Genomic imprinting biomarkers for cervical cancer risk stratification
Xue Xiao, Wei Wang, Peng Bai, Ying Chen, Zhengwen Qin, Tong Cheng contributed equally.
Abbreviations
-
- ACD
-
- Advanced Cell Diagnostics
-
- ACS
-
- American Cancer Association
-
- AGC
-
- Atypical glandular cells
-
- ASC-H
-
- Atypical squamous cells-high-grade cannot be excluded
-
- ASCUS
-
- Atypical squamous cells of undetermined significance
-
- AUC
-
- Area under the receiver operating characteristic curve
-
- BAE
-
- Bi-allelic expression
-
- CI
-
- Confidence interval
-
- CIN
-
- Cervical intraepithelial neoplasia
-
- DAPI
-
- 4',6-diamidino-2-phenylindole
-
- DNA
-
- Deoxyribonucleic acid
-
- FITC
-
- Fluorescein isothiocyanate
-
- GNAS
-
- Guanine nucleotide-binding protein, alpha-stimulating complex locus
-
- HM13
-
- Histocompatibility minor 13
-
- HPV
-
- Human papillomavirus
-
- hrHPV
-
- High-risk human papillomavirus
-
- HSIL
-
- High-grade squamous intraepithelial lesion
-
- IRB
-
- Institutional Review Board
-
- ISH
-
- In-situ hybridization
-
- IQR
-
- Inter-quartile range
-
- LOI
-
- Loss of imprinting
-
- LSIL
-
- Low-grade squamous intraepithelial lesion
-
- MAE
-
- Multi-allelic expression
-
- NBF
-
- Neutral buffered formalin
-
- NILM
-
- Negative for intraepithelial lesion or malignancy
-
- NPV
-
- Negative predictive value
-
- PCR
-
- Polymerase chain reaction
-
- PPV
-
- Positive predictive value
-
- QCIGISH
-
- Quantitative chromogenic imprinted gene in-situ hybridization
-
- RNA
-
- Ribonucleic acid
-
- RT
-
- Room temperature
-
- SCC
-
- Squamous cell carcinoma
-
- SNRPN
-
- Small nuclear ribonucleoprotein polypeptide N
-
- SNU13
-
- Small nuclear ribonucleoprotein 13
-
- STROBE
-
- STrengthening the Reporting of OBservational studies in Epidemiology
-
- TCT
-
- Thinprep cytologic test
-
- TE
-
- Total expression
Cervical cancer remains a significant global public health issue due to its high incidence and mortality. Current clinical guidelines recommend screening for high-risk human papillomavirus (hrHPV)-DNA alongside a Thinprep cytologic test (TCT) before further medical evaluation [1]. The hrHPV-DNA test detects 14 high-risk HPV genotypes including the predominant hrHPV16/18, which can cause cervical abnormalities that may progress to cancer if untreated. TCT is paired with the hrHPV-DNA test to pathologically classify cervical specimens into categories based on increasing malignancy risks. Despite the high sensitivities, both tests have high false positive rates which lead to unnecessary colposcopy while HPV is cleared naturally in most women without progressing into lesions. To reduce overdiagnosis and overtreatment, several DNA methylation detections [2, 3] have been developed for triaging the malignancy risk of hrHPV-positive cervical lesions, but have yet to become clinically available. Here, we proposed an epigenetic biomarker panel based on imprinting alterations as a high-performance triage method to improve cervical cancer risk assessment accuracy in hrHPV-positive women.
Loss of imprinting (LOI), an early molecular event in carcinogenesis, is an epigenetic phenomenon when a normally silenced allele of the imprinted gene is activated and expressed [4]. Using the quantitative chromogenic imprinted gene in-situ hybridization (QCIGISH) to visualize and quantify imprinted genes’ transcription sites in the nuclei, early epigenetic changes through LOI have been shown as effective biomarkers for detecting multiple malignancies [5]. In the present study, we first screened imprinted gene candidates using resected cervical tissue samples and subsequently developed a cancer risk stratification method based on cytological specimens diagnosed by colposcopy and biopsy (Supplementary Figure S1). The diagnostic model was blindly validated in prospectively collected cytological samples by comparing the QCIGISH results with colposcopy biopsy pathology. Full study protocols are detailed in the Supplementary file.
To identify the most efficient biomarker panel for differentiating malignancy in cervical lesions, we evaluated four candidate imprinted genes based on prior research evidence and targeted literature review of female cancers: guanine nucleotide-binding protein, alpha-stimulating complex locus (GNAS) related to thyroid cancer, osteosarcoma, and skin cancer [6], small nuclear ribonucleoprotein polypeptide N (SNRPN) associated with seminoma, yolk sac tumor, and acute myeloid leukemia [7], histocompatibility minor 13 (HM13) linked to breast cancer [8], and small nuclear ribonucleoprotein 13 (SNU13) involved with lung cancer [9]. QCIGISH was applied to 79 formalin-fixed paraffin-embedded samples comprised of 30 benign, 13 cervical intraepithelial neoplasia grade 1 (CIN1), 14 CIN3, and 22 malignant cases (Supplementary Table S1) for all candidates based on visual evaluation using bright field microscopy. We quantitatively analyzed aberrant allelic expression via N0, N1, N2 and N3+ signals, and calculated bi-allelic (BAE), multi-allelic (MAE), and total expression (TE) measurements (Figure 1A,B, Supplementary Figures S2, S3). Histopathological classifications were dichotomized by combining benign with CIN1 cases and combining CIN3 with malignant cases. Significant differences in BAE, MAE, and TE measurements were observed for GNAS, HM13, and SNU13, with higher area under the receiver operating characteristic curve (AUC) values (all P < 0.05), except for SNRPN (Supplementary Figures S4, S5). These findings substantiated the formulation of a three-gene epigenetic imprinting biomarker panel composed of GNAS, HM13, and SNU13 for model development.
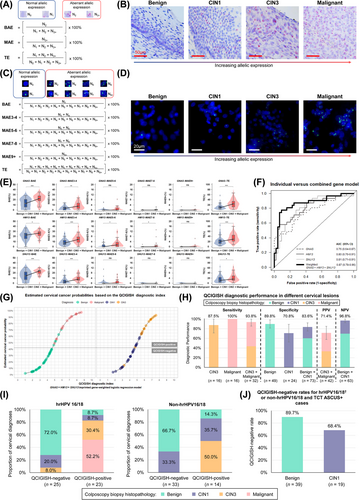
We subsequently performed QCIGISH detection for the three pre-screened imprinted genes on 75 retrospectively collected cytological samples with biopsy-confirmed diagnoses of 29 benign, 15 CIN1, 15 CIN3, and 16 malignant cases to train a cervical cancer risk stratification model. To refine allelic expression measurements for malignancy differentiation, we extended the N3+ measurements to N3-N4, N5-N6, N7-N8, and N9+, allowing for more precise stratification of imprinting alterations to MAE3-4, MAE5-6, MAE7-8, and MAE9+, respectively, and utilized fluorescent microscopy to capture the QCIGISH images (Figure 1C,D, Supplementary Figure S6). Imprinting alterations of all genes were significantly elevated in the CIN3 + malignant group than in the benign + CIN1 group (Figure 1E), showing moderate to high discrimination between groups (Supplementary Figure S7). We applied binary logistic regression to classify patients into cervical cancer low- or high-risk groups, with model development and parameters detailed in Supplementary Figure S8. To accurately estimate the model performance with a limited number of samples, we optimized model parameter settings involving the imprinting alteration biomarkers, gene weight combinations and dichotomization threshold criteria through a 500-cycle internal bootstrap (Supplementary Figure S9, Supplementary Tables S2-S5). The optimal imprinted gene-weighted model improved discrimination, achieving an overall AUC of 0.87 (95% confidence interval [CI], 0.78-0.96) (Figure 1F), including an apparent sensitivity of 87.1% (95% CI, 75.3%-98.9%) and specificity of 75.0% (95% CI, 62.2%-87.8%) (Supplementary Table S5). Logistic regression curves were plotted using the estimated cervical cancer probabilities against the QCIGISH diagnostic indices for each gene and their weighted equivalents (Supplementary Figures S10-S12).
We independently and blindly validated the model in 105 cytological samples diagnosed by colposcopy and biopsy, including 49 benign, 24 CIN1, 16 CIN3, and 16 malignant cases (Figure 1G). The diagnostic sensitivities were 100% for malignant cases and 93.8% (95% CI, 85.4%-100%) for CIN3 and malignant cases combined (Figure 1H). Diagnostic specificities were estimated at 89.8% (95% CI, 81.3%-98.3%) for all confirmed benign cases and 83.6% (95% CI, 75.1%-92.1%) for benign and CIN1 cases combined, which could help improve the accuracy of clinical assessments of cervical lesions when used in combination with hrHPV-DNA tests [10]. For QCIGISH-positive cases, 71.4% (95% CI, 57.8%-85.1%) were histopathologically CIN3 and malignant, while 96.8% (95% CI, 92.5%-100%) of QCIGISH-negative cases were benign and CIN1. Moreover, the QCIGISH positivity rate in CIN3 cases was 87.5% (Figure 1H), demonstrating diagnostic viability among molecular triage methods [2, 3]. The high sensitivity could potentially aid in reducing false negatives during triage, which is crucial for detecting cervical abnormalities early.
The model performance evaluation across different HPV genotypes showed that QCIGISH triage was sufficiently accurate for assessing the malignancy risks among hrHPV-positive women (Figure 1I). Particularly for hrHPV16/18 genotype patients, 92.0% of the QCIGISH-negative cases were confirmed benign or CIN1, while 82.6% of the QCIGISH-positive cases were CIN3 or malignant. Furthermore, 100% of the QCIGISH-negative cases had benign or CIN1 diagnoses for non-hrHPV16/18 genotypes, while 50.0% of the QCIGISH-positive cases were CIN3.
Additionally, we analyzed the QCIGISH triage performance on two groups recommended for colposcopy: (1) hrHPV16/18 genotype patients; and (2) non-hrHPV16/18 genotype patients with TCT grades evaluated as atypical squamous cells of undetermined significance and above (ASCUS+) [1]. Most benign and CIN1 cases were QCIGISH-negative, contributing 89.7% (95% CI, 80.2%-99.3%) and 68.4% (95% CI, 47.5%-89.3%), respectively (Figure 1J). With epigenetic imprinting biomarkers, unnecessary colposcopy referrals could be significantly avoided, further advancing the diagnostic accuracy of HPV and TCT co-testing and ultimately improving the subsequent medical management for these patients (Supplementary Figure S13).
In conclusion, the preliminary findings demonstrated the QCIGISH's robustness as a novel cervical cancer risk assessment test based on aberrant expression of GNAS, HM13, and SNU13 imprinted genes, despite the recognized need for further validation in a larger cohort. These results revealed the high diagnostic sensitivity and specificity of this model, which can be useful when applied adjunctively with HPV and TCT co-testing, allowing physicians to rule out malignancy more confidently while reducing overdiagnosis for benign and low-risk cervical lesions in hrHPV-positive cases. Altogether, QCIGISH is a promising triage alternative, with the potential to improve the clinical diagnostic efficacy and medical management of cervical lesions.
AUTHOR CONTRIBUTIONS
X.X., W.W., T.C., N.Z. and H.L. designed this study. X.X., W.W., P.B., Y.C., Z.Q., J.W., L.X., X.G., H.Z., L.Y. and W.L. collected the clinical samples. T.C., X.L., J.P.P., P.S. and X.W. performed the experiments and collected the data. J.P.P. performed the statistical analysis. W.H., R.S. and C.Y. did the pathological review. X.X., W.W., P.B., Y.C., Z.Q., T.C., X.L. and J.P.P. interpreted the results. X.L. and J.P.P. drafted the manuscript with contributions from all authors, and X.X., C.Y., H.X. and F.B. revised the paper. Y.L., N.Z., Y.Z. and H.L. supervised and coordinated this study. All authors have read and approved the final manuscript.
ACKNOWLEDGMENTS
We thank Hu Zhao (West China Second University Hospital, Sichuan University) for his assistance in collecting the samples and clinical information. We also thank Jia Liu (West China Second University Hospital, Sichuan University) and Kejun Li (Lisen Imprinting Diagnostics Wuxi, Ltd.) for their coordination between hospitals.
CONFLICT OF INTEREST STATEMENT
TC, XL, JPP, PS, XW and NZ are employees of Lisen Imprinting Diagnostics, Inc. No competing interests were reported by the other authors.
FUNDING INFORMATION
This study was supported by the National Natural Science Foundation of China (82071651), Chengdu Science and Technology Bureau (2017-GH02-00030-HZ), National Key Research and Development Program of China (2022YFC3600304, 2022YFC2704700), Science and Technology Department of Sichuan Province (23ZDYF2489), Sichuan Province Cadre Health Care Scientific Research Project (CGY2023-1701) and the Chengdu Technological Innovation R&D Project (2021-YF05-00595-SN). The study was partially sponsored by Lisen Imprinting Diagnostics, Inc.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Institutional Reviewer Boards of Nanjing First Hospital (No. 2017-035R), West China Second University Hospital, Sichuan University (No. 2022-205), Chengdu Women's and Children's Central Hospital (No. 2023-34), and Zigong Maternity and Child Health Care Hospital (No. 2023-12). Written informed consents were received from all patients.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed in this article were included within the article and the supplementary file. The raw data of this article are available from the corresponding authors upon request.




