SYNCRIP promotes cell cycle progression and lung tumorigenesis by modulating AURKB translation
Hyeon Ji Kim and Hye Guk Ryu contributed equally to this work.
Abbreviations
-
- AURKB
-
- Aurora kinase B
-
- CHX
-
- Cycloheximide
-
- FLUC
-
- Firefly luciferase
-
- INCENP
-
- Inner centromere protein
-
- IRES
-
- Internal ribosome entry sites
-
- ITAFs
-
- IRES trans-acting factors
-
- LSCC
-
- Lung squamous cell carcinoma
-
- LUAD
-
- Lung adenocarcinoma
-
- RLUC
-
- Renilla luciferase
-
- SYNCRIP
-
- Synaptotagmin-binding cytoplasmic RNA-Interacting Protein
-
- TRAF6
-
- TNF receptor associated factor 6
Dysregulation of cellular processes, such as cell division and proliferation, is a hallmark of cancer and is driven by the aberrant expression of cell cycle-related genes [1]. Aurora kinase B (AURKB), due to its pivotal role in mitotic progression, has been implicated in various cancers. Overexpression or hyperactivation of AURKB significantly contributes to tumorigenesis and cancer progression [2]. Although mechanisms that enhance AURKB activity, including binding to INCENP, autophosphorylation [3], and ubiquitination by TRAF6 [4], have been extensively investigated, regulation of AURKB synthesis, particularly mRNA translation, remains unclear. The translation of eukaryotic mRNAs typically occurs either through cap-dependent scanning or through direct ribosomal binding to specialized RNA elements known as internal ribosome entry sites (IRES). IRES-mediated translation is strongly influenced by specific RNA-binding proteins, known as IRES trans-acting factors (ITAFs). SYNCRIP (Synaptotagmin-binding cytoplasmic RNA-interacting protein), also known as hnRNP Q, has been identified as an ITAF [5], integrating various aspects of RNA metabolism with key cellular processes. Here, we aim to elucidate the mechanism of AURKB mRNA translation and investigate whether SYNCRIP regulates AURKB mRNA translation in lung cancer.
To investigate the mechanisms underlying AURKB mRNA translation, we analyzed AURKB protein levels following treatment with rapamycin, an inhibitor of eIF4E-mediated cap-dependent translation, and cycloheximide (CHX), an inhibitor of the elongation phase of translation. Treatment with CHX significantly reduced AURKB protein production, whereas rapamycin had no effect, suggesting that AURKB can be translated via a cap-independent mechanism (Figure 1A, Supplementary Figure S1A-B). To further validate this finding, we used a bicistronic reporter containing the 5′-UTR of AURKB mRNA positioned between the coding sequences for Renilla (RLUC) and Firefly (FLUC) luciferases (Supplementary Figure S1C). This experiment confirmed that the AURKB 5’-UTR facilitates translation of a downstream cistron in a cap-independent manner (Figure 1B, Supplementary Figure S1D).
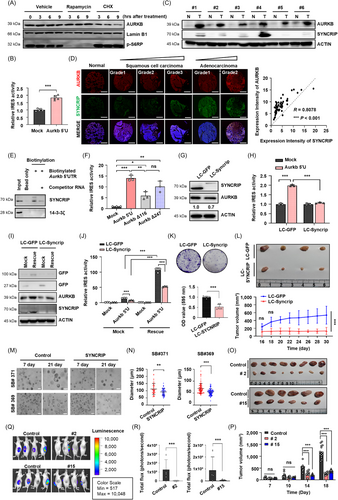
We next examined the correlation between AURKB and SYNCRIP protein expression in human lung cancer tissues. Utilizing the LinkedOmics database, we identified 11,466 genes associated with SYNCRIP in lung adenocarcinoma (LUAD) and 11,928 genes in lung squamous cell carcinoma (LSCC) (Supplementary Figure S2A-B) [6]. Gene set enrichment analysis revealed a strong positive correlation between AURKB and SYNCRIP in both LUAD and LSCC (Supplementary Figure S2C-G). Additionally, immunoblot analysis revealed higher expression levels of both proteins in tumor tissues compared to adjacent normal tissues (Figure 1C). We further validated the correlation between these proteins across various types of lung cancer using lung tissue microarrays (Figure 1D). In LUAD, the expression of AURKB and SYNCRIP significantly increased with cancer progression (Supplementary Figure S3A-B). Previous report has shown that SYNCRIP binds to specific mRNA sequences, such as UAUYRR or AYAAYY (Y = C/U, R = A/G), and affects the translation of target mRNAs [7]. An RNA pull-down assay confirmed the association between SYNCRIP and the Aurkb 5'-UTR (Figure 1E). We identified potential cis-acting elements within the 5′-UTR that are crucial for IRES-mediated translation (Figure 1F, Supplementary Figure S3C-D). These results suggest that SYNCRIP functions as a trans-acting factor in the translation of AURKB mRNA.
To validate SYNCRIP-mediated AURKB expression, we knocked out SYNCRIP using CRISPR/Cas9 (LC-SYNCRIP; Supplementary Figure S4A). SYNCRIP deficiency resulted in a reduction of AURKB protein levels (Figure 1G, Supplementary Figure S4B), while AURKB mRNA levels remained unchanged (Supplementary Figure S4C). We then examined whether SYNCRIP modulates the translational activity of AURKB mRNA in a cap-independent manner. In SYNCRIP-deficient cells, IRES-mediated translation of AURKB mRNA was significantly downregulated (Figure 1H, Supplementary Figure S4D). The absence of SYNCRIP also led to an increased proportion of cells in the G2/M phase (Supplementary Figure S4E-F). Rescue experiments restored AURKB protein levels and IRES activity of the 5′-UTR of Aurkb mRNA in Syncrip-restored LLC cells (Figure 1I-J). These results suggest that SYNCRIP knockout suppresses AURKB protein expression by inhibiting its IRES-mediated translation, resulting in G2/M phase arrest.
SYNCRIP deficiency led to a clear reduction in colony-forming capacity and growth (Figure 1K, Supplementary Figure S4G-H). In a xenograft mouse model, tumors derived from SYNCRIP knockout cells were notably smaller in both volume and size compared to those derived from control cells (Figure 1L). Additionally, adenocarcinoma patient-derived organoids transfected with sgRNA targeting SYNCRIP and Cas9 exhibited reduced growth (Figure 1M-N). These results suggest that SYNCRIP plays a crucial role in the proliferation of lung cancer cells.
To exclude off-target effects of SYNCRIP and confirm the role of the AURKB 5′-UTR in translational regulation, we created a cis-acting region deletion model by removing the 21-221 nucleotide region of the Aurkb mRNA 5′-UTR in LLC cells (Supplementary Figure S5). Deletion of the cis-acting domain in the Aurkb 5′-UTR disrupted cell proliferation and reduced colony-forming ability, similar to SYNCRIP-deficient cells (Supplementary Figure S6A-B). Two randomly selected cell lines (#2 and #15) showed decreased AURKB protein levels and a reduction in phospho-histone H3 (Ser10). Moreover, both cell lines exhibited an increased proportion of cells arrested in the G2/M phase compared to the control group (Supplementary Figure S6C-E). These cell lines also displayed significantly reduced tumor growth in vivo (Figure 1O-R). These results indicate that inhibiting AURKB expression suppresses tumor growth both in vitro and in vivo, underscoring the importance of the AURKB 5′-UTR in regulating its protein expression.
In conclusion, our findings provide new insights into the translational regulation of AURKB in LUAD. The upregulation of AURKB by SYNCRIP not only emphasizes its role in cell cycle progression but also highlights its potential involvement in tumorigenesis. Recently, new functions for SYNCRIP have been identified in various cancers, including leukemia [8] and prostate cancer [9]. The importance of SYNCRIP extends beyond cell cycling to genomic stability and drug resistance [9]. Understanding the detailed mechanisms of SYNCRIP in various cancers could provide crucial evidence for the development of a novel class of anticancer therapeutics in the future.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Study design and conception: Kyong-Tai Kim and Do-Yeon Kim. Data acquisition and analysis: Hyeon Ji Kim, Hye Guk Ryu, Mingyu Kang, Namgyu Lee, Hyo-Jin Kim, Sangjune Kim, Kyung-Ha Lee, Wanil Kim, and Jin-Seok Byun. Technical and material support: Dahye Lee and Chaeuk Chung. Data interpretation and discussion: Hyeon Ji Kim, Hye Guk Ryu, Kyong-Tai Kim, and Do-Yeon Kim. Manuscript writing: Hyeon Ji Kim, Hye Guk Ryu, Kyong-Tai Kim, and Do-Yeon Kim. Manuscript editing and approval of the present version: all authors.
ACKNOWLEDGEMENTS
The Biospecimens used in this study were provided by the Biobank of AJOU University Hospital, a member of Korea Biobank Network.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
FUNDING INFORMATION
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2022R1C1C1006181, 2022R1A6A3A13071217, and RS-2023-00272063). This work was also supported by funds donated by Dr Jae Kyu Lee and Mr Jason Gim.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Male ICR mice aged 10-12 weeks were used, and all animal protocols were approved by Kyungpook National University Animal Care Committee (KNU 2022-0248).
Open Research
DATA AVAILABILITY STATEMENT
The data supporting the results of this study are presented in the online Supplementary Materials. The datasets utilized in this study can be obtained from the corresponding author upon a reasonable request.




