Yap methylation-induced FGL1 expression suppresses anti-tumor immunity and promotes tumor progression in KRAS-driven lung adenocarcinoma
Ji Jiang, Pengfei Ye, and Ningning Sun contributed equally.
Abstract
Background
Despite significant strides in lung cancer immunotherapy, the response rates for Kirsten rat sarcoma viral oncogene homolog (KRAS)-driven lung adenocarcinoma (LUAD) patients remain limited. Fibrinogen-like protein 1 (FGL1) is a newly identified immune checkpoint target, and the study of related resistance mechanisms is crucial for improving the treatment outcomes of LUAD patients. This study aimed to elucidate the potential mechanism by which FGL1 regulates the tumor microenvironment in KRAS-mutated cancer.
Methods
The expression levels of FGL1 and SET1 histone methyltransferase (SET1A) in lung cancer were assessed using public databases and clinical samples. Lentiviruses were constructed for transduction to overexpress or silence FGL1 in lung cancer cells and mouse models. The effects of FGL1 and Yes-associated protein (Yap) on the immunoreactivity of cytotoxic T cells in tumor tissues were evaluated using immunofluorescence staining and flow cytometry. Chromatin immunoprecipitation and dual luciferase reporter assays were used to study the SET1A-directed transcriptional program.
Results
Upregulation of FGL1 expression in KRAS-mutated cancer was inversely correlated with the infiltration of CD8+ T cells. Mechanistically, KRAS activated extracellular signal-regulated kinase 1/2 (ERK1/2), which subsequently phosphorylated SET1A and increased its stability and nuclear localization. SET1A-mediated methylation of Yap led to Yap sequestration in the nucleus, thereby promoting Yap-induced transcription of FGL1 and immune evasion in KRAS-driven LUAD. Notably, dual blockade of programmed cell death-1 (PD-1) and FGL1 further increased the therapeutic efficacy of anti-PD-1 immunotherapy in LUAD patients.
Conclusion
FGL1 could be used as a diagnostic biomarker of KRAS-mutated lung cancer, and targeting the Yap-FGL1 axis could increase the efficacy of anti-PD-1 immunotherapy.
Abbreviations
-
- ATCC
-
- American Type Culture Collection
-
- ChIP
-
- Chromatin immunoprecipitation
-
- CHX
-
- cycloheximide
-
- co-IP
-
- co-immunoprecipitation
-
- CTLA-4
-
- cytotoxic T-lymphocyte-associated antigen 4
-
- DAPI
-
- 4',6-diamidino-2-phenylindole
-
- DMSO
-
- dimethyl sulfoxide
-
- ERK1/2
-
- extracellular regulated kinase 1/2
-
- FBS
-
- Fetal bovine serum
-
- FCM
-
- Flow cytometry
-
- FGL1
-
- fibrinogen-like protein 1
-
- HA
-
- hemagglutinin
-
- IF
-
- Immunofluorescence staining
-
- IHC
-
- immunohistochemistry
-
- KRAS
-
- Kirsten rat sarcoma viral oncogene homolog
-
- LAG-3
-
- Lymphocyte-activation gene 3
-
- LUAD
-
- lung adenocarcinoma
-
- MDSC
-
- modulating myeloid-derived suppressor cell
-
- MHC-II
-
- the major histocompatibility complex class II
-
- NSCLC
-
- Non-small cell lung cancer
-
- PBMCs
-
- peripheral blood mononuclear cells
-
- PD-1
-
- programmed cell death-1
-
- PTMs
-
- Post-translational modifications
-
- SD
-
- Standard Deviation
-
- RT-qPCR
-
- Quantitative Real-Time quantitative PCR
-
- SET1A
-
- phosphorylated SET1 histone methyltransferase
-
- SET1A
-
- SET1 histone methyltransferase
-
- TBE
-
- TEAD-binding sites
-
- TCGA
-
- the Cancer Genome Atlas
-
- TME
-
- tumor microenvironment
-
- Yap
-
- Yes-associated protein
1 BACKGROUND
Lung adenocarcinoma (LUAD) is a leading cause of cancer-related death worldwide, with Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations occurring in approximately 32% of cases, making them prominent oncogenic drivers in this context [1, 2]. Despite notable strides in developing allele-specific KRASG12C inhibitors, direct targeted inhibition of KRAS remains elusive [3]. Furthermore, both the intrinsic and acquired resistance to direct KRAS inhibition observed in the majority of cases underscores the enduring requirement for complementary approaches to counter KRAS-driven oncogenesis effectively [3, 4]. From this perspective, immunotherapy has expanded the array of available treatment options for cancer patients [5]. The successful blockade of immune checkpoint receptors, such as programmed cell death protein1 (PD-1) and cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4), has demonstrated clinical efficacy in several cancer types, including LUAD [6, 7]. However, the immunotherapy response rates of patients with KRAS-mutated cancer remain suboptimal [8]. Therefore, it is imperative to investigate the mechanisms of immune evasion induced by KRAS mutations and explore alternative immune checkpoint pathways.
Fibrinogen-like protein 1 (FGL1) was initially characterized in hepatocytes for its roles in metabolism and liver regeneration [9, 10], and it has recently attracted attention due to its high expression in various types of tumors, especially LUAD [11]. However, its underlying mechanisms and functional implications remain elusive. Lymphocyte-activation gene 3 (LAG-3) is a transmembrane protein found predominantly on activated T cells and has emerged as a pivotal inhibitory receptor in immune regulation [12]. LAG-3 plays a multifaceted role in negatively regulating the proliferation, activation, and homeostasis of both CD4+ and CD8+ T cells [13, 14]. Notably, recent studies have identified FGL1 as a major functional ligand of LAG-3 that acts independently of major histocompatibility complex (MHC) class II (MHC-II) molecules [13, 14]. The interaction between FGL1 and LAG-3 has been shown to have an inhibitory effect on antigen-specific T-cell activation [15-17]. This intriguing interplay between FGL1 and LAG-3 has prompted intense investigation, as it may be the key to understanding how tumors exploit immune checkpoint mechanisms to evade immune surveillance.
Yes-associated protein (Yap) is an important cofactor downstream of the Hippo signaling pathway and is considered a central mediator of lung cancer pathogenesis [18-20]. Yap exhibits pronounced activation in lung cancer and contributes significantly to tumorigenesis and disease progression [21-23]. However, notably, mutations in components of the Hippo pathway, including Yap, are rarely found in lung tumors [24]. This finding suggests that the dysregulation of Yap in the context of cancer is likely orchestrated by Hippo pathway-independent mechanisms. Post-translational modifications (PTMs), such as phosphorylation, ubiquitination, methylation, and acetylation, are crucial regulatory processes governing protein behavior in both physiological and pathological contexts [25]. Recent research has revealed an interesting mechanism of Yap regulation, particularly in lung cancer, where Yap is subjected to SET1 histone methyltransferase (SET1A)-mediated monomethylation, notably at K342 [26]. This modification appears to influence the subcellular localization and transcriptional activity of Yap, further implicating Yap as a critical driver of lung cancer [26]. However, whether KRAS directly modulates Yap activity through the Hippo pathway remains controversial [27-29].
In this study, we aimed to investigate the role and mechanism of FGL1 in the regulation of KRAS-driven lung cancer by Yap and to develop a combination therapy strategy for lung cancer.
2 MATERIALS AND METHODS
2.1 Cell lines and culture conditions
All cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and short tandem repeat genotyping was performed prior to relevant experiments. The human non-small cell lung cancer (NSCLC) cell lines used were A549, H157, H358 and NCI-H1299, and the mouse Lewis lung cancer cell line LLC and human embryonic kidney (HEK)-293T cells were also used.
A549 cells were cultured in Ham's F-12K medium (21127022, Thermo Fisher, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; 04-001-1ACS, Biological Industries, Kibbutz Beit HaEmek, Israel), 100 U/mL penicillin/streptomycin (C0222, Beyotime, Shanghai, China) and 2 mmol/L glutamine (PB180420, Procell, Wuhan, Hubei, China). H157 and H358 cells were cultured in complete Roswell Park Memorial Institute (RPMI)-1640 medium (11875119, Thermo Fisher) supplemented with 10% FBS, 100 U/mL penicillin/streptomycin and 2 mmol/L glutamine. NCI-H1299 cells were cultured in complete RPMI-1640 medium supplemented with 10% FBS, 1 mol/L sodium pyruvate (11360070, Thermo Fisher), 100 U/mL penicillin/streptomycin and 4 mmol/L glutamine. LLC and HEK-293T cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM) (01-100-1A, Biological Industries) supplemented with 10% FBS, 1 mol/L sodium pyruvate, 100 U/mL penicillin/streptomycin and 4 mmol/L glutamine.
All cell lines were cultured under standard conditions at 37°C with 5% CO2. Medium changes and cell passaging were conducted at regular intervals to ensure the optimal growth and health of the cells.
2.2 Human specimens collection and preparation
The lung cancer tissue specimens used in this study were obtained from lung cancer patients who underwent surgical treatment at the First Affiliated Hospital of Anhui Medical University between September 2020 and October 2022, and sample collection was performed according to the guidelines of the Biomedical Ethics Committee of Anhui Medical University (Approval No: 20200462 Hefei, China). All samples were obtained with the patient's informed knowledge and were reviewed by two independent pathologists from the abovementioned hospital.
Following collection, the tissue samples were subjected to paraffin embedding. The paraffin blocks were then sliced into sections using a rotary microtome (HM325, Thermo Fisher).
2.3 Short hairpin RNAs (shRNAs) and lentiviral infection
The targeted shRNA sequences were synthesized and subsequently subcloned by ligating the oligonucleotides into multiple cloning sites of the pLVH1-EF1a-puro vector (SORT-B19, Biosettia, San Diego, CA, USA). To prepare the lentiviruses, this shRNA sequence was co-transfected with the delta 8.9 plasmid and the VSV-G plasmid into HEK-293T cells using Lipofectamine 3000 (L3000015, Invitrogen, Carlsbad, CA, USA). The viral supernatant was harvested 48 to 72 h post-transfection, filtered through a 0.45 µm filter, and diluted 2:3 with fresh medium containing 8 mg/mL polybrene. Target cells at 80% confluence were then infected with the viral supernatant. Western blotting was then performed to determine the transduction efficiency. All the shRNA sequences are listed in Supplementary Table S1.
2.4 Construction of cells with stable overexpression
To construct the plasmids for stable overexpression, the coding sequences of wild-type (WT) KRAS, Gly12Val (G12V) mutant KRAS, pLV3-CMV-Yap-3×FLAG-Neo, pEnCMV-3×FLAG-SETD1A-Neo, and pCMV-FGL1-3×FLAG-Neo were ligated into specific lentiviral vectors (mouse: #H11219; human: #GL122; OBiO Technology, Shanghai, China). Notably, the protospacer adjacent motif region in a single-guide RNA requires synonymous mutations in the corresponding cDNA. Via the lentiviral production and transduction methods described above, we obtained successfully transduced cells. The overexpression efficiency was evaluated using Western blotting.
2.5 Animals and in vivo experiments
C57BL/6JGpt (strain no. N000013), Yap-flox (strain no. T052147), and B6-KrasLSL-G12D/+ mice (strain no. T004551) were obtained from GemPharmatech (Nanjing, Jiangsu, China). Sftpc-Cre mice (strain no. 028054) were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). All studies utilized 4- to 6-week-old sex-matched mice. Mice of the same sex were randomly assigned to the control group or the experimental group. All animal procedures were approved by the Animal Ethics Committee of the Institute of Clinical Pharmacology at Anhui Medical University (Hefei, Anhui, China) (Ethical approval number: PZ-2022-019) and were performed in strict accordance with the stipulated guidelines. The mice were housed in a temperature-controlled sterile room with appropriate humidity and light conditions. The mice were euthanized by asphyxiation via carbon dioxide anesthesia after the experiment.
For in vivo experiments, the animals were randomized. To obtain KrasLSL-G12D/+-Sftpc-Cre double-positive transgenic mice, lung-specific transgenic Sftpc-Cre mice were crossed with KrasLSL-G12D/+ mice. Sftpc-Cre mice were also crossed with Yap-flox (Yapfl/fl) mice to obtain Yapfl/fl-Sftpc-Cre double-positive transgenic mice. Furthermore, Yapfl/fl-Sftpc-Cre mice were crossed with KrasLSL-G12D/+-Sftpc-Cre mice to obtain the Yapfl/fl-KrasLSL-G12D/+-Sftpc-Cre mice. Finally, all transgenic mice were injected intraperitoneally with tamoxifen (HY-13757A, MedChem Express, Monmouth Junction, NJ, USA) (100 mg/kg/day) for 5 consecutive days before use in subsequent experiments. Mice that lost more than 15% of their body weight, showed reluctance to move, or developed an abnormal gait were euthanized.
A mouse subcutaneous tumor model was established in 4- to 6-week-old WT C57BL/6JGpt mice by subcutaneous injection of shFGL1, Yap-oe, Yap-oe + shFGL1, SET1A-oe or shControl LLC cells (5 × 106). Tumor growth was monitored. The tumor dimensions were measured by calipers once every 2-3 days, and the tumor volumes were calculated with the equation V = π/6 × length × width2. Mouse body weights were measured every three days. Two to three weeks after LLC cell injection, the mice were euthanized, and the tumors were collected.
After tumor cell inoculation, when the tumor volume reached 100 mm3, administration of immune checkpoint drugs was commenced. On days 1, 3, 6, 12, 15, and 18, the mice were injected intraperitoneally with 100 µg of an anti-PD-1 monoclonal antibody (mAb) (1:500, BE0146-50mg, BioXCell, Lebanon, NH, USA) and/or an anti-FGL1 mAb (BE0332, BioXCell). Tumors were measured every 3 days starting on day 6 after drug injection. At the end of the experiment, the mice were euthanized, and all the tumors were collected.
A mouse lung metastasis model was established in 4- to 6-week-old WT C57BL/6JGpt mice by tail vein injection of shFGL1 and shControl LLC cells (1 × 106). Then, whole lung tissue was collected from the mice after 30 days.
2.6 Preparation of adeno-associated virus (AAVs)
AAV2/9, a viral vector, shows a degree of organ-targeting specificity because of the stable expression and gene integration capacity of AAV2, and it also shows high affinity for lung and nervous system tissues. AAV-CaMKIIα-mCherry (AAV2/9, 6×1012 vg/mL), AAV-CaMKIIα-mCherry-FGL1 shRNA (AAV2/9, 2×1013 vg/mL) and AAV-CaMKIIα-mCherry-scramble shRNA (AAV2/9, 1×1013 vg/mL) were obtained from Zhi En Biology (Hefei, Anhui, China). KrasLSL-G12D/+-Sftpc-Cre mice were injected with 300 µL of the viral suspension via the tail vein. Animals with unsuccessful injections were excluded from the analysis.
2.7 Western blotting
Cell lysis buffer (comprising 25 mol/L Tris-HCl (pH 7.6), 1% NP-40, 150 mol/L NaCl, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and 0.5 mol/L phenylmethanesulfonyl fluoride) was used to prepare the whole-cell lysates. Adherent cultured cells were subjected to three rounds of rinsing with PBS and subsequent removal of the medium. Following cell lysis with the abovementioned cell lysis buffer, the protein concentration was determined using a bicinchoninic acid (BCA) assay (23227, Thermo Fisher). The samples were loaded onto a 4-12% Bis-Tris sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (89888, Invitrogen), and the proteins were separated by electrophoresis at 150 V until the marker reached the bottom of the gel. Subsequently, the separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (#IPVH00010, Merck Millipore, Billerica, MA, USA). The membrane was blocked for 1 h at room temperature using 5% bovine serum albumin (BSA) (10711454001, Sigma, St. Louis, MO, USA) and then incubated with the primary antibody diluted in 5% BSA at 4°C for 12 h. This step was followed by four washes with Tris-buffered saline (TBS) + 0.5% Tween 20 (TBST). The membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature, followed by another four washes with TBST. Immunoreactions were detected using enhanced chemiluminescence (ECL) reagent (34577, Thermo Fisher), and images were acquired on a Tanon 5200 series (Tanon, Shanghai, China) fully automated chemiluminescence imaging system. The relative abundances of individual proteins were determined by quantifying the intensities of the corresponding protein bands on the Western blots (relative to the intensity of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) band) using ImageJ software (Bethesda, MD, USA). The primary antibodies used were as follows: anti-GAPDH (1:2,000, A19056, ABclonal, Wuhan, Hubei, China), anti-FGL1 (1:1,000, A20335, ABclonal), anti-PD-1 (1:1,000, A23007, ABclonal), anti-CD112 (1:1,500, A9622, ABclonal), anti-phospho-extracellular signal-regulated kinase 1/2 (p-ERK1/2) (1:1,000, sc-81492, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-ERK1/2 (1:1,500, A4782, ABclonal), anti-Granzyme B (GZMB) (1:1,500, 46890, Cell Signaling Technology, Danvers, MA, USA), anti-Perforin (1:1,500, 31647, Cell Signaling Technology), anti-Yap K342me (1:1,000, A18651P, ABclonal), anti-Yap (1:1,000, 14074, Cell Signaling Technology), anti-p-Yap (1:1,500, 13008, Cell Signaling Technology), anti-p-Lats1 (1:1,500, 9157, Cell Signaling Technology), anti-Lats1 (1:1,500, 3477, Cell Signaling Technology), anti-p-Mst1 (1:1,000, PA5-40177, Thermo Fisher), anti-Mst1 (1:1,500, A21842, ABclonal) and anti-SET1A (1:1,000, A18231, ABclonal) antibody. The anti-rabbit secondary antibody (A0423, Beyotime) and anti-mouse secondary antibody (A0473, Beyotime) were used at a 1:5,000 dilution.
2.8 Immunohistochemistry (IHC)
Paraffin sections (5 mm thick) of mouse tumor tissues and human lung cancer tissue samples were dewaxed by immersion in xylene, an alcohol gradient and, finally, double-distilled water. Routine hematoxylin-eosin (HE) staining was then performed. For IHC staining, the dewaxed paraffin sections were treated with citrate buffer at 95°C for 40 min for antigen retrieval. The sections were then blocked with 5% sheep serum for 30 min. The following primary antibodies diluted in 1% sheep serum were used: anti-ki67 (1:200, 12202, Cell Signaling Technology), anti-FGL1 (1:200, A20335, ABclonal), anti-SET1A (1:200, A18231, ABclonal), anti-Yap (1:200, 14074, Cell Signaling Technology) and anti-Yap K342me (1:200, A18651, ABclonal). The sections were incubated with these antibodies at 4°C overnight. Then, the sections were washed three times with PBS and treated with either biotin-labeled anti-mouse IgG (1:200, D110099, Sangon Biotech, Shanghai, China) or biotin-labeled anti-rabbit IgG (1:200, D110066, Sangon Biotech) for 1 h at room temperature. The sections were then washed three times with PBS and incubated for 30 min with streptavidin-HRP conjugates (1:300, B110053, Sangon Biotech). After three washes with PBS, color in the tissue sections was developed using 3,3 N-diaminobenzidine tetrahydrochloride (DAB) solution (A690009, Sangon Biotech), and the sections were then counterstained with hematoxylin. Two pathologists confirmed each sample pathologically. Quantification of positively stained cancer cells as a percentage of total cells was performed using ImageJ software.
2.9 Cell treatments
To evaluate the effect of inhibition of RAS signaling by U0126 (HY-12031A, MedChem Express) on the expression of FGL1, Yap K342me, ERK1/2 and p-ERK1/2 in A549, H157, H358 cells and NCI-1299 cells overexpressing KRASG12V, cells were treated with 1, 5, 10 or 20µmol/L U0126 (a MEK inhibitor) or dimethyl sulfoxide (DMSO, D8371, Solarbio, Beijing, China) for 24 h. To observe the ubiquitination of A549 cells and NCI-1299 cells overexpressing KRASG12V, cells were treated with 2 µmol/L U0126 or DMSO for 24 h.
2.10 Immunofluorescence staining (IF)
Paraffin sections of mouse tumor tissues were deparaffinized and subjected to antigen retrieval in 95°C citrate buffer for 40 min. Blocking was performed with 5% BSA for 30 min prior to incubation with the following primary antibodies diluted in 1% BSA: anti-Yap (1:200, 14074, Cell Signaling Technology), anti-FGL1 (1:200, A20335, ABclonal), anti-CD8a (1:200, ab217344, Abcam, Cambridge, MA, USA), anti-HA-Tag (1:400, M20003, Abmart, Shanghai, China), anti-CD206 (1:200, 24595, Cell Signaling Technology), anti-Foxp3 (1:200, 12653, Cell Signaling Technology), anti-EGF (1:200, A13615, ABclonal), and anti-SET1A (1:200, A18231, ABclonal). The sections were incubated with these primary antibodies overnight at 4°C. After three washes with PBS, the tissue sections were incubated with Alexa Fluor 647-conjugated anti-mouse IgG (M213808, Abmart) or Alexa Fluor 488-conjugated anti-rabbit IgG (M213211, Abmart) as a secondary antibody, along with 4',6-diamidino-2-phenylindole (DAPI) (DB1306, Thermo Fisher), for 1 h. The sections were then rinsed three times with PBS and blocked with Anti-Fade Mounting Medium (E675011, Sangon Biotech).
For cell sample preparation, cells were washed three times with PBS and fixed with 4% paraformaldehyde at room temperature for 15 min. The fixed cells were rinsed with PBS and then permeabilized with 0.2% Triton X-100 (P0096, Beyotime) for 10 min. This step was followed by overnight incubation with primary antibodies at 4°C. Subsequent steps, including secondary antibody incubation and sealing, were similar to those used for IHC staining.
2.11 Ubiquitination assay
For the ubiquitination assay, the Myc-Ub, Flag-SET1A (WT), Flag-SET1A (T1185A), Flag-FGL1 (P58521, Miaoling Biology, Hubei, Wuhan, China) and HA-KRASG12V plasmids were transfected into A549 cells and NCI-1299 cells. A549 cells and NCI-1299 cells (80% confluence) were transfected with the indicated constructs for 36 h, after which the cells were lysed using prechilled 0.5% Tris-NaCl-Tween (TNTE) lysis buffer composed of 0.5% Triton X-100, 50 mol/L NaCl, 1 mol/L ethylenediaminetetraacetic acid (EDTA) (E8040, Solarbio), and 50 mol/L Tris-HCl (pH 8.0). The cell lysates were then subjected to immunoprecipitation using an anti-Flag antibody, eluted by boiling in 1% SDS for 10 min, diluted tenfold with 0.5% TNTE lysis buffer, and subjected to another immunoprecipitation step with the anti-Flag antibody. The presence of ubiquitinated proteins was then assessed using Western blotting.
2.12 Quantitative Real-Time quantitative PCR (RT-qPCR)
Total RNA from mouse lung tumor tissues or A549 cells treated with U0126 was extracted using TRIzol (15596-026, Invitrogen). The RNA concentration was measured with a Nanodrop 1000 spectrophotometer (Thermo Scientific). Subsequently, 1 µg of total RNA was reverse transcribed into cDNA using a PrimeScript RT Kit (RR047A, Takara, Kyoto, Japan). RT-qPCR was performed using SYBR Premix Ex TaqII (RR820A, Takara) on a Bio-Rad iCycler iQ system (Bio-Rad, Hercules, CA, USA). All target gene expression levels were normalized to that of GAPDH, which served as the internal reference gene. Each sample was assayed in triplicate. Relative expression levels were computed using the ΔΔcycle threshold method. The primers used in this study are listed in Supplementary Table S1.
2.13 Luciferase reporter assay
HEK-293T cells were used for the luciferase assay. In 24-well cell culture plates, the p-FGL1-luc reporter (50 ng) containing the FGL1 promoter sequence was co-transfected with the Renilla luciferase plasmid to account for variations in transfection efficiency. This assay was facilitated using the Dual-Luciferase Reporter Assay System (E1910, Promega, Madison, WI, USA). Concurrently, cells were co-transfected with the Yap, p-FGL1-WT and mutant plasmids. To ensure a uniform DNA content across all wells, a basic luciferase reporter vector (PGL3-basic)was added at 1.5 mg per well. Cells intended for use in the luciferase assays were plated at 25% confluence. DNA transfection was performed the next day, and the cells were washed 6 h later. The cells were harvested and lysed after 48 h and were then subjected to a luciferase reporter assay using a dual luciferase reporter kit. Each transfection condition was performed in triplicate, and the entire experiment was repeated independently at least three times.
2.14 Flow cytometry (FCM) analysis
Excised mouse tumor tissues were digested in 1 mg/mL collagenase type I (C8140, Solarbio) and collagenase type IV (17104019, Thermo Fisher) for 30 min at 37°C. The digested tumor tissues were filtered through a 70 µm filter, and erythrocytes were lysed with Red Blood Cell Lysis Buffer (C3702, Beyotime) to prepare cell suspensions. Solutions of primary antibodies against CD3e (145-2C11, BD Biosciences, San Jose, CA, USA), CD8a (553035, BD Biosciences), Perforin (S16009A, BioLegend, San Diego, CA, USA), Interferon-γ (IFN-γ) (48-7311-82, Invitrogen) and tumor necrosis factor-α (TNF-α) (48-7321-82, Invitrogen) were prepared using FCM staining buffer (PBS with 2% FBS). The cells were incubated for 30 min at 4°C in the dark prior to washing with FCM buffer. The cells were then resuspended and analyzed using a flow cytometer (Beckman Coulter, Miami, FL, USA).
In vitro FCM was performed to assess apoptosis in tumor cells. After cell plating, carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) (40715ES25, Yeasen Biotechnology, Shanghai, China) was added for cell labeling. Following the experimental treatment, the culture supernatant was discarded. The cells were washed and permeabilized using Fix/Perm Buffer (GAS004, eBioscience, San Diego, CA, USA) for 20 min at 4°C. Staining with Annexin V (A35110, Thermo Fisher) was then performed. After a 25 min incubation, the cells were resuspended in FCM staining buffer and analyzed using a flow cytometer. The data were analyzed using FlowJo 10 software (Tree Star, Ashland, OR, USA).
2.15 T cell-mediated killing assay
CD8+ T cells were isolated from mouse spleen tissue, single-cell suspensions were prepared using an anti-mouse CD8+ T-cell sorting kit (B90011, Selleck, Houston, TX, USA) through negative sorting, and the cells were then cultured in a coculture system. The CD8+ T cells were activated with 2 µL/mL Dynabeads™ Human T-Cell Activator CD3/CD28 (11161D, Thermo Fisher) for 8 h for improved tumor killing. Then, treated NCI-H1299 cells, LLC cells and primary lung cancer cells isolated from mouse lung tumor tissues were cocultured separately with CD8+ T cells for 24 h. Finally, tumor cell apoptosis was detected by FCM.
2.16 co-immunoprecipitation (co-IP)
A549 cells were lysed in a buffer prior to centrifugation at 16,000 × g for 10 min. Then, 2 µL/mL of IgG and 20 µL/mL of protein A/G agarose (P2055, Beyotime) were added to the supernatants, and the mixtures were incubated at 4°C for 1 h with slow shaking. Then, the indicated antibody (2 µL/mL) was added to each sample, and the mixtures were incubated at 4°C overnight with rotation. Subsequently, the supernatants were incubated with 20 µL/mL protein A/G agarose for 6 h at 4°C. After washing with PBS five times, the beads were boiled in SDS-PAGE sample buffer and subsequently subjected to SDS-PAGE. Then, Western blot analysis was performed using specific primary antibodies and HRP-conjugated secondary antibodies, and protein bands were visualized by chemiluminescence.
2.17 Chromatin immunoprecipitation (ChIP)
The promoter sequence of FGL1 was downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov), and the binding site to the transcription factor TEAD was predicted in the JASPAR database (https://jaspar.genereg.net) and verified by ChIP experiment. Cells were subjected to crosslinking with 1% formaldehyde for 8 min at room temperature, and crosslinking was then quenched with 0.125 M glycine. Lysis was performed using an extraction buffer (10 mol/L NaCl, 0.5% NP40 and protease inhibitor added to 10 mol/L Tris HCl (pH 7.5)). For chromatin preparation, a digestion solution containing 15 mol/L NaCl, 60 mol/L KCl and 1 mol/L CaCl2 was added to the lysate, which was then treated with MNase (NEB, M0247S) at 37°C for 20 min. The resulting solution was combined with 2× Stop/ChIP buffer to generate chromatin fragments of 200-500 bp. The sample was centrifuged at 16,000 × g for 10 min at 4°C to collect the supernatant as the whole-cell input DNA. For immunoprecipitation, protein A/G Dynabeads were preblocked with 1× Stop/ChIP buffer, and this mixture was then combined with 2.5 mg of anti-TEAD4 (ab155244, Abcam) and anti-Yap (1:1,000, 14074, Cell Signaling Technology) antibodies for a 3 h incubation at 4°C. The extracted chromatin was then added to this antibody-bead complex mixture for overnight incubation at 4°C with rotation. The washing steps involved three sequential 5 min washes using wash buffer 1, wash buffer 2 and Tris/LiCl buffer, followed by one wash with Tris/EDTA buffer at room temperature. For elution and reversal of crosslinking, 100 µL of 1× elution buffer was added to the antibody-bead complexes, and 50 µL of 2× elution buffer was added to the input sample; both were then incubated overnight at 65°C. For DNA purification, first, RNaseA was added for a 1 h incubation at 37°C. Proteinase K was then added for a 2 h incubation at 55°C to degrade the proteins. DNA was extracted using phenol, chloroform and isoamyl alcohol, precipitated with ethanol, and finally resuspended in 25 µL of Tris/EDTA buffer. The prepared DNA was suitable for qPCR analysis, as shown in Supplementary Table S1.
2.18 Cycloheximide (CHX) chase assay
HEK-293T cells were transfected with the SET1A and KRASG12V plasmids using Lipofectamine 3000. At 24 h post-transfection, the culture medium was supplemented with CHX at a concentration of 10 mg/mL in complete DMEM supplemented with 10% FBS. The cells were further incubated for 0, 1, 3, 6, or 9 h. After each designated time point, cellular proteins were extracted using the aforementioned protein extraction method. The expression of the extracted proteins was then evaluated using Western blotting.
2.19 Bioinformatic analysis
The UALCAN-LUAD dataset (https://ualcan.path.uab.edu/index.html) was used to compare the expression of FGL1 mRNA and SET1A mRNA in 515 lung cancer patients with that in 59 healthy controls. Kaplan-Meier survival analysis of patients stratified by FGL1 expression was performed via the GEPIA server (http://gepia.cancer-pku.cn/index.html). The LUAD and LUSC databases of GEPIA were used for disease-free survival analysis of lung cancer patients after treatment. Quartile was used as cutoff values to determine low and high protein expression. The dataset GSE26850 (https://www.ncbi.nlm.nih.gov/geo/download/?acc = GSE26850) contains microarray data of mRNA expression profiles of a mouse LUAD model carrying the Kras-G12D mutation. The SOFT and Series Matrix files were downloaded for RMA normalization and subsequently processed with log2 for analysis and the results are presented as volcano plots. The dataset GSE13963 (https://www.ncbi.nlm.nih.gov/geo/download/?acc = GSE13963) contains microarray data of mRNA expression profiles of c-raf transgenic mice. Changes in FGL1 mRNA expression were analyzed after normalization and log2 treatment. Next, standardized pan-cancer expression profiles and corresponding clinical information were downloaded and decompressed from the UCSC Xena data frame (https://xenabrowser.net). The LUAD clinical information and gene annotation files were screened. The coding proteins with expression profiles were extracted, the grouping of lung cancer patients and healthy control samples were done. A total of 483 LUAD patients and 347 control samples were obtained, and then the expression differences of FGL1 between the two groups were viewed and point bar graph statistics were made.
We downloaded The Cancer Genome Atlas (TCGA-LUAD) somatic mutant cancer tissue data from TCGA (https://portal.gdc.cancer.gov), and 16 KRAS-mutated (KRAS+) and 29 KRAS wild-type (KRAS-) LUAD cancer tissue samples were collected. Subsequently, R version 4.3.0 (https://github.com/r-hub/R/releases) was used for statistical analyses. Pheatmap package in R (https://github.com/raivokolde/pheatmap) was applied to heatmap plot.
2.20 Statistics and reproducibility
Each experiment was conducted a minimum of three times, and the presented data are representative of at least three independent experiments. For statistical evaluation, we used Prism 8 (GraphPad) software. The measurement data are presented as the mean ± standard deviation (mean ± SD) values. One-way or two-way analysis of variance (ANOVA), Student's t test and the Wilcoxon rank-sum test were used to determine the significance of differences between experimental groups. P values of < 0.05 were considered significant. The results from all critical experiments demonstrated robust reproducibility. All replicate data and exact P values are explicitly annotated in the figure legends and summarized in the text.
3 RESULTS
3.1 FGL1 expression was elevated and required for cancer formation in KRAS-driven cancer
Research into the mechanisms of immune escape has focused predominantly on tumor-intrinsic factors, including the expression of ligands for immune checkpoint receptors. Thus, to identify immune checkpoint ligands that are crucial in KRAS-driven cancer, we first analyzed microarray data containing mRNA expression profiles from a murine LUAD model harboring the KrasG12D mutation (GSE26850). Among the identified checkpoint ligands (CD112, CD155, CD80, CD86, galectin-9 (Gal-9), programmed death ligand 1 (PD-L1), and FGL1), FGL1 exhibited a significant increase in expression (Figure 1A). To further elucidate whether KRAS can indeed regulate the expression of FGL1, we analyzed FGL1 expression in c-raf transgenic mice (GSE13963), because c-raf is a downstream component of the KRAS signaling pathway [2]. Our results revealed that the expression of FGL1 was significantly increased in mice expressing oncogenic c-raf than in mice expressing WT c-raf (Figure 1B). A series of analyses of lung cancer tissues from KrasLSL-G12D mice and control mice confirmed that FGL1 expression was upregulated in KRAS-driven cancer (Figure 1C-E and Supplementary Figure S1A-C). To further confirm that KRAS mutation can result in FGL1 upregulation, we overexpressed KRASG12V in the NRAS-mutated human lung cancer cell line NCI-H1299. Subsequent experiments showed that the overexpression of KRASG12V in NCI-H1299 cells led to the upregulation of FGL1 (Figure 1F and Supplementary Figure S1D), and treatment with the MEK inhibitor U0126 significantly reversed this effect (Supplementary Figure S1E). Consistent with this finding, U0126 significantly reduced FGL1 levels in KRAS-mutated lung cancer cell lines A549, H157, and H358 (Supplementary Figure S1F-H). These findings suggest that FGL1 is downstream of the KRAS-rapidly accelerated fibrosarcoma (RAF)-MEK-extracellular regulated kinase (ERK) pathway.
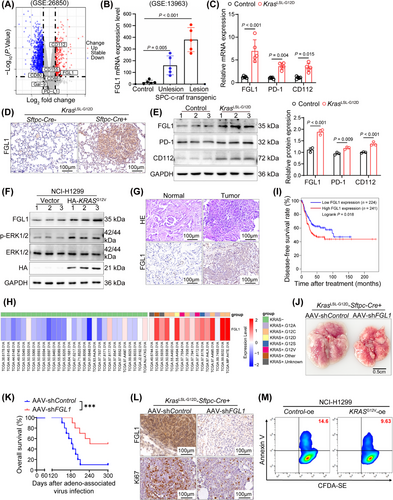
Subsequently, we investigated the effect of FGL1 on LUAD progression. FGL1 expression was increased in the lung cancer tissues of patients with KRAS mutations compared to that in adjacent normal tissues (Figure 1G and Supplementary Figure S1I). Furthermore, analysis of TCGA-LUAD data from the GEPIA server showed increased expression of FGL1 in lung cancer patients with KRAS mutations (Figure 1H and Supplementary Figure S1J). Notably, the disease-free survival time of patients with low FGL1 expression was longer than that of patients with high FGL1 expression (Figure 1I).
To evaluate the function of FGL1 in KRAS-driven cancer, FGL1 was knocked down in the lungs of KrasG12D mice using an AAV, and found that its effect on inhibiting tumor growth lasted up to 6 months. FGL1 knockdown in KRAS-driven lung tumors led to an evident decrease in the tumor burden 5 weeks after AAV-mediated delivery of shFGL1 (Figure 1J and Supplementary Figure S1K-L). Furthermore, this intervention significantly increased the survival rate of the mice (Figure 1K). Additionally, analysis of lung cancer samples revealed a parallel decrease in cell proliferation as FGL1 expression decreased in tumor tissues, consistent with the observed decrease in the tumor size in mice treated with AAV-shFGL1 (Figure 1L and Supplementary Figure S1M-N). In vitro, KRASG12V-overexpressing NCI-H1299 cells were cocultured with CD8+ T cells isolated from mouse spleens, and the results of the assays revealed a decrease in tumor cell apoptosis (Figure 1M and Supplementary Figure S1O), which was attributed to KRAS mutation-driven FGL1 upregulation. Thus, we concluded that the upregulation of FGL1 plays a pivotal role in KRAS-driven lung tumorigenesis.
3.2 FGL1 suppressed the CD8+ T-cell response to LUAD cells in vitro and in vivo
FGL1, which is known to be a ligand for LAG-3, increases tumor growth by suppressing antitumor immune processes in the tumor microenvironment (TME) [30-32]. To confirm the role of FGL1 in antitumor immunity in LUAD, shFGL1 was used to effectively silence FGL1 expression in LLC cells (Figure 2A). The shFGL1-treated tumors exhibited a notable reduction in volume and a decreased percentage of ki67-positive cells compared to control groups (Figure 2B-D). Moreover, FGL1 knockdown notably increased the infiltration of CD8+ T cells within tumor tissues (Figure 2E and Supplementary Figure S2A). In addition, comprehensive flow cytometric analysis revealed that suppression of FGL1 expression led to a significant increase in the secretion of the cytotoxic mediators IFN-γ, TNF-α and Perforin by CD8+ T cells (Figure 2F-G and Supplementary Figure S2B). To further investigate the impact of FGL1 on lung cancer development, a mouse model of lung metastasis was established via intravenous injection of LLC cells with stable FGL1 knockdown (Figure 2H). After 30 days, significant inhibition of lung cancer progression was observed in mice injected with FGL1-silenced cells (Figure 2I). Moreover, silencing FGL1 resulted in significant decreases in the number of M2 macrophages, the number of regulatory T cells, and the expression of EGF in tumor tissues, suggesting that silencing FGL1 can increase anti-tumor immune activity (Supplementary Figure S2C).
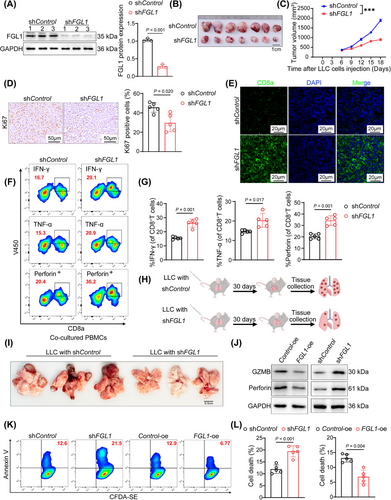
To investigate the immunosuppressive effect of FGL1, we generated A549 cell lines with FGL1 overexpression (FGL1-oe) or FGL1 knockdown (shFGL1). In the in vitro T-cell-mediated cytotoxicity assay, we observed that overexpression of FGL1 rendered the cells insensitive to cytolysis of peripheral blood mononuclear cells (PBMCs) and reduced the expression of cytotoxic molecules in cocultured PBMCs (Figure 2J and Supplementary Figure S2D). Silencing FGL1 had the opposite effects (Figure 2J and Supplementary Figure S2E). Subsequent coculture of CD8+ T cells with FGL1-knockdown A549 cells revealed increased apoptosis in A549 cells, whereas FGL1 overexpression attenuated CD8+ T-cell-induced apoptosis in A549 cells (Figure 2K-L). In conclusion, these findings strongly suggest that the tumor-promoting functions of FGL1 are contingent upon the activation of CD8+ T cells.
3.3 Yap promoted FGL1 expression in LUAD cells and inhibited the CD8+ T-cell response
Yap expression is increased in multiple cancer types, including LUAD and contributes to tumor progression [33]. However, the target genes of Yap in LUAD remain to be explored. Consistent with previous findings, our analysis revealed increased nuclear localization of Yap in LUAD samples from KrasLSL-G12D mice, as well as a substantial increase in the Yap protein abundance within the nucleus (Figure 3A and Supplementary Figure S3A). After KRASG12V overexpression, elevated nuclear retention of Yap was also detected in NCI-H1299 cells (Supplementary Figure S3B). To explore the potential regulation of FGL1 by Yap in KRAS-driven lung cancer, we screened the FGL1 promoter region and identified two putative TEAD-binding elements (TBEs) located within 600 bp upstream of the transcription start site of FGL1 (Figure 3B). These TBEs were confirmed through ChIP assays using anti-Yap and anti-TEAD antibodies, in which the IgG control did not show significant binding (Figure 3C). To further demonstrate the co-transcriptional effect of Yap on FGL1 expression, we inserted the FGL1 promoter sequence into PGL3-basic and found that it was strongly activated by Yap; the results upon single or combined mutations of TBE-1 and TBE-2 indicated the importance of these elements in regulating FGL1 promoter activity (Figure 3D). Moreover, overexpression of Yap in LLC cells caused a notable increase in the FGL1 protein level (Figure 3E). These findings suggest that FGL1 is a direct target gene of Yap.
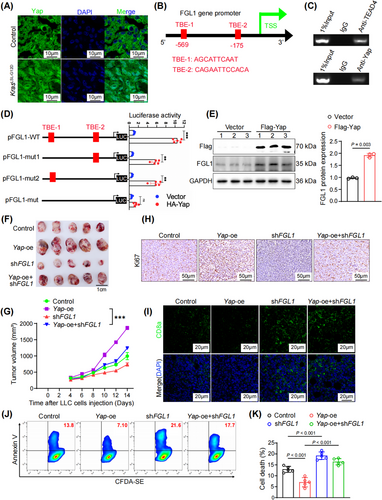
To further explore the role of Yap in FGL1-mediated immune escape in vivo, we inoculated syngeneic C57BL/6 mice with Yap-oe, Yap-oe + shFGL1, shFGL1 or control LLC cells. Notably, FGL1 silencing significantly attenuated the effect of Yap overexpression induced promoting immune escape in lung cancer cells, as evidenced by the reduction of tumor volume (Figure 3F-G and Supplementary Figure S3C-D). The increase in the proportion of ki67-positive cells resulting from Yap overexpression was reversed following FGL1 silencing (Figure 3H and Supplementary Figure S3E). Considering the reinforcing role of Yap in FGL1 transcription, we inferred that Yap is involved in mediating antitumor immune responses in the TME. As anticipated, Yap overexpression inhibited CD8+ T-cell infiltration into tumors, whereas FGL1 silencing counteracted this suppression (Figure 3I and Supplementary Figure S3F). FCM revealed that the decreases in the abundances of IFN-γ, TNF-α, and Perforin resulting from Yap overexpression were reversed after FGL1 silencing (Supplementary Figure S3G-I).
In vitro assays further confirmed that the suppression of FGL1 expression in A549 cells significantly reversed the inhibition of T cell apoptosis induced by Yap overexpression (Figure 3J-K). Taken together, these data confirmed that Yap promoted immune evasion in LUAD in an FGL1-dependent manner.
3.4 KRAS increased Yap activity by regulating Yap methylation, independent of the canonical Hippo pathway
In various human cancers, most components of the Hippo pathway are not synchronously mutated when Yap activity is dysregulated [34, 35]. This finding implies that specific mechanisms might be involved in activating Yap independent of the canonical Hippo pathway during tumorigenesis. Notably, recent studies have indicated that PTMs, such as phosphorylation and methylation, can regulate the localization and activity of Yap [26, 36]. Therefore, we sought to determine whether KRAS influences Yap activity by modulating its PTMs. To this end, we analyzed the phosphorylation and methylation statuses of Yap in mouse lung tumor tissues via Western blotting. Our findings revealed an increase in the methylation of Yap at K342, a highly conserved lysine residue, in KrasLSL-G12D mice compared to WT mice (Figure 4A). Interestingly, our Western blot analysis also revealed that the levels of crucial components of the Hippo pathway, including macrophage-stimulating 1 (Mst1), p-Mst1, large tumor suppressor kinase 1 (Lats1), and p-Lats1, remained largely unchanged (Figure 4A and Supplementary Figure S4A). This observation prompted us to propose that KRAS mutations may influence Yap activity through noncanonical kinase pathways independent of the Hippo signaling cascade. To confirm that Yap activation in KRAS-mutated lung adenocarcinoma is due to its monomethylation at K342, IHC staining was used to evaluate Yap K342 methylation (Yap K342me) in lung cancer tissues from KrasLSL-G12D mice. Our data indicated a pronounced increase in Yap K342me in Kras mutation-driven tumors (Figure 4B and Supplementary Figure S4B). In addition, we conducted separate experiments by overexpressing KRASG12V in NCI-H1299 cells and treating A549, H157, and H358 cells with the MEK inhibitor U0126. Western blot analysis revealed that KRAS overexpression resulted in an increase in Yap K342me (Figure 4C and Supplementary Figure S4C). Conversely, treatment with the MEK inhibitor U0126 led to the downregulation of Yap K342me (Figure 4D and Supplementary Figure S4D-E). Moreover, an increase in Yap K342me was observed in lung cancer tissues compared to adjacent normal tissues in patients with KRAS mutations (Figure 4E and Supplementary Figure S4F). These results demonstrated that KRAS contributed to facilitating Yap nuclear localization, promoting the methylation of Yap at K342 and increasing its activity.
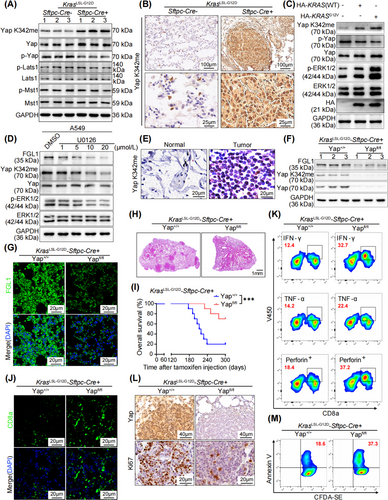
To further investigate the effects of Yap on KRAS-driven LUAD, a Yapfl/fl-KrasLSL-G12D-Sftpc-Cre mouse model was established. Western blot, IF and RT-qPCR analyses revealed a substantial reduction in FGL1 expression upon Yap loss in the lung (Figure 4F-G and Supplementary Figure S4G-I). Importantly, the tumors in Yapfl/fl-KrasLSL-G12D-Sftpc-Cre mice were significantly smaller than those in KrasLSL-G12D-Sftpc-Cre mice (Figure 4H), leading to a marked increase in the survival rate of lung tumor-bearing mice following Yap knockout (Figure 4I). In addition, the absence of Yap led to a significant increase in CD8+ T cell infiltration in tumor tissues (Figure 4J and Supplementary Figure S4J). Moreover, the increase in the number of CD8+ T cells that release IFN-γ, TNF-α and Perforin (Figure 4K and Supplementary Figure S4K) played an essential role in suppressing lung cancer progression in Yap-knockout mice. Consequently, the proportion of ki67-positive cells in mouse lung cancer tissues was significantly reduced upon Yap deletion, demonstrating that Yap deletion effectively hindered the proliferation of lung cancer cells (Figure 4L and Supplementary Figure S4L). These findings suggested that the inhibition of FGL1 transcription attributed to Yap deletion enhanced antitumor immunity in mice in vivo. In vitro coculture of cells isolated from lung cancer tissues with CD8+ T cells demonstrated that Yap knockout significantly increased T-cell-induced lung cancer cell apoptosis (Figure 4M and Supplementary Figure S4M). Collectively, these data suggest that Yap methylation-mediated upregulation of FGL1 facilitates immune evasion and tumor development in KRAS-driven cancer.
3.5 ERK1/2 phosphorylated and stabilized SET1A
Given that the above findings clarify that KRAS mutation leads to increased Yap methylation, we next aimed to identify the underlying mechanism involved. It has been reported that SET1A-mediated monomethylation at K342 regulates Yap activation and promotes tumorigenesis by preventing the nuclear export of Yap [26]. First, we confirmed that the knockdown of SET1A inhibited Yap methylation in KRASG12V-overexpressing NCI-H1299 cells (Figure 5A and Supplementary Figure S5A). Moreover, KRAS mutation increased the protein level and nuclear abundance of SET1A in mouse lung cancer tissues (Figure 5B-C and Supplementary Figure S5B-C). In addition, KRAS overexpression increased SET1A nuclear localization in NCI-H1299 cells (Figure 5D). Furthermore, we performed co-IP experiments and found that SET1A could interact with ERK1/2 in KRAS-mutated lung cancer cells (Figure 5E). To determine whether ERK1/2 regulates SET1A stability through phosphorylation, we conducted Phos-tag SDS-PAGE to examine the phosphorylation status of SET1A in co-transfected cells. Significantly, an upward shift in the band corresponding to SET1A was observed when SET1A was coexpressed with KRASG12V (Figure 5F). Furthermore, inhibiting the kinase activity of ERK1/2 with U0126 resulted in dephosphorylation of SET1A (Figure 5F). These results provided evidence that SET1A was a substrate of ERK1/2. Subsequently, we aimed to identify the specific residues for proline-directed phosphorylation in SET1A related to KRAS-ERK1/2 pathway activation. By analyzing the protein sequence of SET1A, we identified three residues with Pro at the p + 1 position (Thr916, Ser1153 and Thr1185) (Figure 5G), implying that these sites could be targets for phosphorylation by ERK1/2. Moreover, targeted mutation analysis demonstrated that Thr1185 was the specific site at which ERK1/2 phosphorylated SET1A (Figure 5H).
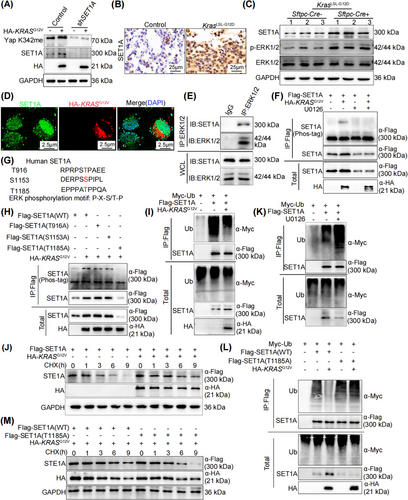
Next, to investigate the effect of ERK1/2-mediated phosphorylation of SET1A on its protein stability, cells were treated with the protein translation inhibitor CHX. SET1A ubiquitination was significantly reduced and SET1A degradation was slowed in cells overexpressing KRASG12V (Figure 5I-J). Conversely, inhibiting ERK1/2 activity with U0126 promoted SET1A ubiquitination and degradation (Figure 5K and Supplementary Figure S5D). In contrast, coexpression of KRASG12V with T1185A-mutated SET1A failed to stabilize the nonphosphomimetic SET1A mutant, which exhibited more rapid ubiquitination and degradation than did the WT SET1A protein (Figure 5L-M). Overall, these results suggest that SET1A is phosphorylated and stabilized via the KRAS-ERK1/2 axis to increase Yap K342me.
3.6 SET1A suppressed antitumor immunity in LUAD
Given the above data showing that KRAS mutation leads to the promotion of SET1A-mediated methylation of Yap, our next goal was to determine the role of SET1A in tumor immunity. First, we analyzed the expression level of SET1A in the TCGA database; SET1A exhibited high expression in LUAD compared to normal tissues (Figure 6A). Further IHC staining confirmed the elevated SET1A expression in lung cancer patients (Figure 6B and Supplementary Figure S6A). To validate the involvement of SET1A in antitumor immunity in LUAD, we established an LLC cell line with SET1A overexpression. Western blotting revealed that overexpression of SET1A significantly increased the level of Yap K342me, further resulting in an elevated FGL1 protein level (Figure 6C and Supplementary Figure S6B). We then inoculated C57BL/6 mice with SET1A-oe or control LLC cells to establish a subcutaneous tumor model. The tumors with SET1A overexpression exhibited a significantly greater volume and a higher percentage of ki67-positive cells (Figure 6D-E). Furthermore, overexpression of SET1A led to a significant increase in the expression of FGL1 within tumor tissues (Figure 6F and Supplementary Figure S6C). On the other hand, overexpression of SET1A led to reduced infiltration of CD8+ T cells into tumor tissues (Supplementary Figure S6D). Comprehensive flow cytometric analysis revealed that SET1A overexpression led to a decrease in the number of CD8+ T cells that secreted the key cytotoxic markers IFN-γ, TNF-α, and Perforin (Figure 6G-H and Supplementary Figure S6E).
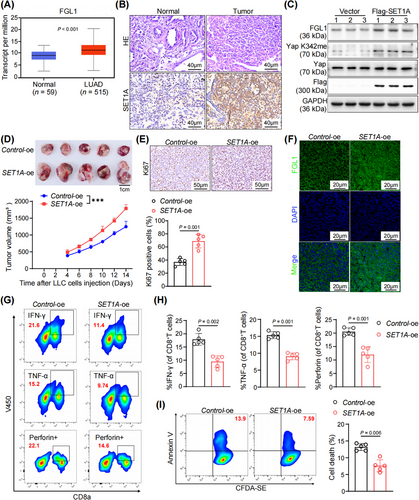
Furthermore, in vitro assays confirmed that the upregulation of SET1A impaired the killing of LLC cells by CD8+ T cells, resulting in reduced apoptosis (Figure 6I). Collectively, these data confirmed that SET1A promoted tumor immune evasion by upregulating FGL1 expression in LUAD.
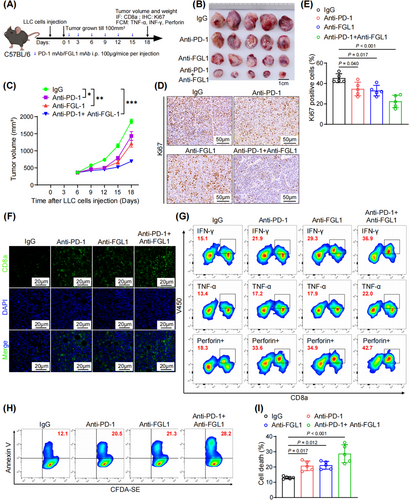
3.7 Combined blockade of PD-1 and FGL1 improved lung cancer treatment outcomes
PD-1 and FGL1 are crucial for T-cell suppression, suggesting that dual blockade of PD-1 and FGL1 may exhibit stronger antitumor efficacy than blockade of either alone. To investigate this hypothesis, mice bearing subcutaneous tumors derived from LLC cells were treated with anti-PD-1 and anti-FGL1 antibodies (Figure 7A). Notably, the combination treatment of anti-PD-1 and anti-FGL1 antibodies significantly inhibited tumor growth in mice (Figure 7B-C and Supplementary Figure S7A-B). Furthermore, compared with either antibody alone, the combined treatment led to a more pronounced reduction in the proportion of ki67-positive cells, indicating a decrease in tumor cell proliferation (Figure 7D-E). Although treatment with either antibody alone increased the infiltration of CD8+ T cells into the tumors, we observed that the combination of the two antibodies could further amplify this effect (Figure 7F and Supplementary Figure S7C). Phenotypic analyses also demonstrated a marked increase in cytotoxic T-cell activity when both antibodies were administered (Figure 7G and Supplementary Figure S7D-E). In in vitro cocultures of CD8+ T cells and LLC cells treated with both antibodies, the apoptosis rate was significantly higher than that in cocultures treated with either single agent (Figure 7H-I). In summary, our findings underscore the substantial improvement in antitumor immunity achieved through dual blockade of PD-1 and FGL1, which surpasses the effectiveness of anti-PD-1 monotherapy.
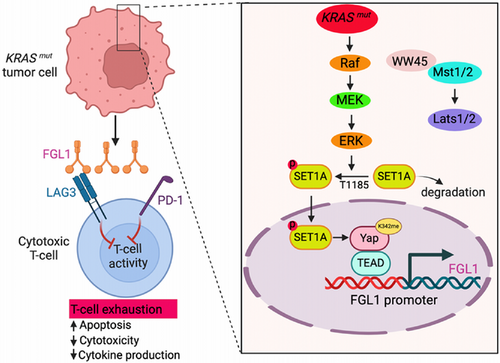
3.8 Deciphering KRAS-driven immune evasion mechanisms in lung cancer
We elucidated the intricate mechanisms by which KRAS mutations contribute to immune evasion in lung cancer cells (Figure 8). In the context of KRAS-mutated lung cancer, aberrant activation of the RAS pathway leads to the phosphorylation of SET1A at T1185 via the MEK/ERK signaling cascade. This phosphorylated form of SET1A exhibits increased stability and increased nuclear retention. Subsequently, SET1A-mediated methylation of Yap at K342 is increased, which further promotes the nuclear entry of Yap. Then, Yap binds to the promoter TEAD region of FGL1 and activates its transcription. The newly translated FGL1 interacts with LAG-3 receptors on the surface of T cells and effectively dampens their immune function. This series of molecular events reduces the infiltration of intratumoral CD8+ T cells and weakens their function as cytotoxic T cells, thereby promoting the immune escape of lung cancer cells and worsening disease prognosis. This molecular landscape highlights the importance of targeting both Yap and FGL1 for therapeutic interventions specifically tailored for KRAS-mutated lung cancer.
4 DISCUSSION
In this study, we identified FGL1 as a novel immunotherapeutic target and showed that genetic ablation or mAb-mediated inhibition of FGL1 enhanced CD8+ T-cell responses and elicited antitumor immune activity in KRAS-driven LUAD. The advent of immune checkpoint-targeted antibodies has offered a glimmer of hope for a selective subset of patients with advanced-stage KRAS-mutated cancer [37, 38]. However, the precise role of KRAS signaling in the TME and the tumor immune response is poorly understood. Recent findings have highlighted the overexpression of FGL1 in numerous solid tumors, in which is correlated with reduced 5-year overall survival rates [39-42]. However, the precise mechanisms governing the increased expression of FGL1 in tumor cells have remained largely elusive. Here, we discovered that KRAS mutations could activate ERK1/2, leading to the phosphorylation of tyrosine 1185 on the methyltransferase SET1A and its subsequent protein stabilization. Furthermore, SET1A methylation results in Yap monomethylation at K342, causing Yap to accumulate in the nucleus and transcriptionally regulate FGL1 expression. Recently, overexpression of FGL1 has been reported in STK11-mutant NSCLC [43]. This phenomenon is linked to an immune-cold TME, and the secretion of FGL1 has been suggested to potentially contribute to the systemic suppression of T-cell activation and the limited infiltration of immune cells into tumors [11, 44]. Previous studies have demonstrated that STK11 could suppress the function of nuclear Yap in response to metabolic stress through the activation of MST1 or Hippo-independent pathways [45-47]. Moreover, mutations in the STK11 gene are often accompanied by KRAS-activating mutations [48]. Thus, future investigations to determine whether both STK11 and KRAS impact the expression of FGL1 through a common molecular mechanism, specifically via modulation of the Hippo-Yap pathway, thereby influencing FGL1 expression in LUAD, are warranted.
Yap overexpression has been linked to tumor formation, progression and survival in lung cancer [19, 49, 50]. However, the precise mechanistic connection between Yap activity and KRAS remains poorly understood. Some studies have suggested that KRAS mutant could activate RASSF1A, leading to Yap downregulation in lung cancer [51]. Additionally, KRAS mutant has been shown to activate the MST2 kinase, which is anticipated to inhibit Yap [52]. Furthermore, increased levels of Yap have been reported in both human NSCLC and mouse models of KRAS-induced NSCLC, but whether tumors could develop without Yap has not been confirmed [53, 54]. Conversely, other studies have shown that the Yap level is not affected by modulation of KRAS mutant expression [27], indicating a complex regulatory relationship that may vary across tumors, stages of tumorigenesis, and specific KRAS mutations. Our study elucidates the mechanism by which KRAS regulates Yap through PTMs. KRAS mutations activate ERK1/2, leading to tyrosine 1185 phosphorylation of the methyltransferase SET1A, thereby stabilizing this protein. Furthermore, SET1A methylation results in Yap monomethylation at K342, resulting in Yap accumulation in the nucleus.
Previous studies have shown that Yap expression in tumor cells directly affects T-cell activity in the TME [55]. Furthermore, Yap promotes antitumor immunity by modulating myeloid-derived suppressor cell (MDSC) differentiation and expansion in the TME, which in turn hampers cytotoxic T-cell functions [56]. However, there is limited functional evidence regarding the role of Yap in immunosuppression in lung cancer in vivo. Recent research has shown that Yap transcriptionally upregulates PD-L1 in NSCLC cells [57]. PD-L1 is an immune checkpoint ligand that is expressed on the surface of tumor cells and is involved in tumor immune evasion [58]. PD-1 binds to PD-L1, acting as a receptor for this ligand. The binding of PD-L1 and PD-1 inhibits the antitumor function of tumor antigen-specific CD8+ T cells to result in tumor immune escape [59]. The identified role of mutated KRAS in orchestrating a TME conducive to immune escape underscores its role in facilitating tumor progression. Notably, favorable clinical outcomes have been associated with an increased intratumoral population of cytotoxic CD8+ T cells [60]. However, the influence of oncogenic KRAS on the fate of these tumor-infiltrating cytotoxic CD8+ T cells remains incompletely understood. Our research highlights Yap as a central regulator of FGL1 expression, identifying it a key player in immune evasion in KRAS-driven LUAD. This discovery not only reveals a potential therapeutic target but also underscores the significance of discerning the precise molecular mechanisms underlying immune escape in distinct subtypes of LUAD.
Cytotoxic CD8+ T cells play a critical role in driving tumor regression. However, their efficacy is often hindered by inhibitory ligand-receptor interactions within the TME. Combination therapy has become a promising approach to improve the efficacy of PD-1/PD-L1 immunotherapy and overcome resistance [47]. The identification of novel immune checkpoint targets has significant implications for cancer treatment. Our research demonstrates that simultaneous blockade of PD-1 and FGL1 effectively alleviates KRAS-mutated lung cancer. In summary, our findings provide valuable mechanistic insights into the elevated FGL1 level in KRAS-mutated cancers. Additionally, our study suggested that FGL1 could be a biomarker for KRAS-mutated cancers and that targeting the Yap-FGL1 axis has potential as an anticancer therapeutic strategy.
This study has several limitations. In KRAS mutation-driven lung cancer, Yap may regulate FGL1 expression through several different mechanisms. Therefore, further studies are needed to obtain a more comprehensive understanding.
5 CONCLUSIONS
In summary, our findings collectively underscored the pivotal role of FGL1 in suppressing CD8+ T-cell responses and promoting immune escape during KRAS-driven lung tumorigenesis. We revealed that KRAS mutations intricately regulated FGL1 expression through SET1A-mediated Yap methylation, independent of the canonical Hippo signaling pathway. Furthermore, we elucidated the critical role of KRAS-ERK1/2-mediated phosphorylation of SET1A in mediating its nuclear localization and protein stability. Most notably, we showed that the application of a combination therapy targeting FGL1 and PD-1 demonstrated significant efficacy in suppressing lung tumor progression. These findings may shed some light on the development of novel interventions that harness the immune system to treat KRAS-mutated lung cancer. The insights gained from this study are promising for improving the clinical management of patients with KRAS-driven LUAD.
AUTHOR CONTRIBUTIONS
Huan Qiu, Shihao Zhang, and Wei Wei conceived the project. Ji Jiang, Pengfei Ye, Ningning Sun, Weihua Zhu, Mei Yang, Manman Yu, Jingjing Yu, Hui Zhang, Zijie Gao, Ningjie Zhang, Cuncun Zhang, Sainan Miao, Mengqi Chai, Wenmin Liu, Yue An, Shijie Guo, Yuru Ji, Siqi Li and Jian Hong performed the experiments. Huan Qiu, Shihao Zhang, and Ji Jiang co-wrote the paper. All authors reviewed the manuscript.
ACKNOWLEDGEMENTS
This work was supported by National Natural Science Foundation of China Youth Project 82002450 (to H.Q.); The Research Program for Higher Education Institutions in Anhui Province 2022AH030081 (to S.Z.); Basic and Clinical Collaboration Enhancement Program of Anhui Medical University 2020xkjT023 (to H.Q.); The Research Program for Higher Education Institutions in Anhui Province 2023AH050656 (to H.Q.).
CONFLICT OF INTEREST STATEMENTS
The authors declare no competing interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All animal procedures had approval from the Animal Ethics Committee of the Institute of Clinical Pharmacology at Anhui Medical University (Ethics number: PZ-2022-019) and were performed in strict accordance with the stipulated guidelines. All procedures involving human participants in this study were approved by the Biomedical Ethics Committee of Anhui Medical University (Approval No: 20200462 Hefei, China), and informed consent from patients was acquired.
Open Research
DATA AVAILABILITY STATEMENT
The high-throughput sequencing data related to this study have been downloaded from the GEO database under accession codes GSE26850 and GSE13963. All other original data supporting this study's findings are available from the corresponding author upon reasonable request.




