USP51/PD-L1/ITGB1-deployed juxtacrine interaction plays a cell-intrinsic role in promoting chemoresistant phenotypes in non-small cell lung cancer
Abstract
Background
Programmed death ligand 1 (PD-L1) has been demonstrated to facilitate tumor progression and therapeutic resistance in an immune-independent manner. Nevertheless, the function and underlying signaling network(s) of cancer cell-intrinsic PD-L1 action remain largely unknown. Herein, we sought to better understand how ubiquitin-specific peptidase 51 (USP51)/PD-L1/integrin beta-1 (ITGB1) signaling performs a cell-intrinsic role in mediating chemotherapeutic resistance in non-small cell lung cancer (NSCLC).
Methods
Western blotting and flow cytometry were employed for PD-L1 detection in NSCLC cell lines. Coimmunoprecipitation and pulldown analyses, protein deubiquitination assay, tissue microarray, bioinformatic analysis and molecular biology methods were then used to determine the significance of PD-L1 in NSCLC chemoresistance and associated signaling pathways in several different cell lines, mouse models and patient tissue samples. Ubiquitin-7-amido-4-methylcoumarin (Ub-AMC)-based deubiquitinase activity, cellular thermal shift and surface plasmon resonance (SPR) analyses were performed to investigate the activity of USP51 inhibitors.
Results
We provided evidence that cancer cell-intrinsic PD-L1 conferred the development of chemoresistance by directly binding to its membrane-bound receptor ITGB1 in NSCLC. At the molecular level, PD-L1/ITGB1 interaction subsequently activated the nuclear factor-kappa B (NF-κB) axis to elicit poor response to chemotherapy. We further determined USP51 as a bona fide deubiquitinase that targeted the deubiquitination and stabilization of the PD-L1 protein in chemoresistant NSCLC cells. Clinically, we found a significant direct relationship between the USP51, PD-L1 and ITGB1 contents in NSCLC patients with chemoresistant potency. The elevated USP51, PD-L1 and ITGB1 levels were strongly associated with worse patient prognosis. Of note, we identified that a flavonoid compound dihydromyricetin (DHM) acted as a potential USP51 inhibitor and rendered NSCLC cells more sensitive to chemotherapy by targeting USP51-dependent PD-L1 ubiquitination and degradation in vitro and in vivo.
Conclusions
Together, our results demonstrated that the USP51/PD-L1/ITGB1 network potentially contributes to the malignant progression and therapeutic resistance in NSCLC. This knowledge is beneficial to the future design of advanced cancer therapy.
Abbreviations
-
- AKT
-
- AKT serine/threonine kinase
-
- BIRC5
-
- baculoviral IAP repeat containing 5
-
- BRCA1
-
- breast cancer gene 1
-
- BRCA2
-
- breast cancer gene 2
-
- Co-IP
-
- coimmunoprecipitation
-
- CHX
-
- Cycloheximide
-
- DAPI
-
- 4’,6-diamidino2-phenylindole
-
- DDP
-
- cisplatin
-
- DHM
-
- dihydromyricetin
-
- DMEM
-
- Dulbecco's Modified Eagle Medium
-
- DMSO
-
- dimethylsulfoxide
-
- DTT
-
- dithiothreitol
-
- EGFR
-
- epidermal growth factor receptor
-
- ERK
-
- extracellular regulated MAP kinase
-
- FBS
-
- fetal bovine serum
-
- GAPDH
-
- glyceraldehyde-3-phosphate dehydrogenase
-
- GSS
-
- glutathione synthetase
-
- GST
-
- glutathione S-transferase
-
- HIF1α
-
- hypoxia inducible factor 1 alpha
-
- JAK
-
- Janus kinase
-
- IFN-γ
-
- interferon gamma
-
- IRF1
-
- interferon regulatory factor 1
-
- ITGB1
-
- integrin beta 1
-
- LC
-
- Lung cancer
-
- MBP
-
- maltose binding protein
-
- MEK
-
- MAP kinse-ERK kinase
-
- mTOR
-
- mechanistic target of rapamycin kinas
-
- NEAA
-
- non-essential amino acids
-
- NF-κB
-
- nuclear factor-kappa B
-
- NSCLC
-
- non-small cell lung cancer
-
- PD-L1
-
- programmed death ligand 1
-
- PI3K
-
- phosphatidylinositol 3-kinase
-
- RIPA
-
- radio immunoprecipitation assay
-
- shRNAs
-
- short hairpin RNAs
-
- SLC31A2
-
- solute carrier family 31 member 2
-
- SPOP
-
- speckle type BTB/POZ protein
-
- SPR
-
- surface plasmon resonance
-
- STAT1
-
- signal transducer and activator of transcription 1
-
- RNF31
-
- ring finger protein 31
-
- TRIM21
-
- tripartite motif containing 21
-
- TNF-α
-
- tumor necrosis factor alpha
-
- TNM
-
- tumor node metastasis
-
- Ub-AMC
-
- C-terminal conjugate of ubiquitin with 7-amino-4-methylcoumarin
-
- UCHL1
-
- ubiquitin C-terminal hydrolase L1
-
- USP7
-
- ubiquitin specific peptidase 7
-
- USP51
-
- ubiquitin-specific peptidase 51
-
- β-TrCP
-
- beta-transducin repeat containing E3 ubiquitin protein ligase
-
- γH2AX
-
- phosphorylated histone H2AX
1 BACKGROUND
Lung cancer (LC) contributes to the largest proportion of global cancer-associated mortality [1]. Non-small cell lung cancer (NSCLC) is the most prevalent form of LC, and makes up almost 85% of all LC cases [2]. Patients suffering from locally advanced or metastatic NSCLC typically obtain platinum-based chemotherapy [3]. Despite a consistent rate of initial responses, LC has relatively poor prognosis owing to drug resistance and tumor recurrence [4]. Therefore, it is urgent and necessary to elucidate the underlying mechanism of action behind chemoresistance in NSCLC in order to develop new and efficacious treatment for NSCLC.
Programmed death ligand 1 (PD-L1), otherwise named B7-H1 and CD274, is a 40 kDa transmembrane protein found on immune cells and multiple cancer cells, such as NSCLC [5, 6]. Recent studies have reported that PD-L1, an immune checkpoint protein, negatively modulates anticancer immunity via association with the programmed cell death protein 1 (PD-1) receptor [7]. Despite multiple investigations examining the pivotal role of PD-L1 to inhibit T-cell or other immune cell functions in immune escape, new reports indicate that cancer cell-intrinsic PD-L1 potentially also modulate immune-independent cellular activities [8-12]. For example, some investigations suggested that cancer cell-intrinsic PD-L1 modulates glucose metabolism in sarcomas [13] and cell growth and autophagy in melanoma, ovarian and prostate cancers [14]. In the aforementioned mechanisms, PD-L1 functions through the phosphatidylinositol 3-kinase (PI3K)/serine/threonine kinase (AKT)/mechanistic target of rapamycin kinase (mTOR) network. Additionally, the aberrant PD-L1 expression has been demonstrated as a promising indicator for prognosis and chemotherapeutic response in patients with breast [15] and colorectal cancers [16]. Thus, targeting PD-L1 is crucial for chemotherapeutic sensitization through the inhibition of DNA damage repair [10]. In line, PD-L1 assessment in NSCLC cells allows to identify a patient population with higher likelihood of recurrence and a decreased responsiveness to chemotherapy [10]. However, the potential mechanisms concerning the cell-intrinsic role of PD-L1 and its aberrant regulation in cancer biology and treatment have not been fully elucidated.
A growing body of evidence have revealed that the PD-L1 content is regulated by multiple cellular factors (e.g., growth factors, chemokines, oncogenes and tumor suppressor genes) at the transcriptional level. For example, interferon gamma (IFN-γ) is a strong activator of PD-L1 expression, and it employs the Janus kinase (JAK)/signal transducer and activator of transcription 1 (STAT1)/interferon regulatory factor 1 (IRF1) pathway in multiple types of human cancers, including NSCLC [17, 18]. Also, activating mutations involving RAS and epidermal growth factor receptor (EGFR) oncogenes are cancer-inducing and thus increase cancer cell-intrinsic PD-L1 levels via the PI3K-AKT and MAP kinase-ERK kinase (MEK)/extracellular regulated MAP kinase (ERK) networks in NSCLC cells [19, 20]. Nevertheless, there are limited reports on the molecular mechanisms behind the posttranslational modifications of PD-L1 [14]. The E3 ligases speckle type BTB/POZ protein (SPOP) and beta-transducin repeat-containing E3 ubiquitin protein ligase (β-TrCP) have been identified to ubiquitinate the PD-L1 protein and bring about its degradation in prostate [12, 21] and breast cancers [22]. Alternately, COP9 signalosome subunit 5 (CSN5), as a deubiquitinase, is upregulated by the tumor necrosis factor alpha (TNF-α)/nuclear factor-kappa B (NF-κB) network in response to chronic inflammation, leading to PD-L1 protein stabilization and impairment of the anticancer therapy in breast cancer cells [23]. Therefore, a comprehensive understanding of the molecular mechanisms of PD-L1 posttranslational modulation can potentially aid in the development of novel and efficacious drugs that diminish PD-L1 protein content and overcome malignancy and therapeutic resistance in human cancers. In the present study, to ascertain whether a cancer cell-intrinsic role of PD-L1 is linked to the chemoresistant phenotypes in NSCLC, we performed a comprehensive study and jointly analyzed data from both in vitro and in vivo experimentations. Moreover, we sought to clarify the molecular processes underlying PD-L1 posttranslational regulation and its clinical relevance in NSCLC occurrence and progression.
2 MATERIALS AND METHODS
2.1 Cell culture
Human NSCLC cell lines A549 and PC9 and human embryonic kidney cell line 293T were acquired from the American Type Culture Collection (Manassas, VA, USA), and cisplatin (DDP)-resistant A549R and PC9R cells from the Tianjin Medical University Cancer Institute and Hospital (Tianjin, China). All cells were verified to be mycoplasma negative via a GMyc-PCR mycoplasma test kit (Yeasen, Shanghai, China). A549 and PC9 cells were grown in RPMI-1640 (Gibco, Waltham, MA, USA), containing 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (ThermoFisher Scientific, Waltham, MA, USA). 293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco), containing 10% FBS, 1% sodium pyruvate (Gibco), 1% non-essential amino acids (NEAA) (Gibco) and 2% glutamine (Gibco). A549R and PC9R cells were cultured in RPMI-1640, containing 10% FBS, 1% penicillin/streptomycin and 2.5 μmol/L DDP (Selleck, Houston, TX, USA).
2.2 Reagents and drugs
PD-L1 neutralizing antibody avelumab (Selleck), the dosages were 2 μg/mL in vitro and 5 mg/kg in vivo; the NF-κB inhibitor Bay 11-7085 (TOPSCIENCE, Shanghai, China), the dosage was 5 μmol/L in vitro; the protein synthesis inhibitor cycloheximide (CHX) (Selleck), the dosage was 100 μg/mL in vitro; the chemotherapeutic drug DDP, the dosages were 5 μmol/L in vitro and 5 mg/kg in vivo; the proteasomal inhibitor MG132 (Selleck), the dosage was 20 μmol/L in vitro; the lysosomal inhibitor chloroquine (CQ) (Selleck), the dosage was 20 μmol/L in vitro.
2.3 Lentivirus-mediated knockdown and expression
Specific short hairpin RNAs (shRNAs) for PD-L1 and USP51 underwent annealing and subcloning into the pLV-H1-EF1α-puro vector (Biosettia, San Diego, CA, USA). The full-length PD-L1 and USP51 as well as their deletion constructs (PD-L1-ET, PD-L1-TC, Flag-PD-L1-E, Flag-PD-L1-C, Flag-PD-L1-C1, Flag-PD-L1-C2, Flag-PD-L1-C3, Myc-USP51-N and Myc-USP51-C) (Sangon Biotech, Shanghai, China) underwent subcloning into the pLV-EF1-MCS-IRES-Bsd vector (Biosettia). The individual mutants of PD-L1 (K263R, K270R, K271R, K280R and K281R) (Sangon Biotech) also underwent subcloning into the pLV-EF1-MCS-IRES-Bsd vector. The constructed and packaged plasmids including gag polyprotein (Gag-Pol), regulator of expression of virion protein (Rev) and vesicula stomatitis virus glycoprotein G (VSVG) (Biosettia) were then co-incorporated into 293T cells with Lipofectamine 2000 reagent (Invitrogen, Waltham, MA, USA) to produce lentivirus particles. The details of primers used can be found in Supplementary Table S1.
2.4 RNA isolation and quantitative RT-PCR (qRT-PCR)
Total RNA was isolated with Trizol (Yeasen), followed by the first-strand cDNA generation using TransScript SuperMix (TransGen, Beijing, China). The chemoresistant genes were specifically amplified using quantitative PCR with SYBR Green Master Mix (Yeasen). The following PCR parameters were used: 95°C for 10 min, then 40 cycles of 95°C for 15 s, and 60°C for 1 min. The value of △△Ct was employed to determine chemoresistant gene relative expression. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a normalization control. The primer details can be found in Supplementary Table S1.
2.5 Western blotting analysis
Total protein isolation and Western blotting was conducted as reported previously [24]. Protein samples were collected in lysis buffer for radio immunoprecipitation assay (RIPA) (Yeasen) with protease and phosphatase inhibitors (Yeasen), then quantified with a bicinchoninic acid (BCA) kit (ThermoFisher Scientific). Protein separation was performed with sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) prior to transfer to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA), then blocked for 1 h in 5% skimmed milk (Yeasen) at room temperature, before overnight incubation with primary antibodies at 4°C with subsequent 1 h incubation in secondary antibodies at room temperature. Lastly, protein visualization employed enhanced chemiluminescence (ECL)-based chemiluminescence (Yeasen). A summary of the employed antibodies is available in Supplementary Table S2.
2.6 Deubiquitination assay
Cells were incorporated with haemagglutinin (HA)-ubiquitin and the previously described plasmids, then treated with MG132 (Yeasen) for 6 h to inhibit ubiquitin-proteasome-dependent dysregulation of protein. Cell lysates were made in RIPA buffer (Yeasen), prior to overnight incubation in anti-Flag antibody or IgG at 4°C. The resulting immunoprecipitates were assayed using an anti-HA-Tag antibody-mediated Western blotting. The details of antibodies used can be found in Supplementary Table S2.
2.7 Protein expression and purification for PD-L1 proteins
Glutathione S-transferase (GST)-tagged PD-L1 and its corresponding truncated mutants (GST-PD-L1-E, GST-PD-L1-IgV, GST-PD-L1-IgC2 and GST-PD-L1-C) (Sangon Biotech) underwent cloning into the pGEX-6p-1 vector (Biosettia) prior to expression in E. coli BL21 cells (WeiDi, Shanghai, China). Cell induction employed 0.5 mmol/L isopropyl β-D-thiogalactoside (IPTG) (Yeasen) in Luria-Bertani (LB) medium (Sigma, St. Louis, MO, USA) at 16°C for 24 h to induce PD-L1 protein expression. The cell pellet was dissolved in lysis buffer (50 mmol/L Tris, 300 mmol/L NaCl), and 2 mg/mL lysozyme (Yeasen) was added and incubated at 4°C overnight, followed by ultrasonication using Ultrasonic Crusher (SCIENTZ, Ningbo, Zhejiang, China), and a 20 min centrifugation at 18,000 × g at 4°C. Following removal of supernatant, the inclusions were collected in solubilization buffer [200 mmol/L Tris, 5 mmol/L dithiothreitol (DTT), 8 mol/L urea, pH = 8.0]. Insoluble material was centrifuged at 18,000 × g for 20 min at 4°C and removed, and the supernatant was placed onto Glutathione Sepharose 4B beads (ThermoFisher Scientific) and equilibrated with binding buffer (50 mmol/L Tris, 300 mmol/L NaCl) overnight. The protein was eluted with elution buffer (50 mmol/L Tris, 300 mmol/L NaCl, 10 mmol/L glutathione, pH = 7.5-8.0) and stored at -80°C.
2.8 Coimmunoprecipitation and GST pulldown assays
Cell lysates were made in RIPA buffer, precleared using desired antibody or IgG at 4°C overnight, with subsequent incubation with protein G Resin (GenScript, Nanjing, Jiangsu, China) for 4 h. For GST pulldown assay, we incubated the cell lysates in GST-tagged proteins and Glutathione Sepharose 4B beads overnight at 4°C. Finally, the immunoprecipitates were separated with Western blotting with corresponding antibodies. The details of antibodies used can be found in Supplementary Table S2.
2.9 In vitro binding assays
The purified proteins of histidine (His)-USP51 and GST-PD-L1 (full-length PD-L1 and its corresponding truncated mutants) were incubated at 4°C overnight. Glutathione Sepharose 4B beads were introduced and maintained for 4 h, before Western blotting of the bound proteins with USP51 antibody (H00158880-A01, Abnova, Taiwan, China).
2.10 In vitro USP51 activity assay
C-terminal conjugate of ubiquitin with 7-amino-4-methylcoumarin (Ub-AMC) was used to measure the deubiquitinase activity of USP51. Briefly, the reaction system contained 50 mmol/L 2-hydroxyethyl, 0.5 mmol/L ethylene diamine tetraacetic acid, 100 mmol/L NaCl,1 mmol/L DTT, 0.1 mg/mL bovine serum albumin, 64 nmol/L USP51 (MERRYBIO, Nanjing, Jiangsu, China) and 5 μmol/L DHM (T2998, TOPSCIENCE) and was incubated at 37°C for 1 h. Then, 250 nmol/L Ub-AMC (U-550, BostonBiochem, Emeryville, CA, USA) was added, and the fluorescence intensity was detected using a microplate reader (BMG Labtech, Offenburg, Germany) with a 345 nm excitation/460 nm emission optic module for 3 h. All experiments had three biological replicates, and the mean value was analyzed.
2.11 Surface plasmon resonance (SPR)
The binding kinetic evaluations were conducted on a Biacore T200 (GE Healthcare, Boston, MA, USA). Briefly, His-tagged USP51 were immobilized in Acetate 4.0 (BR-1003-49, GE Healthcare) on a Biacore Series S NTA chip (CM5) using N-terminal His tag capture, then amine-coupling with 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride/N-hydroxy succinimide/ethanolamine (Sigma) as per kit guidelines. A 7500 response unit immobilization was attempted. Compounds were examined in two-fold dilution series in immobilization buffer containing 5% (V/V) dimethylsulfoxide (DMSO), at 30 μL/min, with 60 s contact time and 120 s dissociation time. We also included a solvent correction curve between 4.5%-5.8% (V/V) DMSO. Ten-point dilution curves before and after compounds were used to confirm surface integrity throughout the experiment. The Biacore T200 Evaluation software (version 3.0, GE Healthcare) was employed with a kinetic fit.
2.12 Cellular thermal shift assay
Cells underwent a 5 h DHM treatment at specified concentrations, then, they were accumulated for 3 min in PBS with a complete protease inhibitor cocktail, and respectively incubated at 40°C, 43°C, 46°C or 49°C. Next, cells were freeze-thawed in liquid nitrogen, and the supernatant was collected. Finally, proteins were assessed using Western blotting assay with USP51 antibody (SAB1305451, Sigma).
2.13 Cell viability assessment
In all, 3,000 cells/well were plated in 96-well plates and treated with different drugs for different time points. Cell survivability was assessed via cell counting kit-8 (Yeasen), as per kit guidelines, using absorbance at 450 nm via a microplate reader (BMG Labtech).
2.14 Colony formation experiment
In all, 750 cells/well were plated in 6-well plates before treatment with different drugs for 10 days. Cells underwent a 30 min fixation in 4% paraformaldehyde (Sigma), and 15 min crystal violet staining. The number of colonies was counted (colony formation rate = colony number/plated cell number).
2.15 Flow cytometric analysis of cell apoptosis
Cells were treated with different drugs for 48 h before apoptotic assessment with the fluorescein isothiocyanate (FITC) Annexin-V/prodium iodide (PI) apoptosis detection kit (Yeasen) as per kit directions. Apoptotic cell percentage was evaluated using flow cytometry (LSR, BD Biosciences, San Diego, CA, USA).
2.16 Immunofluorescence assay
Cells underwent 24-hour treatment with varying drugs before fixation in 4% paraformaldehyde, followed by overnight incubation with phosphorylated histone H2AX (γH2AX) antibody (9718S, Cell Signaling Technology, Boston, MA, USA) at 4°C. The next day, cells were treated for 1 h with DyLight 594-conjugated secondary antibody (ab96885, abcam, Cambridge, Cambs, UK) before a 3 min staining with 4’,6-diamidino2-phenylindole (DAPI) (1μg/mL). The labeled nuclei were detected by Confocal FV1000 (Olympus, Tokyo, Japan). The percentage of cells with ≥10 foci was quantified. At least 100 cells were counted per well.
2.17 Tumor xenograft experiment
All animal protocols received ethical approval from the Medical College of Nankai University (2021-SYDWLL-000040, Tianjin, China). We obtained 6-week-old male NOD-SCID mice from Beijing HFK Bioscience LTD (Beijing, China), and injected cells into their bilateral groins. Once tumors achieved a size of 50 mm3, mice were treated with different drugs for three weeks. When the tumor size reached to approximately 800 mm3, the mice were euthanized following intraperitoneal anesthesia with ketamine and promethazine. The tumor weight was weighed, and volume was calculated as 1/2 (length × width2).
2.18 Tissue microarray and immunohistochemistry (IHC)
Microarrays of human lung adenocarcinoma tissues (HLugA150CS03 and HlugA180Su04) were obtained from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China). Samples underwent USP51, PD-L1 (17952-1-AP, Proteintech, Wuhan, Hubei, China) and ITGB1 (34971T, Cell Signaling Technology) antibody staining for IHC assays with the Envision Kit (Dako) in accordance as per kit protocols. Several pathologists then blindly scored the immunostaining, as described previously [24]. This protocol received ethical approval from the Nankai University (NKUIRB2021010).
2.19 Bioinformatic analysis
The gene expression profiling interactive analysis (GEPIA) database (http://gepia.cancer-pku.cn/) was used to analyze the effect of ITGB1 on clinical survival. JASPAR (http://jaspar.genereg.net/) analysis was employed to identify a series of canonical NF-κB response elements in the promoter region of chemoresistance-related genes.
2.20 Statistical analysis
The experimental data were analyzed by GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA) and SPSS 17.0 (IBM, Chicago, IL, USA) software (Chicago, IL, USA). Data from all experiments are expressed as mean ± standard deviation (SD) of three distinct replicates. Gene profile correlation in various samples were assessed via Spearman's rank correlation test. Inter-group comparison was performed using one-way analysis of variance (ANOVA). Unpaired data assessments were conducted using Student's t-test, as appropriate. P value < 0.05 was set as the significance threshold.
3 RESULTS
3.1 The extracellular domain of PD-L1 was required for the regulation of chemosensitivity in a cell-intrinsic manner
Consistent with previous report showing that abnormal PD-L1 content is involved in chemoresistance development in human cancers [10], our Western blotting and flow cytometry assay results revealed a significant increase in PD-L1 levels in DDP-resistant A549R cells compared to the wild-type A549 cells (Figure 1A). Thus, to further investigate the significance of cancer cell-intrinsic PD-L1 in chemoresistance, specific shRNAs targeting PD-L1 were introduced in A549R followed by treatment with DDP. Based on our cell viability (Figure 1B), colony formation (Figure 1C) and cell apoptosis analyses (Figure 1D), PD-L1 depletion strongly promoted DDP-induced cell proliferation retardation and cell apoptosis. Similarly, the anti-apoptosis protein Bcl-2 was strongly diminished, whereas the pro-apoptosis proteins Bax and cleaved Caspase-3 were enhanced in shPD-L1/A549R cells in response to DDP treatment (Supplementary Figure S1A). We also explored the effect of PD-L1 on DDP-induced γH2AX expression, which is a marker for DNA damage repair [25]. The results confirmed that PD-L1 knockdown significantly enhanced DDP-induced formation of γH2AX nuclear foci (Figure 1E) and the level of γH2AX expression (Supplementary Figure S1B). Consistently, we obtained the similar results in shPD-L1/PC9R cells (Supplementary Figure S2), indicating that cell-intrinsic PD-L1 potentially modulates chemosensitivity in NSCLC cells.
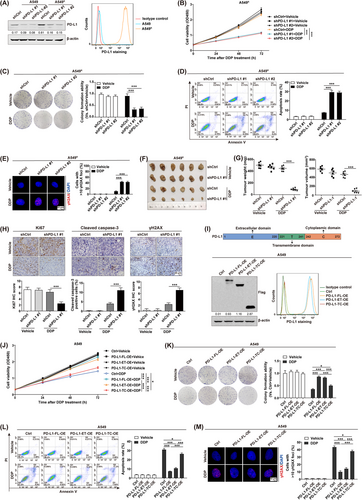
Further, we determined whether aberrant PD-L1 content influenced tumor responsiveness to chemotherapeutic treatment in vivo. To do so, shPD-L1/A549R or shCtrl/A549R cells were subcutaneously injected into BALB/c nude mice to develop a tumor xenograft model, which was then treated with DDP. Based on our observation, DDP strongly suppressed tumor development in BALB/c nude mice with shPD-L1/A549R tumors as compared to shCtrl/A549R tumors (Figure 1F-G). Immunohistochemical staining further demonstrated that DDP treatment resulted in decreased Ki67 expression but increased cleaved Caspase-3 and γH2AX expression in shPD-L1/A549R tumors as compared to shCtrl/A549R tumors (Figure 1H). Thus, the above observations confirmed the involvement of aberrant PD-L1 in NSCLC chemoresistance, independent of the immune system in vitro and in vivo.
Next, to elucidate the functional domain of PD-L1 that contributes to its cell-intrinsic role in chemoresistance, the full-length PD-L1 (PD-L1-FL) and two deletion constructs PD-L1-ET (containing the extracellular and transmembrane domains of PD-L1) and PD-L1-TC (containing the transmembrane and cytoplasmic domains of PD-L1) were generated and overexpressed in A549 cells (Figure 1I). The cell viability (Figure 1J), colony formation (Figure 1K), cell apoptosis (Figure 1L and Supplementary Figure S3A), and DNA damage detection assays (Figure 1M and Supplementary Figure S3B) confirmed that overexpression of PD-L1-FL strongly reduced cell proliferation retardation and cell apoptosis and decreased the formation of γH2AX nuclear foci upon DDP treatment. Notably, we found that overexpression of PD-L1-ET showed a similar effect to attenuate cancer cell responsiveness to DDP. We observed the same alterations in PC9 cells (Supplementary Figure S4), indicating that the PD-L1 extracellular domain conferred the regulation of chemosensitivity through a cell-intrinsic mechanism in NSCLC cells.
3.2 ITGB1 mediated PD-L1-induced chemoresistance
In addition, we moved to identify the downstream signaling pathway that mediated PD-L1-induced chemoresistance in NSCLC cells. Surprisingly, treatment with the PD-L1 neutralizing antibody avelumab did not show a chemosensitizing effect by the analysis of cell viability (Supplementary Figure S5A), colony formation (Supplementary Figure S5B), cell apoptosis (Supplementary Figure S5C-D), and DNA damage detection (Supplementary Figure S5E-F) in A549R cells. Considering that a complex structure analysis revealed an IgV-dominated binding pattern between PD-L1 and its neutralizing antibody [26], these results highlight that the cancer cell-intrinsic role of PD-L1 in chemoresistance might be independent of its interaction with the PD-1 receptor through the IgV domain. Consequently, we performed an endogenous Co-IP combining with mass spectrometry and identified several cell membrane-bound receptors that could potentially interact with PD-L1 in A549R cells (Supplementary Figure S6A). Quantitative RT-PCR further demonstrated a significant upregulation of ITGB1, CD98 and CD44 expression in A549R cells (Supplementary Figure S6B). Of note, the GEPIA database analysis revealed that elevated ITGB1 levels were closely related to shorter overall survival (OS) (Supplementary Figure S6C) and disease-free survival (Supplementary Figure S6D) in NSCLC patients.
Importantly, the Co-IP assay demonstrated an increased physical interaction between PD-L1 and ITGB1 in A549R cells relative to A549 cells (Figure 2A). Moreover, cell viability (Figure 2B), colony formation (Figure 2C), cell apoptosis (Figure 2D and Supplementary Figure S7A), and DNA damage detection assays (Figure 2E and Supplementary Figure S7B) demonstrated that either PD-L1 depletion or ITGB1 neutralization significantly promoted cancer cell chemosensitivity to DDP in A549R cells; however, the combinatorial treatment did not elicit a synergistic anticancer activity. Similarly, we conducted the same investigations in PC9R cells and achieved comparable results (Supplementary Figure S8).
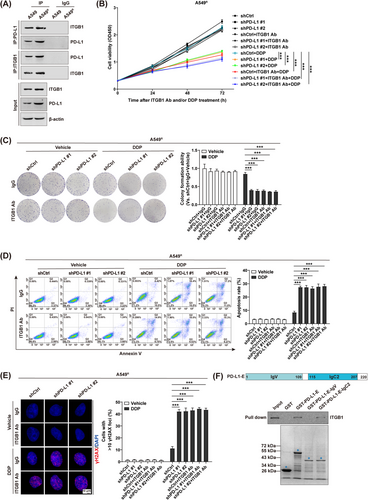
Next, we identified the PD-L1 functional domain that mediates its association with ITGB1. We constructed three GST fusion proteins for the extracellular domain of PD-L1 (PD-L1-E, PD-L1-IgV and PD-L1-IgC2). The results of GST-pulldown assay confirmed that PD-L1 physically interacted with ITGB1 through its extracellular domain (Figure 2F). Of note, the deletion variant PD-L1-IgC2, but not PD-L1-IgV, was able to interact with ITGB1. Taken together, our data proposed that ITGB1 specifically functioned as a membrane-binding receptor for PD-L1 and thus mediates its cell-intrinsic role in chemoresistant NSCLC cells.
3.3 PD-L1 conferred chemoresistance by triggering the ITGB1/NF-κB signaling
Next, we moved to identify the molecular mechanism of PD-L1/ITGB1-regulated cancer cell responsiveness to chemotherapy in NSCLC. Of note, TCGA database analysis revealed that the transcript levels of multiple chemoresistance-related genes, such as baculoviral IAP repeat containing 5 (BIRC5) [27], breast cancer gene 1 (BRCA1) [28, 29], BRCA2 [30, 31], glutathione synthetase (GSS) [32], hypoxia inducible factor 1 alpha (HIF1α) [33], and solute carrier family 31 member 2 (SLC31A2) [34], were positively correlated with those of PD-L1 (Supplementary Figure S9A) and ITGB1 (Supplementary Figure S9B) in NSCLC patients. Quantitative RT-PCR further verified that expression of these chemoresistance-related genes was significantly downregulated by either PD-L1 depletion or ITGB-1 neutralization in A549R cells (Figure 3A). However, the combinatorial treatment did not exhibit a synergistic inhibition, which is consistent with our notion that PD-L1-promoted chemoresistance is mainly mediated through its binding to the ITGB1 receptor on NSCLC cells. Interestingly, analysis using the JASPAR database identified a series of canonical NF-κB response elements in the promoter region of all above chemoresistance-related genes (Supplementary Figure S9C), indicating a potential role of the NF-κB signaling in PD-L1/ITGB1-regulated chemoresistance [35]. Thus, expression of the NF-κB components was assessed via Western blotting in shPD-L1/A549R cells with or without ITGB1 neutralizing antibody. We confirmed that either PD-L1 knockdown or ITGB1 neutralization could reduce the expression of phosphorylated IκBα (Figure 3B), with a simultaneous decrease in the nuclear transfer of the P65 subunit (Figure 3C). However, a synergistic inhibition in the NF-κB signaling was not achieved by the combinational treatment.
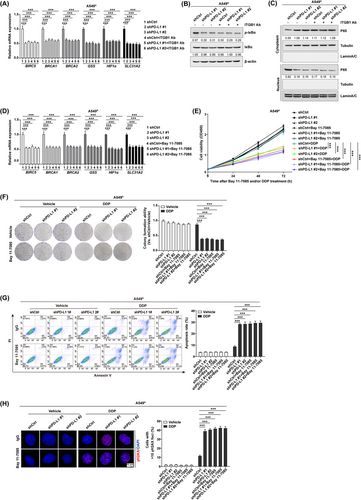
In line, the quantitative RT-PCR (Figure 3D), cell viability (Figure 3E), colony formation (Figure 3F), cell apoptosis (Figure 3G and Supplementary Figure S10A), and DNA damage detection (Figure 3H and Supplementary Figure S10B) assays further revealed that either depletion of PD-L1 or treatment with an NF-κB inhibitor Bay 11-7085 increased A549R cell chemosensitivity to DDP; however, we did not observe a synergistic suppression with the combinational treatment. Similar results were also obtained in PC9R cells (Supplementary Figure S11), confirming that the PD-L1/ITGB1 axis promoted NSCLC cell sensitivity to chemotherapy by triggering the NF-κB signaling.
3.4 USP51 promoted chemoresistance through deubiquitination of PD-L1
To further explore if the abnormal PD-L1 content is related to posttranslational modulation in chemoresistant NSCLC cells, we next treated A549R and A549 cells with the protein synthesis inhibitor CHX. Western blotting analysis demonstrated that the stability of PD-L1 protein was strongly enhanced in A549R cells relative to A549 cells (Supplementary Figure S12A). Moreover, exposure to the proteasomal inhibitor MG132 (Supplementary Figure S12B) and not the lysosomal inhibitor CQ (Supplementary Figure S12C) led to remarked upregulation of PD-L1 protein. The ubiquitination assay data revealed that the polyubiquitination level of PD-L1 protein was strongly decreased upon MG132 treatment in A549R cells compared to that in A549 cells (Supplementary Figure S12D). Also, we confirmed these results in PC9R and PC9 cells (Supplementary Figure S13). These data indicated a ubiquitin proteasome-dependent dysregulation of PD-L1 content in chemoresistant NSCLC cells.
Notably, the analysis of endogenous Co-IP combining with mass spectrometry identified two deubiquitinases [USP51 and ubiquitin C-terminal hydrolase L1 (UCHL1)] and two E3 ubiquitin ligases [ring finger protein 31 (RNF31) and tripartite motif containing 21 (TRIM21)] that could potentially interact with PD-L1 in A549R cells (Supplementary Figure S14A). We further validated the physical interactions of PD-L1 with USP51 (Figure 4A), UCHL1 (Supplementary Figure S14B), RNF31 (Supplementary Figure S14C), and TRIM21 (Supplementary Figure S14D) in PD-L1-overexpressing A549 cells. However, Western blotting and flow cytometry assays demonstrated that the specific interference of USP51 (Figure 4B), but not UCHL1 (Supplementary Figure S14E), RNF31 (Supplementary Figure S14F) and TRIM21 (Supplementary Figure S14G), altered PD-L1 levels in A549R cells. Western blotting data revealed that the expression of USP51 was significantly elevated in A549R cells (Figure 4C). These observations demonstrated a predominant function of USP51 in the posttranslational modulation of PD-L1 in chemoresistant NSCLC cells. In line, the PD-L1 protein stability was strongly decreased upon CHX treatment in shUSP51/A549R cells (Figure 4D). However, USP51-interfered PD-L1 content decrease was strongly weakened with MG132 addition (Figure 4E). Based on our ubiquitination assay, USP51 depletion strongly enhanced PD-L1 protein polyubiquitination in A549R cells (Figure 4F), collectively suggesting that USP51, a deubiquitinase, targeted PD-L1 protein for deubiquitination and stabilization. Similar results were further obtained in PC9R cells (Supplementary Figure S15).
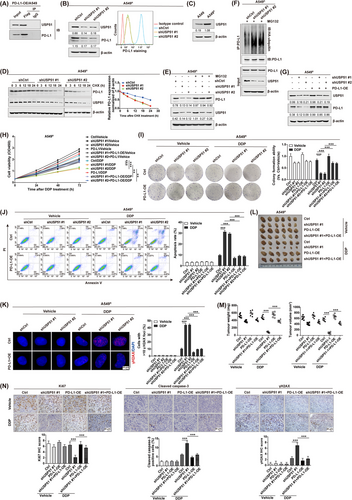
Next, we examined whether USP51 depletion could chemosensitize NSCLC cells through the regulation of PD-L1. To do so, PD-L1 was overexpressed in shUSP51/A549R cells (Figure 4G). The cell viability (Figure 4H), colony formation (Figure 4I), cell apoptosis (Figure 4J and Supplementary Figure S16A), and DNA damage detection assays (Figure 4K and Supplementary Figure S16B) demonstrated that cell responsiveness to chemotherapy was increased in shUSP51/A549R cells; nevertheless, this outcome was strongly diminished in PD-L1-overexpressed cells. We observed comparable results in PC9R cells (Supplementary Figure S17). In line, using BALB/c nude mice xenograft model, we confirmed that USP51 knockdown severely inhibited tumor growth upon DDP treatment, which was suppressed in mice with PD-L1-harboring tumors (Figure 4L-M). Immunohistochemical staining further indicated that the Ki67 content was diminished but the cleaved Caspase-3 and γH2AX contents were increased in tumors from shUSP51/A549R mice when treated with DDP, and this outcome was suppressed in tumors with PD-L1 rescue (Figure 4N). These observations together revealed that USP51 deficiency chemosensitized NSCLC cells via PD-L1 regulation both in vitro and in vivo.
3.5 USP51 deubiquitinated PD-L1 at lysine residues K280/K281
Consequently, we conducted an endogenous Co-IP to validate the USP51 and PD-L1 relationship. The results revealed that USP51 directly associated with PD-L1 in A549 cells, which was further increased in A549R cells (Figure 5A). Importantly, an in vitro binding assay proved that the purified proteins of His-USP51 and GST-PD-L1 could directly associate with one another in cell-free environment (Figure 5B). Moreover, a series of deletion mutants of USP51 (Myc-USP51-N and Myc-USP51-C) and PD-L1 (Flag-PD-L1-E and Flag-PD-L1-C) were generated and overexpressed in A549 cells. Our Co-IP data verified that the deletion variant Myc-USP51-N interacted with the PD-L1 cytoplasmic domain (Figure 5C-D). In addition, three deletion constructs of PD-L1-C were generated to demonstrate that the deletion variant PD-L1-C3, but not PD-L1-C1 and PD-L1-C2, was able to interact with USP51 (Figure 5E), highlighting that the PD-L1C-terminal region (280-290aa) is critical for the USP51-PD-L1 association.
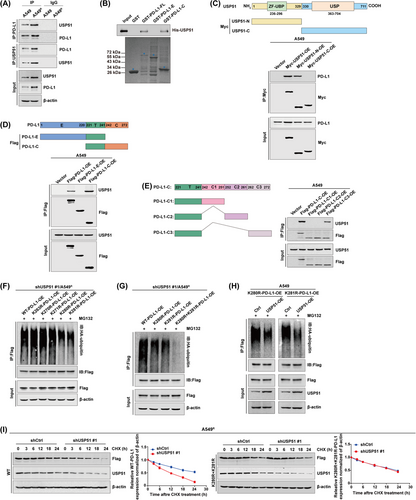
Next, we examined whether USP51 could deubiquitinate the lysine residues of PD-L1-C3. As shown in Figure 5F, the wild-type and individual mutants of PD-L1 (K263R, K270R, K271R, K280R and K281R) were respectively transfected into shUSP51/A549R cells. The ubiquitination assay indicated that the polyubiquitination levels of either K280R or K281R was significantly reduced compared to wild-type PD-L1, however this effect was not shown for the K263R, K270R and K271R mutants. In addition, the simultaneous mutant K280R+K281R totally abolished the polyubiquitination levels of PD-L1 (Figure 5G), identifying lysine residues K280 and K281 as major ubiquitination sites on PD-L1. Indeed, USP51 was able to deubiquitinate PD-L1 at either K280 or K281 lysine residue (Figure 5H). The interference of USP51 led to a strongly reduced stability of wild-type PD-L1 protein; however, this effect was deprived in cells carrying the simultaneous mutant K280R+K281R (Figure 5I). The same experiments were also conducted in PC9R cells and produced comparable results (Supplementary Figure S18). Taken together, our data demonstrated that lysine residues K280 and K281 on the PD-L1 C-terminal region were essential for its deubiquitination and stabilization by USP51 in chemoresistant NSCLC cells.
3.6 The USP51, PD-L1 and ITGB1 contents were positively associated with worse prognosis among NSCLC patients
To further validate the pathological relationship between the USP51/PD-L1/ITGB1 axis and chemoresistant features in human NSCLC, we conducted immunohistochemical staining for USP51, PD-L1 and ITGB1 in 167 samples of primary NSCLC (Figure 6A). We observed direct relationships between USP51, PD-L1 and ITGB1 (Figure 6B). It has been reported that Cyclin D1 [36, 37] and breast cancer gene 1 (BRCA1) [38, 39], which mediate cell proliferation and DNA damage response, respectively, were strongly elevated in NSCLC tissues resistant to genotoxic drugs. Indeed, we revealed that the USP51, PD-L1 and ITGB1 contents were intricately linked to Cyclin D1 (Figure 6C) and BRCA1 (Figure 6D), highlighting the involvement of aberrant USP51/PD-L1/ITGB1 expression in the cellular mechanisms that mediated NSCLC chemoresistance.
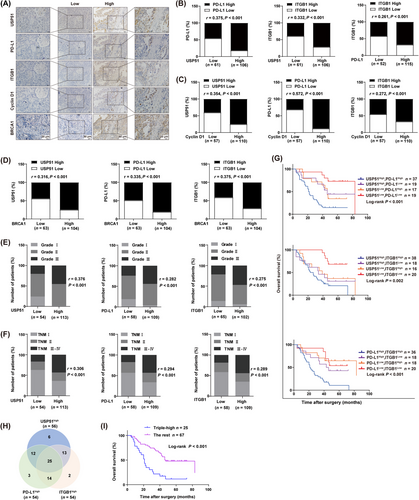
In addition, we observed increased USP51, PD-L1 and ITGB1 levels in high-grade tumors (Figure 6E) and advanced tumor-node-metastasis (TNM) stage (Figure 6F). Of note, the cancer patients with substantially elevated USP51, PD-L1 and ITGB1 levels in tumors exhibited shorter OS relative to patients with reduced USP51, PD-L1 and ITGB1 contents (Figure 6G). We also examined OS of the triple-high group (USP51highPD-L1highITGB1high) compared to the rest (Figure 6H). The results confirmed that patients with concomitantly high expression of USP51, PD-L1 and ITGB1 in tumors had significantly shorter OS (Figure 6I). Taken together, these data revealed that dysregulation of the USP51/PD-L1/ITGB1 axis potentially contributes to the malignant progression and may be employed in the prediction of worse NSCLC patient prognosis.
3.7 Dihydromyricetin acted as a USP51 inhibitor and conferred chemosensitization in NSCLC
Next, we moved to identify the small molecules that could potentially alter USP51 activity. To do so, we used the fluorescent Ub-AMC assay to examine an in-house library containing 188 natural compounds (Figure 7A and Supplementary Table S3). Five compounds, dihydromyricetin (DHM), (-)-epigallocatechin (EGC), gallic acid, linoleic acid, and tannic acid, were shown to significantly inhibit the deubiquitinase activity of USP51 at a concentration of 1 μmol/L (Supplementary Table S4). Further, we conducted a computational molecular dynamics simulation and revealed that three flavonoid compounds DHM, EGC and gallic acid could potentially bind to the catalytic domain of USP51 (Figure 7B). Of note, the analysis of half maximal inhibitory concentration (IC50) indicated that DHM performed a stronger inhibition in USP51 activity than EGC and gallic acid (Figure 7C). In line with this, the SPR binding assay validated a significantly increased binding affinity between USP51 and DHM (Figure 7D). Taken together with the cellular thermal shift assay showing that DHM treatment effectively stabilized USP51 protein in A549R (Figure 7E) and PC9R cells (Supplementary Figure S19), our results indicated that DHM might target USP51 directly.
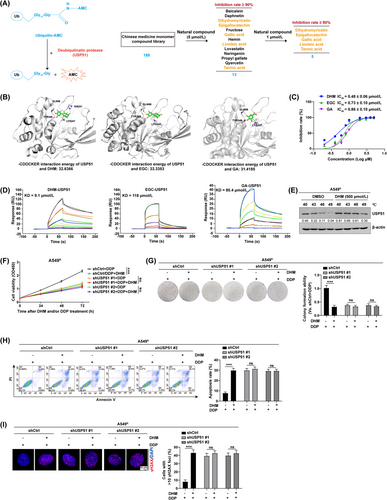
In addition, to examine whether DHM exerted its biological function through a USP51-dependent mechanism, we introduced the full-length USP51 into A549 cells followed by treatment with DHM (Supplementary Figure S20A). Based on our Western blotting results, the PD-L1 content was upregulated in USP51-OE/A549 cells, whereas this effect was significantly abolished by DHM treatment. Consistently, the ubiquitination assay revealed that USP51 deubiquitinase inhibition by treatment with DHM led to increased polyubiquitination of PD-L1 proteins in USP51/A549 cells (Supplementary Figure S20B). The cell viability (Supplementary Figure S20C), colony formation (Supplementary Figure S20D), cell apoptosis (Supplementary Figure S20E-F), and DNA damage detection assays (Supplementary Figure S20G-H) further confirmed that overexpression of USP51 strongly reduced cell proliferation retardation and cell apoptosis but promoted DNA damage repair in response to DDP; however, USP51-induced chemoresistant phenotypes were weakened by the addition of DHM. Similar results were also obtained in PC9 cells (Supplementary Figure S21), highlighting a role of DHM in the regulation of chemosensitization via targeting USP51.
Indeed, treatment with DHM strongly promoted DDP-induced cell proliferation retardation (Figure 7F-G) and cell apoptosis (Figure 7H and Supplementary Figure S22A) in A549R cells, while this effect was impaired by USP51 interference. We also examined the effect of DHM on DDP-induced DNA damage response with or without USP51 depletion (Figure 7I and Supplementary Figure S22B), revealing that DHM conferred responsiveness to chemotherapeutic treatment in a USP51-dependent manner in NSCLC cells. These experiments were further performed in PC9R cells and obtained the similar results (Supplementary Figure S23).
3.8 DHM treatment or ITGB1 neutralization chemosensitized NSCLC
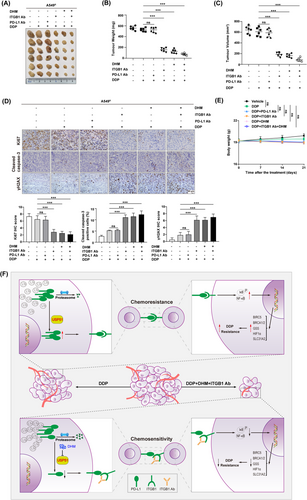
Our study revealed a cell-intrinsic role for USP51/PD-L1/ITGB1 signaling in mediating chemotherapeutic resistance in NSCLC. Therefore, treatment with DHM or anti-ITGB1 might suppress the chemoresistant phenotypes of NSCLC cells and further enhance the efficacy of chemotherapy. To achieve this, A549R cells were subcutaneously injected into BALB/c nude mice to allow the establishment of primary tumors, then they were treated with DHM, anti-ITGB1 or anti-PD-L1 (avelumab) in the presence of DDP (Figure 8A). We noticed that treatment with DHM or anti-ITGB1 effectively promoted the cell responsiveness to DDP, as evidenced by a reduction in tumor volume (Figure 8B) and weight (Figure 8C) by approximately 75%-80%; however, the combinatorial treatment did not exhibit a synergistic inhibition. Of note, the chemosensitizing efficacy was not shown for anti-PD-L1 treatment, which is consistent with our notion that the cell-intrinsic role of PD-L1 in chemoresistance might not achieve by its interaction with PD-1. The immunohistochemical staining also revealed significant reduction in Ki67 content but increase in cleaved Caspase-3 and γH2AX expression in tumors with DHM and/or anti-ITGB1 treatment (Figure 8D). No obvious undesirable side effects, namely reduced body weight (Figure 8E) and toxic pathological alterations occurred in the heart, liver, spleen, lung, and kidney (Supplementary Figure S24). Overall, these results implied that disruption of USP51/PD-L1/ITGB1-deployed juxtacrine interaction by treating cancer patients with the USP51 inhibitor (e.g. DHM) or ITGB1 neutralization might inhibit their chemoresistant properties and thus reduce cancer risk.
4 DISCUSSION
Emerging evidence reported that cancer cell-intrinsic PD-L1 is critical for the development of therapeutic resistance and is intricately linked to worse NSCLC patient prognosis. Hence, identifying the molecular mechanism that contributes to PD-L1 dysfunction can aid in the design of new and highly efficacious anti-neoplastic treatments. Herein, we demonstrated a cancer cell- intrinsic role of PD-L1 in chemoresistant NSCLC cells, which involves the ITGB1/NF-κB axis. More importantly, we recognized USP51 as an original deubiquitinase that marked PD-L1 for deubiquitination and stabilization. The specific inhibition of USP51 activity chemosensitizated NSCLC cells by promoting PD-L1 protein elimination, which have presented a rationale to target the USP51/PD-L1/ITGB1 axis and prevent its tumor-promoting effects in NSCLC (Figure 8F).
Although it is extensively documented that PD-L1 suppresses anticancer immune response, the cancer cell-intrinsic role of PD-L1 and its association with other oncogenic network garnered much interest [14]. For example, aberrant PD-L1 regulates PI3K/AKT signaling to support tumor survival in NSCLC [40], interacts with phosphorylated STAT3 signaling to facilitate tumor necrosis and progression in breast cancer [41], and modulates tumor glucose metabolism by triggering mTOR signaling in sarcoma [42]. Collectively, our analyses indicated that PD-L1 significantly reduced chemotherapy-mediated killing by stimulating the NF-κB axis in NSCLC. These observations clearly identified a non-immune checkpoint function of PD-L1 in axis tumor growth, metastasis and therapeutic resistance in multiple cancer types [11, 43]. Mechanistically, we provided additional evidence that PD-L1 mainly functioned through its extracellular domain to interact with ITGB1 as a membrane-bound receptor in chemoresistant NSCLC cells, which supported the notion that aberrant ITGB1 was linked to various chemoresistance-related events, such as defects in tumor growth, apoptosis and genome integrity [44-46]. Importantly, we found that a distinct fragment PD-L1-IgC2, but not PD-L1-IgV, in the PD-L1 extracellular domain was responsible for its direct binding to ITGB1. Thus, PD-L1 blockade using its neutralizing antibody avelumab, which dominantly bound to the PD-L1-IgV domain, might not be sufficient to interrupt PD-L1/ITGB1 interaction and alter cancer cell sensitivity to DDP in NSCLC. Considering that depletion of the PD-L1-IgV domain effectively deprives the interplay between PD-L1 and PD-1 [47, 48], our observations also highlighted that the cancer cell-intrinsic role of PD-L1 in chemoresistance might be independent of its interaction with PD-1. In line with these, a growing body of evidence have indicated that PD-L1/PD-1 blockage is not completely effective in cancer treatment, as some NSCLC patients remain unresponsive to such treatment [49]. Taken together, these data revealed that a direct correlation between PD-L1 and ITGB1 expression was predominantly present in a subset of advanced NSCLC samples with chemoresistant properties, further detailed investigations on dual blockage of both PD-1 and ITGB1 should be carried out to effectively silence PD-L1 to improve anticancer efficacy.
Several studies have indicated that PD-L1 was highly expressed in numerous solid tumors, particularly NSCLC [50, 51]. Elevated PD-L1 levels are strongly associated with worse clinical outcome in cancer patients [50, 52], which corroborates with the current investigation, which revealed high USP51, PD-L1 and ITGB1 levels in tumors with high tumor grade, advanced TNM stage and poor OS. At the molecular level, mechanisms underlying aberrant PD-L1 expression operate at multiple layers, mainly including genomic alterations, constitutive oncogenic signaling activation, extrinsic factors and epigenetic regulation. For example, PD-L1 gene amplification on chromosome 9p24.1 has been detected in 5.3% of NSCLC cases [53]. Since the janus kinase 2 (JAK2) gene also resides in the 9p24.1 region, increased JAK2 gene copy number also upregulates PD-L1 levels via the JAK-STAT axis in NSCLC [54]. In addition, a number of cellular stimuli, such as TNF-α [23] and interleukin 17 (IL-17) [55], have been demonstrated to induce PD-L1 expression mainly through interacting with NF-κB. Several chemotherapeutic agents, such as paclitaxel, can also promote PD-L1 activation via NF-κB signaling in cancer cells [56]. Here, we extended that study that the cancer cell-intrinsic role of PD-L1 in regulating chemoresistance was predominantly mediated through NF-κB signaling in NSCLC cells. These data collectively proposed that the constitutive oncogenic NF-κB signaling activation might create a positive feedback loop to trigger PD-L1 stimulation in malignant progression of human cancers. Indeed, inhibition in NF-κB activity disrupts this positive feedback loop, which eventually depletes PD-L1 expression and the aggressiveness of NSCLC in vitro and in vivo [35]. On the other hand, both histone acetylation and histone methylation in the PD-L1 gene promoter region are necessary for abnormal PD-L1 content in solid tumors [57, 58], highlighting the involvement of epigenetic mechanisms in PD-L1 regulation. Considering that histone deacetylase inhibitors exhibited anticancer effect by altering PD-L1 expression in pre-clinical models [59], a combinatorial treatment with histone deacetylase inhibitors and PD-L1 suppression may be a synergistic approach in anticancer therapy.
Besides, the present study expanded on the underlying mechanism by which the PD-L1 content was also regulated posttranslationally in chemoresistant NSCLC cells. We validated that USP51 was an original deubiquitinase that marks the PD-L1 protein for deubiquitination and stabilization. At the molecular level, USP51 directly bound and deubiquitinated PD-L1 at lysine residues K280 and K281. The dominant negative mutants of PD-L1, K280R and/or K281R disrupted the USP51catalytic activity, but did not alter PD-L1 protein stability. Thus, USP51 was crucial to the induction of chemoresistance through stabilizing PD-L1 protein in NSCLC. In a subset of human NSCLC cell lines and patient samples, USP51 was directly associated with PD-L1 content. Of note, the USP51 and PD-L1 association was strongly evident in NSCLC patients with malignant properties and poor prognosis. More recently, it was reported that ectopic expression of USP51 was able to induce chemoresistant phenotypes, such as DNA damage response, in certain cancer cells [60]. In addition, another deubiquitinase ubiquitin specific peptidase 7 (USP7) had been identified to regulate PD-L1 protein stabilization in gastric cancer [61]. Targeting USP7 with its small molecule inhibitors sensitized cancer cells to T cell-induced destruction by diminishing cell surface PD-L1 content while decreasing its association with PD-1. Hence, small molecule immune checkpoint inhibitors of PD-1/PD-L1 were shown to be highly beneficial in anticancer therapy [12]. These findings also supported our observation that disruption of USP51/PD-L1-deployed juxtacrine interaction with a USP51 inhibitor DHM showed a higher chemotherapeutic efficacy in NSCLC in vitro and in vivo, suggesting that chemo-immunotherapy or a combined treatment of anti-PD-1/PD-L1 with their specific inhibitors could potentially serve a therapeutic role in future anticancer therapies.
The current study had certain limitations. Firstly, the KRAS mutations account for approximately 30% of lung adenocarcinomas in Western countries and for 10%-15% of cases in Asia [62]. For example, the KRASG12C mutation is highly prevalent in patients suffering from lung adenocarcinoma (13% of total lung adenocarcinoma) and account for approximately 50% of all KRAS mutant cases [63]. The present study was lack of NSCLC cell lines with the KRASG12C mutation, such as H23 and H358 cells [64]. Thus, further studies are required to obtain a more comprehensive understanding. Secondly, due to the low water solubility and chemical stability of DHM, additional research should be carried out to investigate the characteristic and mechanism for inclusion of DHM and evaluate the effects of complexation on antitumor activities of DHM by targeting USP51.
5 CONCLUSIONS
Herein, we proposed a novel signaling network involving the USP51/PD-L1/ITGB1 axis that modulates NSCLC chemoresistance, which supplements the immune-free and cancer cell-intrinsic roles of PD-L1. Given that deubiquitinases are susceptible to pharmacology-mediated suppression, targeting USP51 using small molecule inhibitor (e.g., DHM) to reduce PD-L1 stability in conjunction with anti-PD-1 and/or anti-ITGB1 treatment represents a new therapeutic strategy that could efficiently impair PD-L1 and thus limit local tumor growth and aggressive progression in NSCLC.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Conception and design: JL, XX, GY and SY; Development of methodology: JL, XX, YO, LC, MG, HW, GY and SY; Analysis and interpretation of data (for example, statistical analysis, biostatistics, computational analysis): JL, XX, CQ, ZW, YL, QS, PS, YS, GY and SY; Writing, review of the manuscript: JL, XX, PS, YS, GY and SY.
ACKNOWLEDGMENTS
This work was supported by grants from the International Science and Technology Cooperation Project of China (No. 2022YFE0133300) and National Natural Science Foundation of China (No. 82172801 and No. 81972454).
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Ethics Committee of Nankai University (NKUIRB2021010). The tissue samples were obtained with written informed consent from each patient. All of the experimental procedures involving animals were performed in accordance with a protocol that was approved by the Ethics Committee for Animal Use at the Medical College of Nankai University (2021-SYDWLL-000040).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




