaining data are available within the Article, Supplementary Information, or available from the authors upon request.




