Tryptophan 2,3-dioxygenase-positive matrix fibroblasts fuel breast cancer lung metastasis via kynurenine-mediated ferroptosis resistance of metastatic cells and T cell dysfunction
Yongcan Liu and Shanchun Chen contributed equally to this work.
Abstract
Background
Tumor metastasis is a major threat to cancer patient survival. The organ-specific niche plays a pivotal role in tumor organotropic metastasis. Fibroblasts serve as a vital component of the metastatic microenvironment, but how heterogeneous metastasis-associated fibroblasts (MAFs) promote organotropic metastasis is poorly characterized. Here, we aimed to decipher the heterogeneity of MAFs and elucidate the distinct roles of these fibroblasts in pulmonary metastasis formation in breast cancer.
Methods
Mouse models of breast cancer pulmonary metastasis were established using an in vivo selection method of repeated injections of metastatic cells purified from the mouse lung. Single-cell RNA-sequencing (scRNA-seq) was employed to investigate the heterogeneity of MAFs. Transgenic mice were used to examine the contribution of tryptophan 2,3-dioxygenase-positive matrix fibroblasts (TDO2+ MFs) in lung metastasis.
Results
We uncovered 3 subtypes of MAFs in the lung metastatic microenvironment, and their transcriptome profiles changed dynamically as lung metastasis evolved. As the predominant subtype, MFs were exclusively marked by platelet-derived growth factor receptor alpha (PDGFRA) and mainly located on the edge of the metastasis, and T cells were enriched around MFs. Notably, high MF signatures were significantly associated with poor survival in breast cancer patients. Lung metastases were markedly diminished, and the suppression of T cells was dramatically attenuated in MF-depleted experimental metastatic mouse models. We found that TDO2+ MFs controlled pulmonary metastasis by producing kynurenine (KYN), which upregulated ferritin heavy chain 1 (FTH1) level in disseminated tumor cells (DTCs), enabling DTCs to resist ferroptosis. Moreover, TDO2+ MF-secreted chemokines C-C motif chemokine ligand 8 (CCL8) and C-C motif chemokine ligand 11 (CCL11) recruited T cells. TDO2+ MF-derived KYN induced T cell dysfunction. Conditional knockout of Tdo2 in MFs diminished lung metastasis and enhanced immune activation.
Conclusions
Our study reveals crucial roles of TDO2+ MFs in promoting lung metastasis and DTCs’ immune evasion in the metastatic niche. It suggests that targeting the metabolism of lung-specific stromal cells may be an effective treatment strategy for breast cancer patients with lung metastasis.
List of abbreviations
-
- AKR1C1
-
- aldo-keto reductase family 1 member C1
-
- APC
-
- allophycocyanine
-
- CCL8/9/11/19
-
- C-C motif chemokine ligand 8/9/11/19
-
- CD31
-
- cluster of differentiation 31
-
- CM
-
- conditioned medium
-
- CNV
-
- copy number variation
-
- CTLA4
-
- cytotoxic T-lymphocyte associated protein 4
-
- DEGs
-
- differentially expressed genes
-
- DTCs
-
- disseminated tumor cells
-
- FACS
-
- fluorescence-activated cell sorting
-
- FBS
-
- fetal bovine serum
-
- Fth1
-
- ferritin heavy chain 1
-
- GFP
-
- green fluorescent protein
-
- GSVA
-
- gene set variation analysis
-
- GZMB
-
- granzyme B
-
- HMOX1
-
- heme oxygenase 1
-
- Ido1
-
- indoleamine 2,3-dioxygenase 1
-
- iDTR
-
- inducible diphtheria toxin receptor
-
- IFN-γ
-
- interferon-gamma
-
- IHC
-
- immunohistochemistry
-
- IVIS
-
- in vivo imaging system
-
- KYN
-
- kynurenine
-
- Luc
-
- luciferase
-
- Macro
-
- macro-metastatic lung
-
- MAFs
-
- metastasis-associated fibroblasts
-
- MEMα
-
- minimum essential medium alpha
-
- MFs
-
- matrix fibroblasts
-
- Micro
-
- micro-metastatic lung
-
- MxIF
-
- multiplex immunofluorescence
-
- MYH11
-
- myosin heavy chain 11
-
- MyoF1/2
-
- myofibroblasts1/2
-
- MyoFs
-
- Myofibroblasts
-
- N
-
- normal lung
-
- NRF2
-
- nuclear factor erythroid 2-related factor 2
-
- PBS
-
- phosphate buffered saline
-
- PD-1
-
- programmed cell death protein-1
-
- PDGFRA
-
- platelet-derived growth factor receptor alpha
-
- PD-L1
-
- programmed cell death 1-ligand 1
-
- PE
-
- phycoerythrin
-
- Pre
-
- pre-metastatic lung
-
- Ptgs2
-
- prostaglandin-endoperoxide synthase 2
-
- RSL3
-
- (1S,3R)-2-(2-Chloroacetyl)-2,3,4,9-tetrahydro-1-[4-(methoxycarbonyl)phenyl]-1H-pyrido[3,4-b]indole-3-carboxylic acid methyl ester
-
- scRNA-seq
-
- single-cell RNA sequencing
-
- shFTH1
-
- short hairpin RNA of FTH1
-
- shNC
-
- negative control short hairpin RNA
-
- TDO2/Tdo2
-
- tryptophan 2,3-dioxygenase
-
- UMAP
-
- dimensional reduction analysis of uniform manifold approximation and projection
-
- VDAC2
-
- voltage dependent anion channel 2
1 BACKGROUND
Metastasis is responsible for the majority of cancer deaths [1]. Many solid tumor types and subtypes, including breast cancer, display a non-random distribution of metastasis to distant organs, which is known as organotropism [2]. This phenomenon has long attracted extensive interest for oncologists. Lung is one of the most common sites of breast cancer metastasis, yet our understanding of the fundamental processes remains inadequate [3].
Among the steps involved in metastasis, colonization is considered the most challenging for disseminated tumor cells (DTCs) [4]. Unlike primary mammary or subcutaneous cancers, DTCs will face many obstacles in the lung, such as higher oxidative stress and ferroptosis threat [5, 6]. Thus, the support of the lung metastatic microenvironment is indispensable [7]. As the most abundant cell type in the tumor stroma, the role of metastasis-associated fibroblasts (MAFs) has become a new focus [8-11]. However, few previous studies have considered the heterogeneity of fibroblasts when studying the interactions between MAFs and DTCs or the immune-regulatory effect of MAFs. Recently, a metastasis-promoting lung fibroblast population with high prostaglandin-endoperoxide synthase 2 (Ptgs2) expression, named Ptgs2hi fibroblasts, was identified in breast cancer, which remodels the local immune microenvironment and facilitates pre-metastatic niche formation [12]. However, no study to date has examined how lung fibroblast subtypes and their function changes over time with metastasis progression. Moreover, the underlying mechanisms by which MAF subsets help DTCs adapt to high stress environments need further clarification.
Here, we aimed to characterize the heterogeneity of MAFs and elucidate roles of specific subtype in favoring lung metastasis formation, which may offer valuable understandings into the mechanisms underlying metastatic progression and serve as useful targets when developing future therapies against metastatic disease.
2 MATERIALS AND METHODS
2.1 Cell culture
Murine mammary carcinoma cell 4T1 (American Type Culture Collection, Manassas, VA, USA) and E0771 (CH3 Biosystems, Amherst, NY, USA) were cultured in RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% streptomycin/penicillin (C0222, Beyotime, Shanghai, China). Possible mycoplasma contamination was routinely and regularly monitored. 4T1/E0771 cells were retrovirally infected with a firefly luciferase reporter (for bioluminescence tracking) and selected with puromycin (1 µg/mL, 540222, Sigma Aldrich, St. Louis, MO, USA) for 14 days, named Luc-4T1 and Luc-E0771. Luc-4T1/E0771-LM3 cells were retrovirally infected with a green fluorescent protein (GFP) reporter and selected by fluorescence-activated cell sorting (FACS), named Luc-GFP-4T1-LM3 and Luc-GFP-E0771-LM3. All the cell lines used in the present study were verified by short tandem repeat genotyping prior to the start of the experiments.
2.2 Mice
All animal experiments were approved by the Ethics Committee of Chongqing Medical University and conducted according to the guidelines for the Care and Use of Laboratory Animals of Chongqing Medical University (2021084). Female BALB/c mice (6 weeks; injected with 4T1 cells) and C57BL/6 mice (6 weeks; injected with E0771 cells) were purchased from the laboratory animal center of Chongqing Medical University, which were housed in specific pathogen free environment. MMTV-PyMT mice were a kind gift from Prof. Suling Liu (Fudan University). Pdgfra-cre transgenic mice, inducible diphtheria toxin receptor (iDTR) transgenic mice and Tdo2 flox/flox (Tdo2fl/fl) mice were from Cyagen Bioscience (Suzhou, Jiangsu, China). Mice were euthanized via cervical dislocation, when weakness or body weight loss (70% of control littermates) was observed.
To establish organotropic metastasis models, Luc-4T1 cells (1 × 106) or Luc-E0771 cells (1 × 106) were suspended in Phosphate Buffered Saline (PBS, 100 µL) and subsequently orthotopically implanted into mice. Mice were anaesthetized and intraperitoneally injected with D-luciferin (150 mg/kg, MX4603-100MG, Maokangbio, Shanghai, China) to monitor metastasis by bioluminescence imaging (BLI) with the in vivo imaging system (IVIS) (Caliper Life Sciences, Hopkinton, Massachusetts, USA). Lungs with metastasis were dissociated by type I collagenase (SCR103, Sigma Aldrich), and inoculated to obtain DTCs. With repeated fat pad injection of DTCs, organotropic metastasis tumor cells (Luc-4T1-LM3 and Luc-E0771-LM3) were selected by three rounds of selection in vivo.
For depletion of PDGFRA+ MFs, Pdgfra-cre mice and iDTR mice were crossed to generate Pdgfra-DTR knock-in mice. Thereafter, mice were intraperitoneally injected with 25 ng/g of diphtheria toxin (DT, D0564, Sigma-Aldrich) twice per week to generate C57BL/6 iDTR+ mice. Pdgfra-cre mice were further crossed with Tdo2fl/fl mice for generating C57BL/6 Tdo2cKO mice. The same method was used to obtain BALB/c iDTR+ mice and BALB/c Tdo2cKO mice. MMTV-PyMT mice were crossed with C57BL/6 Tdo2cKO mice for generating MMTV-PyMT Tdo2cKO mice.
To compare the metastasis burden in iDTR+ and iDTR− mice (or Tdo2fl/fl and Tdo2cKO mice), 4T1-LM3 or E0771-LM3 (1 × 106) cells were injected (via the tail vein) into mice. When Tdo2fl/fl mice showed signs of weakness, mice were anaesthetized and lungs were imaged ex vivo by IVIS. Lung metastasis burden of MMTV-PyMT Tdo2cKO mice were assessed 5-6 weeks after primary tumor formation. To detect the role of TDO2 and KYN in resistance to ferroptosis, immune escape and metastasis formation of DTCs, 680C91 (TDO2 inhibitors, HY-108681, MedChem Express, Monmouth Junction, NJ, USA) was administrated intraperitoneally once a day at a concentration of 15mg/kg; KYN (HY-104026, MedChem Express) was administered intraperitoneally every other day at a dose of 100 mg/kg; liproxstatin-1 (CAS 950455-15-9, TargetMol, Shanghai, China) was administered intraperitoneally once per day at the dose of 20 mg/kg. Dosing was started from 3 days after inoculation and stopped when control mice showed signs of weakness or imminent death.
2.3 Primary cell isolation
To isolate primary lung stromal fibroblasts, lungs were harvested from 4 stages (N stage: normal; Pre stage: 2 weeks after inoculation; Micro stage: 3 weeks after inoculation; Macro stage: 6 weeks after inoculation) and minced into small pieces, then cultured with Minimum Essential Medium alpha (MEMα, ThermoFisher, Waltham, MA, USA). Primary lung stromal fibroblasts from Tdo2fl/fl or Tdo2cKO mice were isolated as mentioned above. All fibroblasts used in the experiments were less than 6 passages.
Splenic CD4+ and CD8+ T cells were isolated by MojoSort™ Mouse CD4 T (480006, Biolegend, San Diego, CA, USA) or MojoSort™ Mouse CD8 T (480008, Biolegend) Cell Isolation Kit, respectively.
2.4 Lung fibroblasts (LFs) function assay
To assay the function of LFs in DTCs colonization, primary N-LFs (LFs from normal lung), Pre-LFs (LFs from pre-metastatic lung), Micro-LFs (LFs from micro-metastatic lung) and Macro-LFs (LFs from macro-metastatic lung) were isolated and co-injected with 4T1-LM3 or E0771-LM3 cells in a 1:5 ratio (1 × 104 LFs and 5 × 104 tumor cells) via the tail vein, respectively. After 14 days, mice were euthanized (as described in Section 2.2) and lung tissues were collected. To assay the function of LFs in DTCs metastasis formation, 4T1-LM3 or E0771-LM3 (5 × 105) cells were orthotopically implanted into mice. After 10 days, N-LFs, Pre-LFs, Micro-LFs, and Macro-LFs (1 × 105) were administered through the tail vein. Mice were euthanized (as described in Section 2.2) on day 30. Lung tissues were collected and immediately stained with Bouin's fixative solution (PH0976, Phygene Scientific, Fuzhou, Fujian Province, China), counting lung nodules directly. Subsequently, the lungs were gently washed with running water and transferred to 70% ethanol to remove the fixative solution and used for further paraffin embedding.
2.5 Conditioned medium (CM) and collagen contraction assay
To acquire tumor cell-/MF-derived CM, sorted tumor cells/MFs were inoculated in FBS-free 1640/ MEMα medium (37°C, 36 h). Then the supernatant was centrifuged (2,000×g, 4°C, 5 min) and filtered (0.22 µm filter), saved as CM.
Lung stromal fibroblasts (2 × 105) were suspended in 100 µL medium, which was mixed with 100 µL of Collagen Type I Rat Tail (containing 68.75 µL medium; Corning, NY, USA). The mixture was added to l well of 24-well plates in triplicate. After solidification, CM was added and incubated for 24 h, the gels were photographed. Image J (https://imagej.net/software/fiji/downloads) evaluated gel contraction.
2.6 Single-cell RNA sequencing (scRNA-seq)
Lung samples (from N, Pre, Micro and Macro stages) were washed with Dulbecco's phosphate-buffered saline (DPBS, BL1109A, Biosharp, Hefei, Anhui, China) containing 10% streptomycin/penicillin and dissociated using a Tumor Dissociation Kit (130-096-730, Miltenyi Biotec, Bergisch Gladbach, Germany). Single cell suspensions were then filtered by a 70 µm (BS-70-CS, Biosharp) and a 40 µm cell filter (BS-40-CS, Biosharp) subsequently, lysed with Red Cell Lysis Solution (Biosharp), and washed with DPBS with 2% FBS. Hematopoietic (CD45+), epithelial (CD326+), endothelial (CD31+) and tumor cells (GFP+) were excluded by FACS to enrich CD326−CD45−CD31−GFP− populations (fibroblast-enriched cells). Subsequently, fibroblast-enriched cells were used for scRNA-seq, and scRNA-seq profiles were generated by 10× Genomics sequencing.
With the GemCode Technology, single cells were encapsulated into nanoliter-sized GEMs (Gel Bead in emulsion). Post reverse transcription-GEMs were cleaned up, and cDNA was amplified. cDNA was fragmented with repaired fragments end, and the adaptors were ligated to fragments which were double sided solid-phase reversible immobilization (SPRI) selected. Another double sided SPRI selecting was carried out after sample index PCR (reaction volume: 100 µL; temperature and time: 98°C, 45 s; 98°C, 20 s; 54°C, 30 s, 11 cycles; 72°C, 20 s; 72°C, 1 min; 4°C). After checking fragments size distribution and quantifing the library, scRNA-seq was performed using the MGISEQ 2000 platform (BGI, Shenzhen, Guangdong, China).
2.7 scRNA-seq data analysis
Raw scRNA-seq data needed map to the mouse reference genome (GRCm38) using the Cell Ranger Single Cell Software Suite (https://support.10xgenomics.com /single-cell-gene-expression/software/downloads/latest). R package Seurat (v 3.1.0) was used for next analysis. Cells with poor quality (expressing less than 200 genes, containing >90% of the maximum genes and >15% mitochondrial genes) were discarded. In total, 42,348 cells were obtained and assigned to 23 clusters. Copy number variation (CNV) analysis can find the contamination of malignant cells. Raw gene counts were normalized and the most highly variable genes were identified. Two-dimensional visualization was displayed by dimensional reduction analysis of uniform manifold approximation and projection (UMAP). The top differential genes were identified by FindConservedMarkers and served as marker genes (log fold change [logFC] > 0.25 and minimum percent > 0.25). Then, cell type annotation was carried out according to Cell Marker database (http://biocc.hrbmu.edu.cn/CellMarker/). R package Monocle2 was utilized for Pseudotime trajectory analysis. Intercellular interaction analysis was done using CellPhoneDB (https://github.com/ventolab/cellphonedb-data). Gene set variation analysis (GSVA) was performed by R (v 1.40.1). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of scRNA seq were performed on the Dr. Tom analysis system (BGI).
2.8 Bulk RNA sequencing
Total RNA extracted from the tissues/cells was qualified using a Nano Drop and Agilent 2100 bioanalyzer (Termo Fisher Scientifc, Waltham, MA, USA). Subsequent RNA sequencing was carried out by BGI. Differentially expressed genes (DEGs) were selected based on ǀlog2FCǀ > 2. All subsequent analyses (unless noted) were performed on the Dr. Tom analysis system.
2.9 Clinical tissue specimens
Lung metastasis specimens of breast cancer patients (5 cases) were from the First Affiliated Hospital of Chongqing Medical University (Chongqing, China). The specimens were obtained from previous clinical diagnosis by biopsy. All patients were informed and consented, which was approved by the Ethics Committee of Chongqing Medical University (2021084). The clinical information of all patients is listed in Supplementary Table S1.
2.10 Clinical parameter analysis
Gene expression profiles of breast cancer patients (GSE14020, GSE14018, GSE209998 and GSE5327) were normalized using R v.4.3.1. For analysis of the relationship between MFs gene expression signatures and overall survival (time from diagnosis to death) as well as other clinical parameters (metastasis survival [time from lung metastasis to death] and lung metastasis-free survival [time from diagnosis to lung metastasis]), we stratified patients from GSE209998, GSE14020 and GSE14018 based on MFs signatures expression (log2 [FPKM_Value + 1]), using the median as cut-off. For analysis of the relationship between the TDO2 level and lung metastasis-free survival, we stratified patients of GSE5327 based on TDO2 expression, using the median as cut-off. Sangerbox 3.0 (http://sangerbox.com/home.html) was used for Pearson correlation analysis. Gene set enrichment analysis (GSEA, v4.3.2) in GSE14020 and GSE14018 was used to analyze the association between TDO2 expression and lipid peroxisomal metabolism in each group. All analyses (unless noted) were performed using R (v.4.3.1).
2.11 Flow cytometry
To profile LFs along metastatic stages, lungs were collected from mice (orthotopic injection of cancer cells) at weeks 2 (Pre), weeks 3 (Micro) and weeks 6 (Macro). The collected lungs were used to prepare single cell suspension. CD16/32 (101302, Biolegend) was added into cells (1×106) and incubated on ice (10 min). Next, the respective antibodies were added and incubated on ice (30 min). Cells were then washed and re-suspended in FACS buffer (2% FBS in DPBS), adding 4',6-diamidino-2-phenylindole (DAPI) (3 µg/mL, 422801, Biolegend). LFs were analyzed and sorted by FACS using two positive selection markers, CD140a and CD140b, and negative selection markers (CD326, CD45 and CD31). Briefly, CD326−CD45−CD31−CD140a+/CD140b+ cells were regarded as fibroblasts. Numbers of LFs were counted by multiplying the total cells of lung with fibroblasts percentage. FlowJo V10 was used in data analysis (https://www.flowjo.com/).
To verify the dynamic shift in MAFs composition along metastatic stages, lungs were collected from mice at 2 weeks (Pre), 3 weeks (Micro) and 6 weeks (Macro) after orthotopic injection of cancer cells. After excluding hematopoietic (CD45+), epithelial (CD326+), endothelial (CD31+) and tumor cells (GFP+), the remaining cell population was regarded as the fibroblast-enriched cells. Subsequently, CD140b was used to identify fibroblasts; CD140a and CD146 were used to identify MFs and Myofibroblasts-2 (MyoF2) respectively, while the CD140b+CD140a−CD146− cells were regarded as Myofibroblasts-1 (MyoF1).
To elucidate the immune suppressive function of Tdo2high MFs, T cell function was analyzed. The isolated CD4+ T and CD8+ T cells were co-cultured with Tdo2fl/fl-MFs, Tdo2cKO-MFs with or without KYN (0.1 mmol/L) treatment. The system was also supplemented with anti-CD3 (250 ng/mL, 100238, Biolegend)/anti-CD28 antibodies (250 ng/mL, 102116, Biolegend) and interleukin-2 (IL-2, 10 ng/mL, 575404, Biolegend). Flow cytometry analyses of T cells was performed after 48 h. For in vivo analysis, single cell suspensions of lungs from Tdo2fl/fl and Tdo2cKO mice were prepared. Surface marker was stained for 0.5 h on ice. For intracellular cytokine staining, fixed cells were permeabilized with Cyto-FastTM Fix/Perm Buffer Set (426803, Biolegend). For intra-nuclear staining, cells were permeabilized with True-NuclearTM Transcription Factor Buffer Set (424401, Biolegend), and then respective antibodies were added. Flow cytometry was performed using the BD FACSCantoTM plus machines (BD, Franklin Lakes, NJ, USA) and analyzed using FlowJo V10. The following antibodies were used in the study: phycoerythrin (PE) anti-mouse CD45 (2.5 ng/µL,103106, Biolegend), PE anti-mouse CD31 (10 ng/µL, 102408, Biolegend), PE anti-mouse CD326 (10 ng/µL, 118206, Biolegend), allophycocyanine (APC) anti-mouse CD140a (10 ng/µL, 135908, Biolegend), Brilliant Violet 421™anti-mouse CD140a (5 ng/µL, 135923, Biolegend), APC anti-mouse CD140b (10 ng/µL, 136008, Biolegend), PE/Cyanine7 anti-mouse CD146 (2.5 ng/µL, 134714, Biolegend), APC anti-mouse CD3 (5 ng/µL, 100236, Biolegend), fluoresceine isothiocyanate (FITC) anti-mouse CD4 (2.5 ng/µL, 100405, Biolegend), PE anti-mouse CD4 (2.5 ng/µL, 100512, Biolegend), FITC anti-mouse CD8 (10 ng/µL, 100706, Biolegend), PE anti-mouse CD8 (2.5 ng/µL, 100707, Biolegend), Brilliant Violet 421™anti-mouse FOXP3 (5 ng/µL, 126419, Biolegend), FITC anti-human/mouse granzyme B (GZMB, 5 µL per million cells in 100 µL staining volume, 396404, Biolegend), PE anti-mouse CD366 (Tim-3, 2.5 ng/µL, 119703, Biolegend), PE anti-mouse programmed cell death protein-1 (PD-1, 10 ng/µL, 135205, Biolegend).
2.12 Hematoxylin-eosin (H&E) staining and immunohistochemistry (IHC)
The paraffin-embedded lungs were sectioned, de-paraffinized and rehydrated. The slices were stained with H&E staining kit (Solarbio Life Sciences, Beijing, China). For IHC, after antigen retrieval, the sections were incubated with primary antibodies Ki67 (1:100, AF02778, Aifang Biotechnology, Changsha, Hunan, China), PTGS2 (1:100, SC56-06, HUABIO, Hangzhou, Zhejiang, China), TDO2 (1:100, 15880-1-AP, Proteintech, Wuhan, Hubei, China), fibronectin (FN, 1:300, 66042-1-Ig, Proteintech), periostin (POSTN, 1:4000, 66491-1-Ig, Proteintech) or alpha smooth muscle actin (α-SMA, 1:300, GB111364-100, Servicebio, Wuhan, Hubei, China), indoleamine-2,3-dioxygenase 1 (IDO1, 1:100, 13268-1-AP, Proteintech), ferritin heavy chain 1 (FTH1, 1:200, ET1610-78, HUABIO) overnight at 4°C. Subsequently, secondary antibody was incubated for 60 min. Stained with diaminobenizidine for 3 min, the slices finally stained with hematoxylin. The slices were scanned by 3DHISTECH Pannoramic MIDI or KFBIO KF-FL-020. Score IHC staining as follows: 0, no staining; 1, weak; 2, moderate; 3, strong. Score the percentage of stained cells as follows: 0, no staining; 1, < 10%; 2, 10-50%; and 3, > 50%. The IHC score was calculated by multiplying the staining score with the percentage score.
2.13 Picrosirius red staining
To show the collagen fiber in macro-metastatic lung tissues, the de-paraffinized tissue sections at 4 µm thickness were stain with Modified Sirius Red Stain Kit (G1473-100ml, Solarbio Life Sciences) according to the manufacturers’ instructions. Briefly, the tissue sections were stained with Iron Hematoxylin Staining Solution for 5-10 min, and washed with distilled water and tap water for 10-20 s and 5-10 min, respectively. Subsequently, Sirius Red Staining Solution was used for drip dyeing for 5-10 min. Thereafter, the slices were rinsed slightly with running water, and then dehydrated by series of ethanol and transparent by xylene, finally sealed with neutral gum and observed by optical microscope (Nikon, Tokyo, Japan).
2.14 Multiplex immunofluorescence (MxIF)
For MxIF staining, the slices were stained with a 4-color Fluorescence kit (Aifang Biotechnology). Lung sections were de-paraffinized, rehydrated and permeabilized, followed by 15 min of citric acid antigen retrieval under high temperature and high pressure (100°C, 70-80 kPa). After natural cooling, the slides were blocked and incubated with primary antibodies for 17 h (4°C), and secondary antibodies and Opal TSA dyes for 50 min (25°C) and 10 min (25°C), respectively. The next rounds of staining consisted of antigen retrieval (15 min, 100°C), primary antibodies (16 h, 4°C), secondary antibodies (50min, 25°C), and Opal TSA dyes (10 min, 25°C). Afterwards, the slices were stained with DAPI (G1012, Servicebio) and finally sealed. The following antibodies were used: rabbit anti-mouse/human myosin-11 (MYH11, 1:50, 21404-1-AP, Proteintech), rabbit anti-mouse/human PDGFRA (1:200, ab203491, Abcam), rabbit anti-mouse/human TDO2 (1:200, 15880-1-AP, Proteintech), rabbit anti-mouse/human CD4 (1:50, ab288724, Abcam, Cambridge, MA, USA), rabbit anti-mouse/human CD8 (1:100, ab237709, Abcam), goat anti-rabbit polymer-horseradish peroxidase (HRP) secondary antibody (Aifang Biotechnology). The fluorescence signal was captured by 3DHISTECH Pannoramic MIDI II or KFBIO KF-FL-020. Image analysis of the MxIF was assessed by 2 certified pathologists.
2.15 Cell counting kit-8 (CCK-8) assay
Erastin (HY-15763, MedChem Express) and (1S,3R)-2-(2-chloroacetyl)-2,3,4,9-tetrahydro-1-[4-(methoxycarbonyl)phenyl]-1H-pyrido[3,4-b]indole-3-carboxylic acid methyl ester (RSL3, HY-100218A, MedChem Express) were used to induce ferroptosis in tumor cells. 4T1-LM3 (5,000 cells per well) and E0771-LM3 (1× 104 cells per well) were added in 96-well plates (treated with erastin at concentrations of 0, 0.1, 0.5, 1, 2, 5 µmol/L or RSL3 at concentrations of 0, 0.2, 0.3, 0.5, 1, 2 µmol/L), with/without CM/KYN (0.1 mmol/L)/liproxstatin-1(1 µmol/L) treatment for 48 h. For determination of cell viability, CCK-8 (MRC-C23009-500T, Miracle, Chengdu, Sichuan, China) solution was added and cultured 2 h. Optical density (OD) at 450 nm was measured using an Eon microplate reader (BioTek, Biotek Winooski, Vermont, USA).
2.16 qRT-PCR
The extracted RNA was reverse-transcribed into cDNA using PrimeScript RT Reagents Kit (RR036A, Takara). Gene expression was detected using TB GreenTM Premix Ex TaqTM II (RR820A, Takara), and β-Actin was the internal control. PCR conditions: 30 s at 95°C for the preliminary denaturation, and 39 cycles of 5 s at 95°C, 30 s at 58°C, 20 s at 72°C for the amplification, and 10 min at 72°C for the extension. Primer sequences used are shown in Supplementary Table S2.
2.17 Western blotting assay
The proteins were extracted by RIPA lysates (R0010, Solarbio Life Sciences) and quantified by BCA kit (P0010S, Beyotime). Then, proteins were transferred onto PVDF membrane and blocked for 60 min. PVDF membranes were co-incubated with primary antibody (FTH1, 1:2000, ET1610-78, HUABIO) overnight at 4°C. Washed with PBS, the membranes were incubated with appropriate HRP-conjugated secondary antibodies (Biosharp) for 60 min at 25°C, and the ECL kit (1705061, Bio-Rad, Hercules, CA, USA) was used to visualized proteins. After visualization, Image J software was used to quantify the gray value representing the protein expression levels.
2.18 Establishment of stable expression cell lines
shFTH1 and control shRNA were synthesized by GenePharma (Shanghai, China) and cloned into PGLVH1/GFP vector. The sequences of shRNA were listed in Supplementary Table S3. Luc-4T1/E0771-LM3 cells were infected by the packaged lentivirus and selected by FACS to establish stable expression cells.
2.19 Reactive oxygen species (ROS) detection
For ROS detection, 4T1-LM3/E0771-LM3 cells were incubated with BODIPY-C11 (2 µmol/L, thermo.D3861, Invitrogen, Carlsbad, CA, USA) for 30 min (37°C), trypsinized and washed, then analyzed by flow cytometer. For GFP-labeled tumor cells, erastin (1 µmol/L)/deferoxamine mesylate (DFOM, 100 µmol/L, S5742, Selleck, Shanghai, China)-treated 4T1-LM3/E0771-LM3 cells were incubated with Dihydroethidium (DHE, 2 µmol/L, S0063, Beyotime) for 30 min at 37°C, and other steps were consistent with BODIPY-C11.
2.20 Determination of intracellular free Fe2+
For Fe2+ determination, cells were seeded in 48-well plates and incubated with ferro-orange (F374, DOJINDO, Kumamoto, Japan). The cell nuclei were stained by Hoechst 33342 (IH0070, Solarbio Life Sciences). Images were captured by Leica DMi8 microscope (Leica microsystems, Wetzlar, Germany).
2.21 High performance liquid chromatography (HPLC)
To measure KYN concentrations, cells were homogenized with RIPA lysis buffer, and total protein was quantified as mentioned above. The culture medium was centrifuged and filtered. Medium or cell lysis (100 µL) was mixed with 12% perchloric acid (100 µL) and vortex and centrifuged (10,000 × g, 5 min, 4°C). The protein-free supernatants were determined by HPLC assay as described in previous studies [13].
2.22 High content real-time imaging
CD140a+ MFs freshly isolated by flow cytometry were incubated (3,000 cells per microplate) in 96-well microplates (CellCarrier-96 Ultra, PerkinElmer, Waltham, MA, USA) and cultured for 6 h. CD3+ T cells were sorted from spleen by MojoSort™ Mouse CD3 Selection Kit (480100, Biolegend) and subsequently incubated with carboxyfluorescein diacetate succinimidyl ester (CFSE, C34570, Invitrogen), then CD3+ T cells were added to the (10,000 cells per well) microplates and followed by incubation in a high-content imaging system (Operetta CLS, PerkinElmer), and images were acquired every 0.5h for 8 h.
2.23 Enzyme-linked immunosorbent assay (ELISA) analysis
To detect IFN-γ from CD4+ T or CD8+ T cells, the collected supernatants were centrifuged (2,000×g, 4°C, 5 min) and filtered. The levels of IFN-γ were measured using a mouse IFN-γ ELISA Kit (EMC101g.96, NeoBioscience, Shenzhen Guangdong, China).
2.24 Statistics
Statistical analysis was performed by GraphPad 8.0.2 software (https://www.graphpad.com). Experiments were repeated at least three times. Comparisons between two groups were performed using a two-tailed Student's t-test, and multiple groups were compared using a one-way ANOVA. Pearson correlation analysis was employed to identify the correlation between variables. Statistical significance was defined as *P < 0.05; **P < 0.01; ***P < 0.001.
3 RESULTS
3.1 Activated lung fibroblasts promoted lung metastasis of breast cancer
Stromal fibroblasts are the major cells in the lung microenvironment [14]. It has been reported that parental cells (human triple-negative breast cancer MDA-MB-231 cells) and their highly metastatic cancer cells (MDA-MB-231-LM2) have similar functions to induce LF activation, but MDA-MB-231-LM2 cells function earlier and more strongly [9].
To better explore roles of local LFs in breast cancer lung metastasis, we generated lung-tropic metastasis derivatives 4T1/E0771-LM3 by repeated fat pad injection and metastatic clone selection, using luciferase-labeled murine breast cancer cell lines 4T1 and E0771 (Supplementary Figure S1A). The metastatic ability and incidence of lung-tropic metastasis increased with the number of selection and expansion rounds, confirmed by BLI (Supplementary Figure S1B-C). Compared with CM from parental cells, LFs stimulated with CM from LM3 cells showed a significant increase in collagen gel contraction (Supplementary Figure S1D).
Metastatic progression in mice could be divided into three stages: Pre, Micro (when lungs harbor DTCs to form primary micro-metastases) and Macro stages (when macro-metastases are prominent and widespread) [9, 15]. Extracellular matrix (ECM) remodeling is a hallmark of the metastatic niche [16], as demonstrated by elevated FN, POSTN expression and picrosirius red staining in both 4T1-LM3 model (Figure 1A-B) and E0771-LM3 model (Supplementary Figure S2A-B). Fibroblasts are the major players in ECM production [17]. Enhanced expression of α-SMA (a marker of activated fibroblasts) (Figure 1A, Supplementary Figure S2A) suggested the activation of LFs during metastasis progression [9]. Sorted by FACS using two positive selection markers, CD140a (PDGFRA) and CD140b (PDGFRB), and a group of negative selection markers (GFP, CD326, CD45 and CD31) (Supplementary Figure S2C), activated LFs were found to be obviously increased accompanying metastasis progression (Figure 1C). Next, we isolated normal and activated LFs from the lungs of BALB/c and C57BL/6 mice at different stages, named N-LFs, Pre-LFs, Micro-LFs, Macro-LFs, respectively. To understand whether Pre-LFs, Micro-LFs and Macro-LFs had an effect on lung metastasis of breast cancer, DTC colonization and metastatic nodules were evaluated. After co-injection of 4T1-LM3 or E0771-LM3 cells with LFs via the tail vein, Pre-LFs, Micro-LFs and Macro-LFs strikingly promoted lung colonization of DTCs in comparison with N-LFs (Figure 1D, Supplementary Figure S2D). 4T1-LM3 or E0771-LM3 cells were injected into mice mammary gland fat pad to generate primary tumor, and LFs were administered through the tail vein. Injection of Pre-LFs, Micro-LFs and Macro-LFs clearly increased lung metastatic nodules to a greater extent than N-LFs (Figure 1E, Supplementary Figure S2E). Together, these data substantiated that lung stromal fibroblasts acquired metastasis-promoting property.

3.2 Distinct fibroblast subtypes in breast cancer lung metastasis ecosystem
Recent studies reported that primary tumors harbor different fibroblast subtypes with diverse origins and functions [18, 19]; however, the roles of fibroblast subtypes need to be determined in the metastatic microenvironment. To identify fibroblast subtypes associated with breast cancer metastasis progression, we sampled LFs along time points of metastasis development (Pre, Micro and Macro stages) with the normal lung from naive mice as controls, generating scRNA-seq profiles by 10× Genomics sequencing (Figure 2A). We applied a negative selection gating strategy to exclude hematopoietic (CD45+), epithelial (CD326+), endothelial (CD31+) and tumor cells (GFP+). The remaining CD326−/CD45−/CD31−/GFP− cell population was regarded as fibroblast-enriched cells and used for scRNA-seq analysis [20] (Supplementary Figure S3A). A total of 42,348 high-quality cells were retained (Figure 2B), which could be assigned to 23 clusters through Seurat analysis (Figure 2C). Thereafter, CNV analysis was employed to exclude the contamination of malignant cells (Supplementary Figure S3B). According to marker gene expression, clusters were identified as fibroblast, endothelial cell, mesothelial cell, immune cell, mesenchymal progenitor, pericyte, ciliated cell and clara cell (Figure 2D, Supplementary Figure S4A-B). Cluster 19 was not defined due to the lack of known marker gene expression and functional pathways, and was eliminated from further analysis.
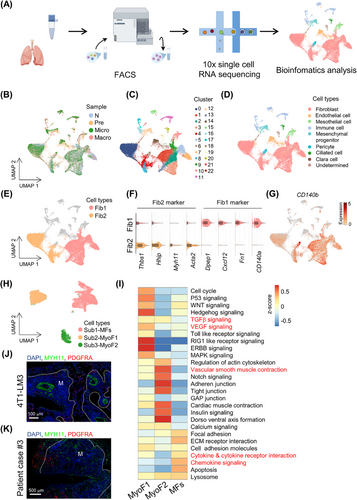
Interestingly, fibroblasts could be obviously partitioned into two broad classes (Figure 2E-F): fibroblast 1 (Fib1, CD140a+; 18,338 cells; clusters 1, 2, 3, 5, 7, 9, 13) and fibroblast 2 (Fib2, Myh11+; 13,518 cells; clusters 0, 4, 8). Both fibroblast subtypes highly expressed canonical fibroblast markers such as CD140b, secreted protein acidic and rich in cysteine (Sparc) and vimentin (Vim) [17] (Figure 2G, Supplementary Figure S4C). However, fibroblast UMAP plot exhibited 3 re-clustered subtypes (Figure 2H). Each subtype of fibroblasts displayed distinct transcriptional profiles (Supplementary Figure S4D). Subtype 1 (corresponding to Fib1 in Figure 2E; named matrix fibroblasts [MFs, CD140a+]) was characterized by high ECM signatures, including collagen molecules (collagen type V alpha 2 chain [Col5a2], collagen type VI alpha 3 chain [Col6a3]), fibronectin 1 (Fn1), lumican (Lum) and versican (Vcan), as well as inflammatory chemokines (C-X-C motif chemokine ligand 12 [Cxcl12], interleukin 6 [Il6], C-C motif chemokine ligand 7 [Ccl7], C-C motif chemokine ligand 2 [Ccl2]) and complement genes (complement C3 [C3] and complement C7 [C7]) (Figure 2F, Supplementary Figure S5A). Simultaneously, terms of GO and KEGG analysis enriched for MFs were related to ECM and collagen fibril organization, inflammatory response regulation and amino acid/cytokine response (Supplementary Figure S5B-C), suggesting that this subtype may participate in immune regulation. Subtypes 2 and 3 (corresponding to Fib2 in Figure 2E) mainly expressed known marker genes for myofibroblasts [21], including actin alpha 2 (Acta2), Myh11, transgelin (Tagln) and hedgehog interacting protein (Hhip) (Supplementary Figure S4D). Moreover, GSVA of Subtypes 2 and 3 showed significant enrichment for the transforming growth factor beta (TGFβ) signaling pathway, the vascular endothelial growth factor (VEGF) signaling pathway and vascular smooth muscle contraction (Figure 2I), indicating their microvascular signature. Accordingly, Subtypes 2 and 3 were named Myofibroblasts-1 (MyoF1) and Myofibroblasts-2 (MyoF2), respectively (Figure 2H). To validate the main fibroblast subtypes described above, MxIF staining was performed in lung sections (harboring 4T1-LM3 metastases), and PDGFRA and MYH11 could obviously distinguish MFs and MyoFs (Figure 2J). Notably, PDGFRA+ fibroblasts were mainly localized in the invasive front of or around metastases; conversely, MYH11+ fibroblasts were mainly localized in the tumor core and microvascular region. Furthermore, morphological differences between MFs and MyoFs (sorted by FACS) in the in vitro culture system were also observed: MFs were plump and irregular; MyoFs were long and spindly (Supplementary Figure S5D). To test the robustness of the fibroblast classification, we repeated the same MxIF staining in lung metastases paraffin sections of two other murine models (E0771-LM3 and MMTV-PyMT transgenic mice model) (Supplementary Figure S5E-F) and breast cancer patients (Figure 2K); classification and location of LFs were consistent with the 4T1-LM3 model, suggesting their conservative character of fibroblast heterogeneity. Together, these data demonstrated mutually exclusive, morphologically distinct fibroblast subtypes in the breast cancer lung metastasis ecosystem.
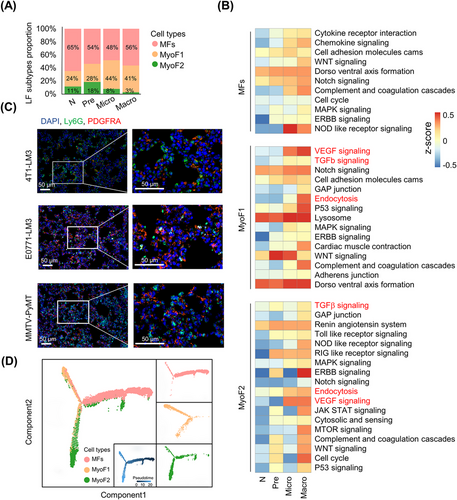
3.3 Composition and transcriptional program of fibroblast subtypes were dynamically reshaped
Accumulating evidence indicates that the tumor microenvironment changes dynamically with tumor progression [22, 23]. Comparably, we hypothesized that lung stromal heterogeneity may become more complicated during tumor metastasis. As expected, each fibroblast population was reproducible across metastasis stages; however, fibroblast composition varied extensively at different time points. MFs accounted for the predominant population at any stage; the proportion of MyoF1 increased gradually, especially at the Micro stage; the abundance of MyoF2 increased at the Pre stage but decreased in subsequent stages (Figure 3A). The dynamic shift in fibroblast subtypes was clearly unveiled by FACS (Supplementary Figure S6A). To determine whether LFs phenotypically evolved as metastases progressed, we characterized transcriptional program changes of fibroblast subtypes with GSVA. Most pathways were shared but diverse degrees of activation in the Pre, Micro, and Macro stages were observed (Figure 3B). For example, chemokine signaling and inflammatory response signatures were observed in MFs since pre-metastases and were further enriched in macro-metastases (Figure 3B). Mobilization and recruitment of neutrophils in the pre-metastatic niche have been widely recognized [24]. Interestingly, we found that neutrophils were enriched around MFs at the Pre stage in multiple mice models (Figure 3C). Here, dynamic changes in gene expression (e.g., C3, Cxcl12) and transcription profile of MFs might provide a reasonable explanation. TGFβ, VEGF signaling and endocytoses were gradually enriched in MyoF1 and MyoF2 (Figure 3B). Consistently, MyoFs were abundant around endothelial cells (Supplementary Figure S6B). To expand our findings, we analyzed metabolism-related biological changes in MAF subtypes. Compared with MyoFs, metabolic programs of MFs displayed stage-dependent characteristics. For instance, amino acid metabolism (e.g., tryptophan [Trp] metabolism) of MFs was instigated early in metastatic stage and functionally persisted throughout metastatic progression, while fatty acid metabolism was enriched since the Micro stage (Supplementary Figure S6C). These data revealed that fibroblast activation-related genes and signaling pathways were gradually enriched since the Pre stage to support tumor-promoting abilities of fibroblast subtypes. To explore whether there is mutual transformation among fibroblast subtypes, as previously reported [25], the Monocle2 algorithm was employed to observe the developmental trajectories of three subtypes. We found a clear tree structure beginning with MFs and ending with MyoF1 and MyoF2 (Figure 3D), suggesting an evolution from MFs towards MyoFs. Together, the composition and transcriptional program of fibroblast subtypes were dynamically reshaped and may contribute to lung-tropic metastasis.
3.4 MF depletion decreased lung metastasis progress
The huge proportion of immune cells in lungs with manifest metastasis may hamper the capture of other cells in scRNA-seq (Supplementary Figure S7A). To avoid interference from abundant immune cells, we removed immune cells (CD45+) by FACS and used the remnant cells from macro-metastatic lung tissue for scRNA-seq, yielding data that could better explore the interactions between DTCs and niche cells in the lung. After cell type identification (Supplementary Figure S7B-D), CellPhoneDB was used to examine intercellular interactions based on ligand-receptor pairs [20]. Interestingly, fibroblasts were the predominant cell population interacting with tumor cells, and among these, MFs had the most ligand-receptor interactions with DTCs (Figure 4A-B). Meanwhile, a high MF signature was associated with decreased cumulative survival in breast cancer patients with lung metastases (Figure 4C). Importantly, a high MF signature also revealed a strong trend toward worsened metastasis survival and lung metastasis-free survival in breast cancer patients with lung metastases (Supplementary Figure S7E).
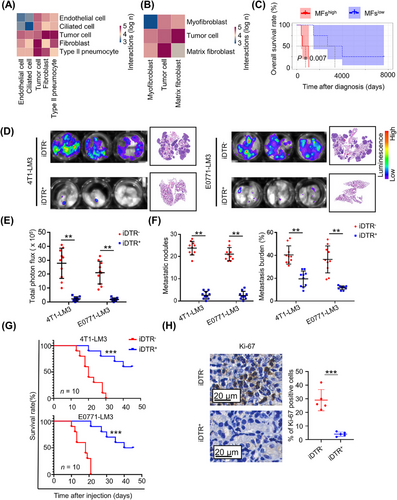
Given the specific expression of PDGFRA in MFs, we explored the functions of MFs on lung metastasis formation using conditional knockout mice to selectively deplete PDGFRA+ MFs. By crossing Pdgfra-cre transgenic mice with iDTR transgenic mice, we established conditionally depleted MFs in the lung (Supplementary Figure S7F). As expected, the deficiency in MFs was more than 85% following the administration of DT (Supplementary Figure S7G). Surprisingly, intravenous injection of 4T1-LM3 or E0771-LM3 exhibited notable reduction of DTC colonies in the lung of iDTR+ mice compared with iDTR− mice (Figure 4D-E). Moreover, iDTR+ mice had less metastasis burden and much longer survival than iDTR− littermates (Figure 4F-G). Simultaneously, dramatically decreased Ki-67 was detected in metastatic nodules of iDTR+ mice in comparison with iDTR− mice (Figure 4H). Collectively, these data showed that selective ablation of PDGFRA+ MFs led to a marked and sustained decrease in metastasis burden.
3.5 TDO2 was highly expressed exclusively in MFs
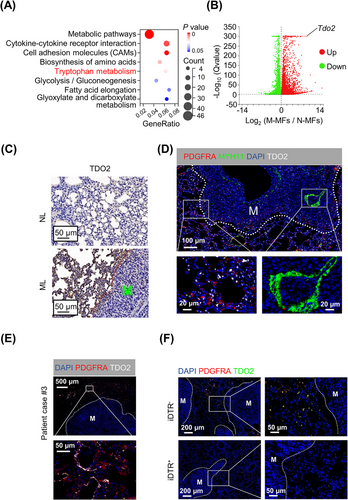
The significant reduction in metastasis burden caused by PDGFRA+ MF ablation led us to investigate how MFs contribute to DTC growth and metastasis formation in the lung. To address this question, bulk RNA-seq of purified fibroblast subtypes was carried out. Gene expression data displayed a high correlation (R > 0.97) between bulk and cognate single-cell profiles (Supplementary Figure S8A). We then tried to select all DEGs between metastatic MFs (M-MFs) and normal MFs (N-MFs) using the bulk RNA-seq data based on ǀlog2FCǀ > 2. Given the gradually activated transcriptional program in MFs in the metastasis process, we reasoned that the genes significantly upregulated in M-MFs (1,109 genes) and those with higher expression in M-MFs than in metastatic MyoFs (M-MyoFs) (420 genes) would be the viably pro-metastatic candidates (Supplementary Figure S8B). Notably, among the 420 genes, there were robust metabolism and cytokine-cytokine receptor interaction biology processes enriched in KEGG analysis of M-MFs (Figure 5A), which replicated the analysis of scRNA-seq data (Figure 3B, Supplementary Figure S6C). Among these signaling processes, Trp metabolism caught our attention, not only because it was instigated early and persisted during advanced metastatic disease (Supplementary Figure S6C) but also because that Tdo2 (encoding tryptophan 2,3-dioxygenase, TDO2 [26]; a key enzyme in catalytic conversion of L-tryptophan [L-Trp] into kynurenine [KYN]) was one of the most upregulated genes in M-MFs (Figure 5B). Moreover, UMAP plot and qRT-PCR analysis showed that Tdo2 was exclusively or highly expressed in MFs rather than in other cell types (e.g., epithelial cells, immune cells and endothelial cells) (Supplementary Figure S8C-D). Indeed, TDO2 was abundant in lungs harboring metastases compared with healthy lungs; more importantly, TDO2 was mainly expressed in the lung stroma but not metastatic nodules, which was similar to MFs’ localization (Figure 5C); and DTCs in metastatic foci were TDO2-negative (Figure 5C). Similar results were obtained in the E0771-LM3 model (Supplementary Figure S8E). More interestingly, the predominant expression of TDO2 was in PDGFRA+ MFs, but not in MYH11+ MyoFs (Figure 5D, Supplementary Figure S8D); accordingly, Tdo2high MFs (M-MFs) produced more KYN than N-MFs (Supplementary Figure S8F). In addition, co-localization of TDO2 with PDGFRA was observed in lung metastatic tissues of breast cancer patients (Figure 5E). Conditional depletion of MFs in iDTR+ mice notably decreased TDO2 levels in lung metastatic foci (Figure 5F). Previous research showed that tumor cells themselves could promote tumor progression by producing large amounts of KYN dependent on the Trp-IDO1 axis (IDO1, the other rate-limiting enzyme in Trp metabolism) [27]. IHC staining showed extremely low IDO1 expression in both primary and metastatic sites of mice and breast cancer patients (Supplementary Figure S8G). Accordingly, KYN production of tumor cells (primary tumor cells and DTCs) was significantly lower than that of MFs during the whole metastasis process (Supplementary Figure S8H). In short, TDO2 was exclusively expressed in MFs.
3.6 Tdo2high MFs fostered lung metastasis
The TDO enzyme is mainly expressed in the liver, whose activity balances the total amount of Trp in the body [28]. Trp enters the liver, where most is oxidized to acetoacetyl-coenzyme A and used for the synthesis of nicotinamide adenine dinucleotide. Extrahepatic organs that metabolize Trp along the KYN pathway (KP) contribute most to circulating levels of KYN and KP metabolites [29]. To determine the function of TDO2 in MFs, we crossed Tdo2fl/fl mice with Pdgfra-Cre mice to generate Tdo2cKO mice, resulting in > 95% reduction of Tdo2 mRNA and TDO2 protein in MFs of lung tissue (Supplementary Figure S8I). Encouragingly, no apparent compensatory effect of indoleamine-2,3-dioxygenase 1 (Ido1, encoding IDO1) was observed (Supplementary Figure S8J); consistently, MFs isolated from Tdo2cKO mice had minimal KYN production (Supplementary Figure S8K). In line with iDTR+ mice, Tdo2 deficiency in Pdgfra+ fibroblasts dramatically mitigated metastasis burden in both 4T1-LM3 and E0771-LM3 metastatic mice models (Figure 6A-D). Correspondingly, tumor cells from metastatic foci in Tdo2cKO mice showed a significant reduction of cell proliferation (Figure 6E). Moreover, Tdo2cKO lung metastasis-burdened mice had much longer survival than Tdo2fl/fl littermates (Figure 6F). However, the growth of primary tumors was not significantly different between Tdo2fl/fl and Tdo2cKO mice (Supplementary Figure S8L-M). Tdo2 deficiency dramatically reduced the lung metastatic burden of parental cells (4T1 and E0771) while there was no significant change in liver, bone, and brain metastatic burden (Supplementary Figure S8N), which indicated that the MF-TDO2 axis was specific in the lung. To better recapitulate the role of Tdo2high MFs in lung metastasis of patients with breast cancer, we crossed the Tdo2cKO strain with MMTV-PyMT mice, evaluating the effects of MF-specific Tdo2 deficiency on spontaneous lung metastasis in this model. Consistent with the aforementioned findings, deletion of Tdo2 in MFs significantly reduced spontaneous lung metastases in MMTV-PyMT mice (Figure 6G). Furthermore, we also observed a trend toward worsened lung metastasis-free survival in breast cancer patients with higher TDO2 expression (Figure 6H). These data demonstrated that TDO2-dependent MF program promoted lung metastasis formation.
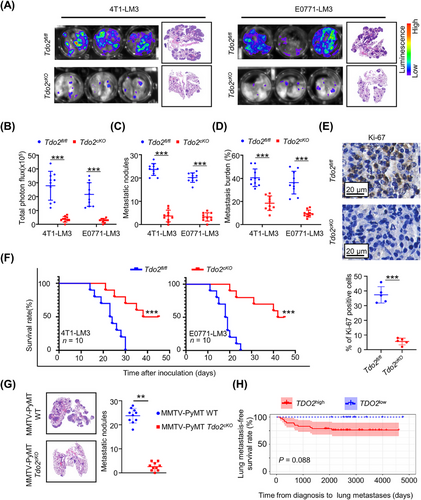
3.7 TDO2-KYN protected DTCs from ferroptosis
To further understand how Tdo2high MFs affect the fate of DTCs, we performed unbiased RNA sequencing in Tdo2high MF-CM-treated 4T1-LM3 cells. Using KEGG analysis, metabolic pathways and ferroptosis were markedly enriched in Tdo2high MF-CM-treated DTCs (Figure 7A). The lung metastasis microenvironment exhibits higher ferroptotic stress compared to primary mammary cancers [30]; thus, developing resistance to ferroptosis is necessary for DTCs. It is reported that aberrant amino acid metabolism played crucial roles in ferroptosis. In particular, the Trp metabolite KYN could protect tumor cells from ferroptosis and promote tumor growth [31]. We therefore wondered whether Tdo2high MFs could decrease the sensitivity of DTCs to ferroptosis. TDO2 and ferroptosis suppressors (nuclear factor erythroid 2-related factor 2 [NRF2] and heme oxygenase 1 [HMOX1]) expression levels showed positive correlations in lung metastases of breast cancer patients; consistently, TDO2 and ferroptosis driver (voltage dependent anion channel 2 [VDAC2]) levels presented a negative correlation (Supplementary Figure S9). Interestingly, the positive correlations between TDO2 and ferroptosis suppressors FTH1 and aldo-keto reductase family 1 member C1 (AKR1C1) were also replicated in lung metastases of osteosarcoma patients (Supplementary Figure S9A). Liproxstatin-1 treatment, a ferroptosis antagonist, reduced erastin-induced DTC death. DTCs, co-cultured with Tdo2high MFs, developed resistance to erastin-induced ferroptosis, whereas 680C91 (a TDO2 inhibitor) treatment significantly ameliorated DTCs’ ferroptosis resistance (Figure 7B). Consistently, CM from Tdo2cKO MFs failed to protect tumor cells against erastin- or RSL3-induced cell death in comparison with CM from Tdo2fl/fl littermates (Figure 7C, Supplementary Figure S10A). These data suggested that Tdo2high MFs played an important role in protecting DTCs from ferroptosis. Ferroptosis is an iron-dependent form of regulated cell death, characterized by altered iron homeostasis, reduced defense against oxidative stress, and abnormal lipid peroxidation [32]. Checked by BODIPY-C11 staining, DTCs cultured with Tdo2high MF-CM had lower lipid ROS levels, while 680C91 treatment increased lipid ROS of DTCs (Figure 7D). Besides, accumulation of lipid ROS induced by erastin was decreased in DTCs cultured with Tdo2high MF-CM rather than in DTCs cultured with 680C91-treated Tdo2high MF-CM (Figure 7D). Remarkably lower cellular Fe2+ was detected in DTCs cultured with Tdo2high MF-CM; besides, Tdo2high MF-CM could rescue the erastin-induced augmentation of Fe2+, while CM from 680C91-treated Tdo2high MFs failed to maintain low cellular Fe2+ level (Figure 7E, Supplementary Figure S10B).
Recent reports showed that KYN-derived metabolites activated anti-ferroptotic pathways [31]. KYN can be converted to anthranilic acid by kynureninase (KYNU) and to kynurenic acid by kynurenine aminotransferases (kynurenine aminotransferase 1 [Kyat1, KATI], aminoadipate aminotransferase [Aadat, KATII], kynurenine aminotransferase 3 [Kyat3, KATIII]). Kynurenine monooxygenase (KMO) controls the conversion of KYN to neuroactive and neurotoxic KP metabolites, including quinolinic acid [29]. To distinguish the forms in which KYN functioned in our study, we examined kynurenine aminotransferase expression in both Tdo2high MFs and DTCs (4T1-LM4 and E0771-LM4 cells). However, low gene expression of KATI–KATIII was observed in both Tdo2high MFs and DTCs (Supplementary Figure S10C). In addition, Tdo2high MFs and DTCs also showed extremely low Kynu and Kmo expression in scRNA-seq (Supplementary Figure S10D). Therefore, directly treating DTCs with KYN, which also markedly elevated resistance to erastin- or RSL3-induced ferroptosis, decreased lipid peroxidation and cellular Fe2+ level as Tdo2high MF-CM done (Supplementary Figure S10E-H).
We next assessed TDO2-KYN-modulated DTC ferroptosis resistance in vivo. Firstly, we checked the levels of PTGS2 [33], a ferroptosis downstream marker, in lung metastases from Tdo2cKO mice and Tdo2fl/fl littermates. There was higher PTGS2 expression in the lung metastases from Tdo2cKO mice than Tdo2fl/fl mice; treatment with KYN remarkably decreased PTGS2 in Tdo2cKO mice. Conversely, treating Tdo2fl/fl mice with 680C91 dramatically increased PTGS2 levels in lung metastases (Figure 7F). More importantly, in breast cancer patients, lung metastases with higher TDO2 had fewer lipid peroxisomal metabolism-associated genes (Figure 7G), which supported the above findings in mice. To determine whether TDO2-KYN-modulated ferroptosis contributed to metastatic outgrowth, DTCs were co-cultured with Tdo2cKO MFs, then treated with liproxstatin-1. DTCs in Tdo2cKO MF co-culture system were more sensitive to erastin compared with DTCs in Tdo2fl/fl MF co-culture system; however, liproxstatin-1 supplement to DTCs and Tdo2cKO MFs co-culture system attenuated erastin-induced ferroptosis (Figure 7H). Tdo2 depletion-induced decrease in metastases (Figure 7I) and corresponding Ki-67-positive tumor cell ratio (Supplementary Figure S10I), and increase of PTGS2 levels in the lung metastases of Tdo2cKO mice (Supplementary Figure S10J) were all effectually blunted by liproxstatin-1 supplement. These findings suggested an essential role of TDO2-KYN in protecting DTCs against ferroptosis to foster lung metastasis.
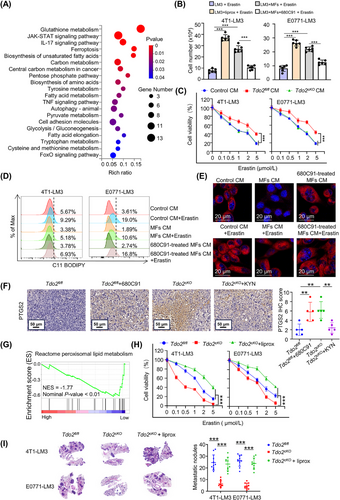
To define the mechanism by which Tdo2high MF-secreted KYN protects DTCs from ferroptosis, the transcriptome of 4T1-LM3 cells treated with CM from M-MFs or N-MFs were analyzed, and the DEGs involved in ferroptosis were determined (Figure 8A). Fth1 was one of the most upregulated genes in M-MF-CM-treated 4T1-LM3 cells, which was confirmed by qRT-PCR (Figure 8B). The enhanced FTH1 protein was further confirmed in 4T1/E0771-LM3 cells co-cultured with CM from Tdo2high MFs, while 680C91-treated Tdo2high MF-CM failed to elevate the FTH1 protein level (Figure 8C). We further demonstrated that KYN could stimulate FTH1 expression by supplementing KYN in the medium (Figure 8D). Besides, FTH1 level in lung metastases was lower in Tdo2cKO mice than in Tdo2fl/fl mice (Figure 8E). Correspondingly, more free Fe2+ accumulated in DTCs derived from Tdo2cKO mice in comparison with Tdo2fl/fl mice (Figure 8F).
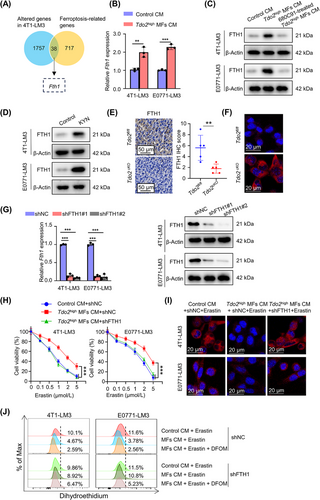
FTH1, heavy chains of ferritin, is an iron storage protein, which combines and stores ferric irons, thus limiting the generation of destructive redox species [34]. Excess ferrous ions (Fe2+) generate abundant toxic ROS through the Fenton reaction [35]. To ascertain whether FTH1 was required for DTCs’ ferroptosis resistance mediated by Tdo2high MFs, FTH1 was knocked down by shRNA in 4T1-LM3 and E0771-LM3 cells, respectively (Figure 8G). CM from Tdo2high MFs could protect 4T1/E0771-LM3 cells, but not FTH1-depleted 4T1/E0771-LM3 cells, against erastin-induced cell death (Figure 8H). The intracellular accumulation of Fe2+ was accordingly decreased in the parental 4T1/E0771-LM3 cells rather than FTH1-depleted 4T1/E0771-LM3 cells (Figure 8I). Moreover, after adding Tdo2high MF-CM, erastin-induced ROS accumulation was accordingly reduced in parental 4T1/E0771-LM3 cells rather than FTH1-depleted 4T1/E0771-LM3 cells. However, treatment of FTH1-depleted 4T1/E0771-LM3 cells with DFOM (the iron chelator) restricted ROS level in the presence of erastin (Figure 8J), suggesting that KYN could reduce ROS level in DTCs by upregulating FTH1 level. Collectively, these results demonstrated that Tdo2high MF-derived KYN upregulated FTH1 level in DTCs, enabling DTCs to resist ferroptosis.
3.8 Tdo2high MFs mediated immune suppression in lung metastasis microenvironment
The aforementioned data demonstrated that the TDO2-KYN axis could directly impact metastasis formation of DTCs. Re-analyzing the scRNA-seq and bulk-seq data of MFs, we noted a significant inflammatory response signaling in MFs (Figure 2I and Figure 3B, Supplementary Figure S5B-C). Moreover, TDO2 and immunity suppressors (PD-1, programmed cell death-ligand 1 [PD-L1], cytotoxic T-lymphocyte associated protein 4 [CTLA4], IL-10) showed positive correlations in lung metastases of breast cancer patients, which was also replicated in lung metastases of colorectal cancer patients (Supplementary Figure S11). There was enhanced expression of T cell chemokine genes (Ccl8, Ccl11) in Tdo2high MFs (Figure 9A), indicating potential T cell recruitment capacity of Tdo2high MFs. Indeed, co-culturing T cells with N-MFs and M-MFs, M-MFs had stronger chemoattractant ability to T cells (green) than N-MFs (Figure 9B; Supplementary videos). Both CD4+ and CD8+ T cells were significantly enriched around MFs, locating on the periphery of metastatic foci (Figure 9C).
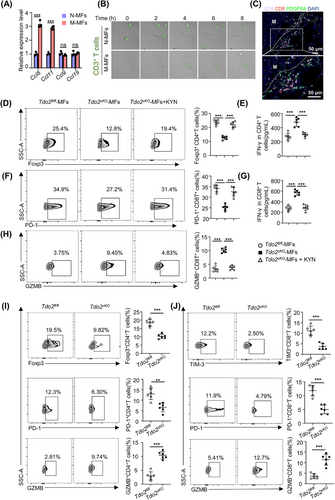
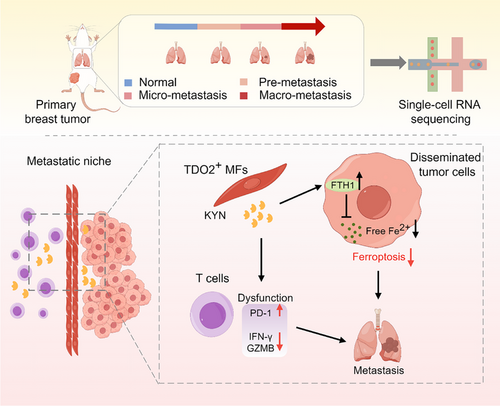
Triple-negative breast cancer is known as a “cold” tumor, characterized by the absence or exclusion of T cells in the tumor parenchyma (“immune-desert”/“immune-excluded”) [36]; and Trp catabolism has been discovered to affect functions of tumor-infiltrating T cells [37]. These clues raised another exciting scientific hypothesis that Tdo2high MFs may participate in the formation of “cold tumor” by preventing T cells from accessing metastatic foci and mediating T-cell suppression in the metastatic niche. To address this intriguing issue, we tried to ascertain whether Tdo2high MFs play immunomodulatory roles in infiltrating T cells. Co-culture of MFs from Tdo2cKO mice with T cells resulted in Foxp3 downregulation and IFN-γ increase in CD4+ T cells (Figure 9D-E); consistently, PD-1 of CD8+ T cells was decreased (Figure 9F), while the secretion of IFN-γ and GZMB was increased in CD8+ T cells (Figure 9G-H). However, KYN treatment reversed these phenotypes of infiltrating T cells (Figure 9D-H), suggesting that Tdo2high MFs potentially suppressed T cell function via KYN. Furthermore, checking the immune-regulatory effects of Tdo2high MFs in vivo using lung metastasis mouse models, we found more abundant regulatory T cells (Foxp3+CD4+) in Tdo2fl/fl mice than in Tdo2cKO mice (Figure 9I). Notable downregulation of PD-1 and increased GZMB were observed in CD4+ T cells from Tdo2cKO mice (Figure 9I). Consistently, CD8+ T cells isolated from Tdo2cKO mice displayed a dramatic downregulation of TIM-3 and PD-1, and enhanced GZMB (Figure 9J), indicating that depleting Tdo2 in MFs can partially reverse the exhausted state of intralesional CD8+ T cells and restore their cytotoxic functions. Taken together, these data revealed that TDO2-mediated Trp catabolism in MFs suppressed T cell functions and established a “barrier” to shield tumor cells from T cell attacks, supporting DTCs’ immune escape. The schematic diagram of TDO2+ MF-mediated DTCs’ ferroptosis resistance and T cell dysfunction is displayed in Figure 10.
4 DISCUSSION
Organotropic metastasis threatens the lives of breast cancer patients, but the underlying mechanisms remain elusive [38]. Indeed, DTCs and its niche may show interdependent evolutionary paths with more certainty than primary tumors [39, 40], highlighting the important role of metastatic niche in metastasis growth. Here, we used scRNA-seq to illuminate the dynamic characteristic changes of MAF subtypes. Combining various mouse models with survival analysis in patient cohorts, we demonstrated crucial roles of specific crosstalk at metastatic niches among DTCs, T cells, and TDO2+ MFs at single-cell resolutions. The up-regulated expression of Tdo2 in activated MFs led to abundant secreted KYN production which promoted metastasis growth by significantly improving ferroptosis resistance of DTCs and impairing T cell function.
Stromal heterogeneity possesses the characteristics of time dependence. Our data uncover the temporal molecular features of 3 distinct local lung fibroblast subtypes. Furthermore, signaling pathways associated with fibroblast activation were instigated in a stage-specific manner. Previous studies raised the concept of dynamic tumor microenvironment: genomically stable cells changed their transcriptional program to track the evolving tumor niche. For example, at breast cancer primary sites, CAF subtypes appear to transition from an early immunoregulatory transcriptional program to a late antigen-presentation program [18]. Current understanding of fibroblast activation is mainly limited to primary tumors, and the focus on MAFs is relatively small [41]. Our findings expand this concept to the lung niche. Interestingly, we observed that the endocytosis programs of LF subtypes (MyoF1 and MyoF2) were instigated early in metastatic stages and persisted functionally, especially in MyoF2. This may explain how breast cancer cells (at the primary site) educated distant resident cells and created a metastases-permissive microenvironment (pre-metastatic niche). For example, in colorectal cancer, primary tumors released extracellular vesicles into the circulation to activate lung fibroblasts [42]; furthermore, exosomes derived from Lewis lung carcinoma (LLC) cells were mainly engulfed by LFs and led to NF-κB activation of fibroblasts [43]. These studies supported our hypothesis about how distant stromal cell subtypes were activated before tumor cells’ arrival. Once DTCs arrive at second sites, fibroblasts and immune cells will be further educated and help DTCs settle in the new environment [44, 45], which promotes the generation of heterogeneity and dynamic change of metastatic microenvironment.
Heterogeneous LFs are hijacked by DTCs to facilitate metastasis. ECM remodeling in the metastatic niche was one of the earliest events in metastasis formation and was correlated with immune cell motility in melanoma [46, 47]. Factors secreted by MAFs (e.g., hyaluronic acid) can promote DTCs proliferation [48, 49]. Additionally, MAFs maintain DTCs ‘‘stemness’’ potential by augmenting WNT signaling in metastatic niche [10]. However, these studies have not considered the heterogeneity of fibroblasts. This may explain why CAFs/MAFs targeting has presented positive results in pre-clinical researches while subsequent clinical studies have yielded conflicting results. Therefore, more efforts are needed to describe aspects of their biology to inform treatment decisions [41]. Our results suggested that lung MAFs included three subtypes and that the tumor promoting effects of MFs were multifaceted. Our study provided insights into the versatility of a single subtype, revealing a possible mechanism of “cold tumor” formation in breast cancer.
Growing evidence indicates that metabolic reprogramming of tumors occurs not only within tumor cells, but also in the stromal cells [50, 51]. Because tumor cells are often in a hypoxic and nutrient-deficient environment, tumor cells are mainly anabolic, while CAFs are mainly catabolic [52]. The current study demonstrated that TDO2-mediated Trp catabolism in MFs regulated DTCs to adapt to microenvironment with higher ferroptotic stress and support the formation of immunosuppression. The conclusion is supported by previous work, suggesting that the catabolism of CAFs and the tumor cell metabolism are coupled and that catabolism of CAFs can provide important metabolic substance for tumor cell growth [53, 54]. Tumor cells and CAFs mutually promote metabolic reprogramming, establishing a symbiotic relationship. Thus, targeting metabolic regulation of MAF subtypes may serve as a promising approach to suppress metastasis formation.
This study also had certain limitations. For instance, we did not explore the mechanisms underlying the upregulation of Tdo2 in MFs. Moreover, how KYN mediates T cell dysfunction needs further investigation. Besides, more studies are needed to determine MyoFs’ functions in metastatic progression.
5 CONCLUSIONS
In conclusion, our findings delineate a multifaceted cellular crosstalk within growing lung metastases of breast cancer. The results show a crucial role for Tdo2high MFs during metastatic progression and emphasize the role of Trp catabolism in DTC survival and immunosuppression formation. These interactions in metastatic nodules may serve as useful targets when developing future therapies against metastatic disease.
AUTHOR CONTRIBUTIONS
Yongcan Liu, Shanchun Chen, Yong Teng, Xueying Wan and Manran Liu designed this study; Yongcan Liu, RuiWang, Haojun Luo, Chao Chang, Peijin Dai, Yubi Gan, and Yuetong Guo conducted the experiments; Yongcan Liu, Yixuan Hou and Yan Sun analyzed the data; Shanchun Chen drafted the initial manuscript; Yubi Gan, Xiaojiang Cui and Manran Liu revised this manuscript; all authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
Special thanks to Prof. Suling Liu from Fudan University for help with animal experiments. This work was supported by National Key Projects of Ministry of Science and Technology of China (MOST 2018YFE0113700), National Natural Science Foundation of China (NSFC82173155, NSFC81874199), and the Outstanding Professorship Program of Chongqing Medical University (2019-R10005) to Manran Liu. This work was also supported by the Outstanding Postgraduate Fund of Chongqing Medical University (BJRC202021, BJRC202025) and the Chongqing Graduate Research and Innovation Project of the Chongqing Education Committee (CYB22218) for Shanchun Chen.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Sample collection and animal experiments were approved by the Ethics Committee of Chongqing Medical University and conducted according to the guidelines for the Care and Use of Laboratory Animals of Chongqing Medical University (2021084).
Open Research
DATA AVAILABILITY STATEMENT
The data and materials could be available from the corresponding author upon reasonable request. Bulk RNA-seq data in the study was deposited into GEO with accession number of GSE269817 and GSE270152 (https://www.ncbi.nlm.nih.gov/geo/). scRNA-seq data have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA014233) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.




