Combining radiation and the ATR inhibitor berzosertib activates STING signaling and enhances immunotherapy via inhibiting SHP1 function in colorectal cancer
Abstract
Background
Immune checkpoint inhibitors (ICIs) targeting programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1) have shown a moderate response in colorectal cancer (CRC) with deficient mismatch repair (dMMR) functions and poor response in patients with proficient MMR (pMMR). pMMR tumors are generally immunogenically “cold”, emphasizing combination strategies to turn the “cold” tumor “hot” to enhance the efficacy of ICIs. ATR inhibitors (ATRi) have been proven to cooperate with radiation to promote antitumor immunity, but it is unclear whether ATRi could facilitate the efficacy of IR and ICI combinations in CRCs. This study aimed to investigate the efficacy of combining ATRi, irradiation (IR), and anti-PD-L1 antibodies in CRC mouse models with different microsatellite statuses.
Methods
The efficacy of combining ATRi, IR, and anti-PD-L1 antibodies was evaluated in CRC tumors. The tumor microenvironment and transcriptome signatures were investigated under different treatment regimens. The mechanisms were explored via cell viability assay, flow cytometry, immunofluorescence, immunoblotting, co-immunoprecipitation, and real-time quantitative PCR in multiple murine and human CRC cell lines.
Results
Combining ATRi berzosertib and IR enhanced CD8+T cell infiltration and enhanced the efficacy of anti-PD-L1 therapy in mouse CRC models with different microsatellite statuses. The mechanistic study demonstrated that IR + ATRi could activate both the canonical cGAS-STING-pTBK1/pIRF3 axis by increasing cytosolic double-stranded DNA levels and the non-canonical STING signaling by attenuating SHP1-mediated inhibition of the TRAF6-STING-p65 axis, via promoting SUMOylation of SHP1 at lysine 127. By boosting the STING signaling, IR + ATRi induced type I interferon-related gene expression and strong innate immune activation and reinvigorated the cold tumor microenvironment, thus facilitating immunotherapy.
Conclusions
The combination of ATRi and IR could facilitate anti-PD-L1 therapy by promoting STING signaling in CRC models with different microsatellite statuses. The new combination strategy raised by our study is worth investigating in the management of CRC.
List of abbreviations
-
- CRC
-
- colorectal cancer
-
- mCRC
-
- metastatic colorectal cancer
-
- PD-1
-
- programmed cell death protein 1
-
- PD-L1
-
- programmed death-ligand 1
-
- ICI
-
- immune checkpoint inhibitor
-
- MSI-H
-
- microsatellite instability-high
-
- dMMR
-
- mismatch repair gene-deficient
-
- MSS
-
- microsatellite stable
-
- pMMR
-
- mismatch repair gene-proficient
-
- TIL
-
- tumor-infiltrating lymphocyte
-
- MHC-I
-
- major histocompatibility complex class-I
-
- Treg
-
- regulatory T cells
-
- MDSC
-
- myeloid-derived suppressor cell
-
- TAM
-
- tumor-associated macrophage
-
- TGF-β
-
- transforming growth factor β
-
- cGAS
-
- cyclic GMP-AMP synthase
-
- STING
-
- stimulator of interferon genes
-
- DC
-
- dendritic cell
-
- ATM
-
- ataxia telangiectasia mutated gene
-
- ATR
-
- ataxia telangiectasia mutated and Rad3-related
-
- IHC
-
- immunohistochemistry
-
- IF
-
- immunofluorescence
-
- dsDNA
-
- double stranded DNA
-
- DEG
-
- differentially expressed genes
-
- GO
-
- gene ontology
-
- GSEA
-
- gene-set enrichment analysis
-
- co-IP
-
- co-immunoprecipitation
-
- IR
-
- radiation
-
- cGAMP
-
- cyclin GMP-AMP
-
- TBK1
-
- TANK binding kinase 1
-
- IRF3
-
- interferon regulatory factor 3
-
- ISG
-
- interferon stimulated gene
-
- type I IFN
-
- type I interferon
-
- TIDC
-
- tumor-infiltrating DC
-
- PTPN6
-
- protein tyrosine phosphatase non-receptor type 6
-
- SHP1
-
- Src homology region 2 domain-containing phosphatase 1
-
- TRAF6
-
- TNF receptor associated factor 6
-
- SUMO1
-
- small ubiquitin-like modifier 1
-
- SAE2
-
- SUMO1 activating enzyme subunit 2
-
- Ubc9
-
- E2-ligase ubiquitin conjugating enzyme E2 I
-
- ZNF451
-
- E3-ligase zinc finger protein 451
-
- STAT3
-
- signal transducer and activator of transcription 3
-
- LARC
-
- locally advanced colorectal cancer
1 BACKGROUND
Colorectal cancer (CRC) is the third most common cancer type and one of the major causes of cancer-related death [1, 2]. Approximately 20% of patients with CRC have metastatic tumors (mCRC) at diagnosis, making most of the lesions unresectable [3]. Over the past few decades, advances in non-surgical techniques and expanded targeted therapies have greatly improved the survival of patients with mCRC. One of the most revolutionary methods is immunotherapy, especially immune checkpoint blockade therapy targeting programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1) [4]. Immune checkpoint inhibitors (ICIs) have shown a moderate response in CRCs and have become the first-line therapy for patients with microsatellite instability-high (MSI-H) or mismatch repair-deficient (dMMR) signatures, which, unfortunately, only consist of approximately 5% of all mCRCs [5-8]. Most patients are microsatellite stable (MSS) or mismatch repair proficient (pMMR), responding poorly to ICIs [9]. Therefore, there is an urgent need to improve the response to ICIs, especially in MSS/pMMR CRCs.
MSS/pMMR tumors generally have low levels of tumor immunogenicity and tumor-infiltrating lymphocytes (TILs), presenting a “cold” microenvironment, underlying potential combinational strategies to turn the cold tumor hot to overcome ICI resistance [10, 11]. As a universal standard of care for many cancer types, radiotherapy plays a complex role in altering the tumor microenvironment [12]. The pro-immunogenic role lies in its ability to promote T cell priming by inducing tumor-associated antigens, upregulating major histocompatibility complex class-I (MHC-I) expression and generating an in situ vaccine [12-14]. In contrast, immune suppressive cells and molecules such as regulatory T cells (Tregs), myeloid-derived suppressor cells (MDSCs), tumor-associated macrophages (TAMs), and transforming growth factor β (TGF-β) are stimulated by irradiation (IR) [15-18]. In addition, accumulating evidence demonstrates that IR-induced cytoplasmic DNA could activate the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway, promoting interferon signaling and dendritic cell (DC) activation, which links DNA damage to innate immune activation and the adaptive antitumor response [19, 20]. This role of IR-induced DNA damage in promoting anti-tumor immunity raises a new perspective on combining radiotherapy with immunotherapy.
Cells harbor complex mechanisms in response to DNA damage, in which ataxia telangiectasia mutated kinase (ATM) and ataxia telangiectasia mutated and rad3-related kinase (ATR) serve as key players in addressing IR-induced DNA double-strand breaks. A Previous study illustrated that ATM inhibition promoted both canonical and non-canonical STING activation to enhance the efficacy of ICIs along with radiotherapy [21], yet the impact of ATR inhibition on STING signaling is not fully understood. Meanwhile, recent studies have unveiled different roles of ATR inhibitors (ATRi) in improving antitumor immunity in various tumor models [22, 23], but the synergy in combination with IR and ICIs in CRCs has not been tested. Hence, the present study aimed to identify the role of ATR inhibition in cGAS-STING signaling and explore the antitumor efficacy of combining ATRi, IR and ICI therapy in colorectal tumors, which could directly inform ICI therapy strategies for patients with CRC.
2 MATERIALS AND METHODS
2.1 Cell culture
Human CRC cell lines HCT116 and SW480 and murine CRC cell lines CT26 and MC38 were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). HCT116 cells were cultured in McCoy's 5A modified medium (HyClone, ThermoFisher, Waltham, MA, USA) with 10% fetal bovine serum (FBS, Gibco, ThermoFisher). SW480 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-high glucose (HyClone, ThermoFisher) with 10% FBS. CT26 and MC38 cell lines were cultured in Roswell Park Memorial Institute (RPMI)-1640 (HyClone, ThermoFisher) with 10% FBS. All cell lines were cultured under 5% CO2 and proper humidity at 37°C in an incubator.
2.2 Reagents and antibodies
The ATRi berzosertib (VE-822, HY-13902) and Src homology 2 domain containing protein tyrosine phosphatase-1 (SHP1) agonist SC43 were purchased from MedChemExpress (HY-136657; Monmouth Junction, NJ, USA). Anti-PD-L1 was purchased from BioXCell (BE0101, clone 10F.9G2, West Lebanon, NH, USA).
2.3 Establishment of mouse CRC models and treatments
Female (5-6 weeks) C57/B6J and Balb/C mice were purchased from Hunan SJA Laboratory Animal Co., Ltd. (Changsha, Hunan, China) and raised in a specific pathogen-free laboratory. Animal experimental procedures were approved by the Laboratory Animal Welfare & Ethics Committee of Tongji Hospital Huazhong University of Science and Technology (permit No. TJH-201910007, Wuhan, Hubei, China). MC38 (1 × 106 cells/100 µL) and CT26 (1 × 106 cells/100 µL) cells were injected subcutaneously into the flanks of the C57/B6J and Balb/C mice. Tumor volume was measured every three days using a caliper and was calculated by V = [length × width2] × 0.5. For irradiation, mice were anesthetized with 1.5% pentobarbital sodium and received a single fraction of 5 Gy X-ray dose to the area of the subcutaneous tumor using the RS2000 X-ray Biological Research Irradiator (160 kV, 25 mA, Source Technologies Inc., Suwanee, GA, USA). Berzosertib (60 mg/kg) was administered by gavage 2 h before IR and consecutively for the next three days. Anti-PD-L1 (10 mg/kg) was administered intraperitoneally every three days, starting on the day of IR. When the tumor volume reached 2000 mm3, the mice were euthanized by CO2 inhalation as a humane endpoint.
2.4 Immunohistochemistry (IHC)
Subcutaneous tumors were harvested, fixed with 4% paraformaldehyde overnight and then embedded in paraffin. Subsequently, tissue sectioning was performed, and sections were deparaffinized, rehydrated, blocked of endogenous peroxidase and subjected to antigen retrieval. Antigen retrieval was performed by microwaving the sections in PH 6.0 citrate buffer (Beyotime, Shanghai, China) at 95°C for 20 minutes, followed by incubation with 5% bovine serum albumin (BSA) (Beyotime) at room temperature for 1 h. Sections were then incubated with primary antibodies anti-CD3e (1: 1:200, ab251607, Abcam, Cambridge, MA, USA) and anti-CD8a (1:200, ab237723, Abcam) overnight at 4°C, followed by PBS wash and secondary antibody (1:5000, Aspen, Wuhan, Hubei, China) incubation at room temperature for 1 h. 3,3'-Diaminobenzidine (DAB; Aspen) was then used for staining, and hematoxylin (Aspen) was used for counter staining. The slides were visualized using a NanoZoomer S360 (Hamamatsu, Japan). The number of CD3+ and CD8+ cells was counted manually in each 10 × 20 field for multiple randomly selected fields. All antibodies and diluted concentrations are listed in the Supplementary Table S1.
2.5 Flow cytometry analysis
Tumors were harvested and prepared as single-cell suspensions using a tumor dissociation kit (130-096-730; Miltenyi Biotec, San Diego, CA, USA) and gentleMACS Dissociator (Miltenyi Biotec) following the manufacturer's protocols. Red blood cells (RBCs) were removed using RBC lysis (Solarbio Life Sciences, Beijing, China). FVS780 (565388, 1:1000; BD Biosciences, Franklin Lakes, NJ, USA) was used for live/dead staining for 5 min, followed by PBS washing and cell marker staining for 1 h. Cells were then washed with PBS and detected using a CytoFLEX LX cytometer (Beckman Coulter, Pasadena, CA, USA). Gating was performed using FlowJo 10.3 (FlowJo LLC, Ashland, OR, USA), and the gating strategies for TILs and DCs are demonstrated in Supplementary Figure S1. Briefly, CD45-PE (1:100), CD3e-BV510 (1:100), CD4-BV421 (1:100), and CD8a-FITC (1:100) were used for tumor-infiltrating T cell staining. CD45-PE (1:100), CD11c-PerCP-Cy5.5 (1:100), I-A/I-E-Alexa Fluor 647 (1:50), CD8a-FITC (1:100), and CD86-PE-Cy7 (1:100) were used for mature DC and tumor-infiltrating DC staining. CD16/32 (1:100) was used for blocking. All the antibodies, their suppliers, and the clone numbers used for flow cytometry are listed in Supplementary Table S1.
2.6 Cell viability assay
Cell viability was evaluated using the Cell Counting Kit-8 (CCK-8, MedChemExpress). Briefly, 5 × 103 cells per well were disseminated onto the 96-well plates. Twenty-four hours later, ATRi was delivered at concentrations from 0 µmol/L to 2 µmol/L, followed by 5 Gy irradiation. Twenty-four hours after IR, cells were incubated with 10% CCK-8 at 37°C for subsequent measurement of absorbance at 450 nm using a microplate reader (Synergy2, BioTek, Winooski, VT, USA).
2.7 Cell cycle analysis
ATRi (1 µmol/L) was added 2 h before IR and remained within the medium until harvest. Cells were harvested 0 h, 4 h, 8 h, and 12 h after IR, fixed using 70% ethanol overnight. The next day, cells were washed with ice-cold PBS and then incubated with RNase (Beyotime, 50 µg/mL) at 37°C for 30 min, followed by Propidium Iodide (PI, 50 µg/mL) staining at 4°C for 30 min. The DNA contents of the cells were analyzed using a CytoFLEX LX cytometer (Beckman Coulter). Phospho-histone H3-Alexa Fluor 488 (1:100, #3465, Cell Singling Technology, Beverly, MA, USA) staining was performed following instructions described by Shen et al. [24] and detected using the CytoFLEX LX cytometer.
2.8 Immunofluorescence microscopy and micronuclei assay
Cells were seeded into a 24-well plate containing coverslips before receiving IR. Twelve hours after 5 Gy radiotherapy, the cells were harvested, fixed with 1% paraformaldehyde, permeabilized with 0.5% Triton, blocked with 5% BSA and then incubated with primary antibodies at 4°C overnight. The primary antibodies were anti-double stranded DNA (dsDNA; 1:200, ab27156, Abcam), anti-cGAS (1:200, #79978, Cell Singling Technology) and anti-NF-κB P65 (1:200, A2547, ABclonal, Wuhan, Hubei, China). The next day, the cells were incubated with secondary antibodies for 1 h at room temperature followed by 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI; Abbkine, Beijing, China) staining for 5 min. The secondary antibodies were Cy3, goat anti-rabbit IgG (1:500, A22220, Abbkine, Beijing, China), DyeLight 488 goat anti-mouse IgG (1:500, A23240, Abbkine), and DyeLight 488 goat anti-rabbit IgG (1:500, A23220, Abbkine). Cover slides were then mounted for microscopy. For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay, sections were stained using the TUNEL staining kit (11684817910, Roche, Germany) according to the manufacturer's protocols. For the micronuclei assay, cells were stained with DAPI with antifade reagent for microscopy and scoring. At least 150 nuclei were counted in 10 fields.
2.9 dsDNA quantification
dsDNA quantification was performed using Quant-iT Pico-Green dsDNA reagent and kits (P11496, ThermoFisher) following the manufacturer's instructions. Briefly, cells were harvested 12 h after IR, seeded into a 96-well plate and incubated with Picogreen for 20 min at room temperature. Then, the fluorescence of each well was detected using a microplate reader (Synergy2) and calculated into dsDNA concentrations based on the standard curve.
2.10 Immunoblotting
Total protein was extracted using Radio Immunoprecipitation Assay (RIPA) lysis buffer (Beyotime, Wuhan, Hubei, China) supplemented with phenylmethylsulfonyl fluoride (PMSF) and phosphatase inhibitor cocktail (HY-K0021, MedChemExpress). Immunoblotting was performed using a BioRad Gel System (Hercules, CA, USA). The proteins were loaded into 10% Sodium Dodecyl Sulfate PolyAcrylamide Gel Electrophoresis (SDS-PAGE) gels, followed by transfer of the proteins to 0.45 µm polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). BSA was used for blocking for 1 h, and the membrane was incubated with primary antibodies overnight (4°C). The membranes were then incubated with secondary antibodies for 1 h at room temperature. Subsequently, the immunoblots were analyzed using SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher) in a BOX Chemi X system (Syngene, Cambridge, UK). The antibodies anti-histone H2A.X (1:1000, #7631), anti-phospho-Checkpoint kinase 1 (CHK1)-Ser345 (1:1000, #2348), anti- TANK binding kinase 1 (TBK1)-Ser172 (1:1000, #5483), anti-TBK1 (1:1000, #3504), anti-phosho-interferon regulatory factor 3 (IRF3)-Ser396 (1:1000, #29047), anti-IRF3(1:1000, #4302), anti-phoshpho-stimulator of interferon genes (STING)-Ser365 (1:1000, #72971), anti-STING (1:1000, #50490), and anti-phospho-signal transducer and activator of transcription 1 (STAT1)-Tyr701 (1:1000, #9167) were purchased from Cell Singling Technology. The primary antibodies anti-CHK1 (1:1000, ab32531) and anti-SHP1 (1:1000, ab227503) were purchased from Abcam. The antibodies anti-p65 (1:1000, A19653), anti-TNF receptor associated factor 6 (TRAF6; 1:1000, A0973), and anti-small ubiquitin like modifier 1 (SUMO1; 1:1000, A19121), anti-signal transducer and activator of transcription 3 (STAT3; 1:1000, A19566), and secondary antibodies HRP-conjugated goat anti-mouse IgG (1:10,000, AS003) and HRP-conjugated goat anti-rabbit IgG (1:10,000, AS014) were from Abclonal. Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:10,000, 1E6D9) was purchased from Proteintech (Wuhan, Hubei, China). All antibodies and their applications are listed in Supplementary Table S1.
2.11 RNA isolation, reverse transcription, and real-time qPCR (RT-qPCR)
RNA was isolated using the FastPure Cell/Tissue Total RNA Isolation Kit V2 (#RC112, Vanzyme, Nanjing, Jiangsu, China) following the manufacturer's protocols. For reverse transcription, PrimeScript RT Master Mix (#RR036A, Takara, Kusatsu, Japan) was used. RT-qPCR was performed using ChamQ Universal SYBR qPCR Master Mix (#Q711-02, Vanzyme) in the ABI-7900HT Sequence Quantification System (Applied Biosystems, Foster, CA, USA). 2−ΔΔCT was calculated as the relative level of mRNA expression. Primer sequences are listed in Supplementary Table S2.
2.12 Cell transfection
The plasmids expressing Flag-Negative control (Flag-NC), Flag-STING, Flag-SHP1, and Flag-SHP1-ΔK127R-mutant and the virus shSTING, shSting, and scramble viruses were purchased from GeneChem (Shanghai, China). The RNAi sequences used in viruses are listed in Supplementary Table S3. The knockdown efficiency was evaluated using immunoblotting. HCT116 cells transiently expressing Flag-NC (control vector), Flag-STING, Flag-SHP1, or Flag-SHP1-ΔK127R-mutant were obtained by transfection of the indicated plasmids using Lipo3000 (ThermoFisher) following the manufacturer's protocols. Viral particles and polybrene (6 µg/ml, GeneChem) were mixed with cell culture medium and added to HCT116 or CT26 cells to establish stable cells with negative control (shNC), shSTING-HCT116 cells and shSting-CT26 cells. Puromycin (10 µg/mL, GeneChem) was used to screen cells with successful transfection.
2.13 Co-immunoprecipitation
Cells lysates were prepared by using NP40 lysis buffer (Beyotime) containing PMSF and phosphatase inhibitor cocktail. Anti-SHP1 (1:100) or anti-Flag (1:100, Sigma, St. Louis, MO, USA) were pre-mixed with protein A/G magnetic beads (HY-K0202, MedChemExpress) for 2 h at 4°C. Cell lysis and magnetic beads were then mixed for 4 h at 4°C, followed by thorough washing and subsequent elution according to the manufacturer's protocols. Samples were then detected with indicated antibodies by immunoblotting.
2.14 Liquid Chromatography Coupled to Tandem Mass Spectrometry (LC-MS/MS)
HCT116 cells were treated with 1 µmol/L ATRi 2 h before IR and remained in the medium until harvest. Twelve hours after radiotherapy, HCT116 cells were lysed, and proteins were harvested for immunoprecipitation using anti-SHP1 (1:100) and subsequent mass spectrometry. LC-MS/MS data acquisition was carried out on a Q Exactive Plus mass spectrometer (ThermoFisher) coupled with an EASY-nLC 1200 system (ThermoFisher) following the manufacturer's protocols. Raw MS data were processed with MaxQuant software (V1.6.6, Munich, Germany) according to the Andromeda database search algorithm, and the spectra files were searched according to the UniProt human proteome database (https://www.uniprot.org/). False discovery rate was set at 1% for filtering search results at both protein and peptide levels.
2.15 Prediction of SUMOylation sites
Three online tools were used to predict the consensus SUOMylation sites: SUMOplotTM Analysis Program (https://www.abcepta.com.cn/sumoplot), GPS-SUMO [25] (https://sumo.biocuckoo.cn/) and JASSA [26] (http://www.jassa.fr/) The protein sequence of SHP1 (P29350) was downloaded from the UniProt human proteome database (https://www.uniprot.org/) and submitted for online prediction.
2.16 RNA sequencing
CT26 tumors were harvested three days after IR. Total RNA isolation was performed using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and library preparation was performed using VAHTS Stranded mRNA-seq Library Prep Kit for Illumina V2 (NR612-01; Vanzyme). Briefly, mRNA was purified from total RNA, fragmented, and cDNA were obtained by two rounds of reverse transcription according to the protocol provided. Adapter ligation and size selection were then performed, followed by amplification of the cDNA library. Sequencing was performed on the Illumina HiSeq platform (San Diego, CA, USA). Library preparation, sequencing, and data processing were accomplished by GENEWIZ (South Plainfield, NJ, USA). Differentially expressed genes (DEGs) were set when the gene expression in the two compared groups was of >1.5-fold difference with an adjusted P value <0.05. DEGs with the log2FoldChange were used through the R package clusterProfiler for Gene ontology (GO) analysis and Gene-set Enrichment Analysis (GSEA), and the results were visualized through the R package ggplot2.
2.17 Statistical analyses
Statistical graphs and analysis were generated and performed using GraphPad Prism 8.0 (San Diego, CA, USA) or R software (version 4.1.2, https://www.r-project.org/). Comparisons of multiple groups were performed by using one-way Analysis of Variance (ANOVA) or two-way ANOVA (group with magnitude change) with Tukey's post hoc test. Comparison between only two groups was performed using unpaired t test or unpaired t test with Welch correction if variances were significantly different. Survival analysis of tumor-bearing mice was performed using the Kaplan-Meier method and compared by the log-rank test. Statistical significance was shown as *P < 0.05, **P < 0.01, and ***P < 0.001.
3 RESULTS
3.1 IR + ATRi enhanced T cell infiltration in CRC mouse models
The ATRi berzosertib has been shown to elicit an immune-modulatory effect in the TC-1 pancreatic mouse model [22, 27]. To assess the effect of the ATRi on CRC, we used two syngeneic tumors MC38 (dMMR, high immunogenicity) [28] and CT26 (pMMR, low immunogenicity) [29] with different microsatellite statuses and evaluated the tumor microenvironment after berzosertib treatments (Figure 1A). Compared to IR alone, the combination of IR and ATRi (IR + ATRi) showed delayed tumor growth (Figure 1B). IHC analysis revealed significant increase of CD3+ and CD8+ cells in both MC38 (Figure 1C-D) and CT26 tumors (Figure 1E-F) in mice treated with IR + ATRi. These results were confirmed by flow cytometry analysis, with the combination therapy exhibiting the highest proportions of CD45+CD3+ T cells and CD8+CD4− T cells and decreased proportion of CD4+CD8− T cells (Figure 1G-J). Taken together, we found that IR + ATRi was able to elicit a favorable immune profile with enhanced TILs in CRC with different immunogenicity statuses.
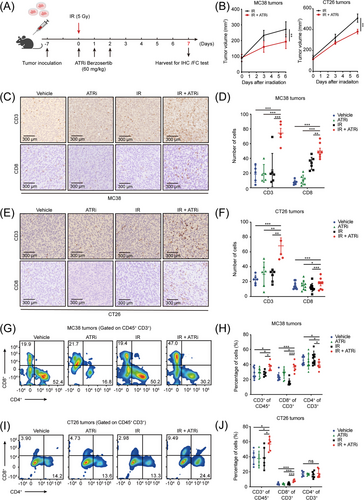
IR +ATRi increases TILs in MC38 and CT26 tumors (A) Treatment schedule for IHC and FC analysis. (B) Tumor volume curve of MC38 and CT26 tumors treated with IR or IR + ATRi. (C-D) Representative IHC images of CD3 and CD8 staining (C) and statistical analysis (D) of MC38 tumors. (E-F) Representative IHC images of CD3 and CD8 staining (E) and statistical analysis (F) of CT26 tumors. (G-H) Representative flow cytometry images (G) and quantitative analysis (H) of TILs in MC38 tumors. (I-J) Representative flow cytometry images (I) and quantitative analysis (J) of TILs in CT26 tumors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Abbreviations: IR, radiation; IHC, immunohistochemistry; FC, flow cytometry; TIL, tumor-infiltrating lymphocyte.
3.2 ATRi sensitizes CRC cells to IR and promotes mitotic progression after IR
For a comprehensive understanding of the mechanisms underlying the pro-inflammatory effect induced by IR + ATRi, we initially investigated ATRi's impact on cell proliferation, DNA damage and cell cycle distribution. As expected, IR + ATRi reduced the proliferation of multiple CRC cell lines, including human CRC cell lines HCT116 and SW480 and mouse CRC cell lines CT26 and MC38, indicating the radio-sensitization effect of ATRi (Figure 2A). In response to IR-induced DNA damage, ATR kinase is activated and promotes G2/M phase arrest to facilitate repair of DNA damage. Immunoblotting and immunofluorescence analysis confirmed the role of ATRi in DNA damage repair, as IR + ATRi treatment resulted in highest levels of γH2AX expression and γH2AX foci (Figure 2B, Supplementary Figure S2). Meanwhile, pCHK1-serine 345 was extensively abrogated after treatment with ATRi or IR + ATRi, indicating the on-target effect of the inhibitor (Figure 2B, Supplementary Figure S3). Subsequent cell cycle distribution analysis revealed sharp increases 8 h and 12 h after IR, indicating IR-induced G2/M phase arrest. In contrast, the accumulation of cells in G2/M phase was remarkably abrogated in HCT116 and CT26 cells after IR + ATRi treatment (Figure 2C-D). The effect was confirmed with IR + ATRi exhibiting increased phospho-histone H3-positive (pH3+) cells 12 h after IR (Figure 2E-F). Histone H3 is specifically phosphorylated during mitosis, indicating that ATRi disrupts the cell cycle checkpoint to promote mitotic entry after DNA damage. Combining these results, it can be concluded that ATRi impaired G2/M checkpoint initiation and maintenance.
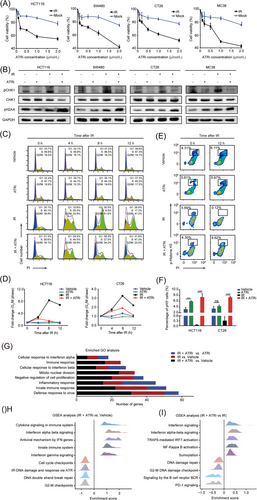
ATRi sensitizes CRC cells to IR, promotes mitotic reentry, and enhances innate immune signaling (A) Cell viability assay of multiple CRC cell lines treated with ATRi and IR +ATRi. (B) Immunoblot analysis of γH2AX and pCHK1 in multiple CRC cell lines treated with different regimens. (C-D) Cell cycle distribution analysis of HCT116 and CT26 cells treated with IR and IR +ATRi. (E-F) Representative of flow cytometry images (E) and quantitative analysis (F) of phospho-histone H3+ cells in HCT116 and CT26 cells. (G) Number of DEGs in GO analysis of the RNA-seq results. (H) GSEA of DEGs in the IR +ATRi group compared to the vehicle group. (I) GSEA of DEGs in the IR +ATRi group compared to the IR group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Abbreviations: CRC, colorectal cancer; IR, radiation; CHK1, checkpoint kinase 1; pH3, phospho-histone H3; RNA-seq, RNA-sequencing; DEG, differentially expressed gene; GO, gene ontology; GSEA, gene-set enrichment analysis.
3.3 RNA-seq revealed enhancement of innate immune signaling following IR + ATRi treatment
Next, CT26 tumors under different regimens were harvested for RNA-seq analysis for a comprehensive understanding of the in vivo effect of IR + ATRi. DEGs were evaluated based on the functional classification of GO terms. Compared to the vehicle control, mice treated with IR and the combination therapy had the most profound changes in “defense response to virus”, “innate immune response” and “inflammatory response” processes, suggesting alteration of the innate immune signaling induced by IR + ATRi (Figure 2G). GSEA also verified interferon-related signaling activation, including interferon (IFN)-alpha beta and IFN-gamma signaling, as well as DNA damage response downregulation via ATR kinase (Figure 2H). Consistently, IR + ATRi treatment also exhibited a gene signature with enhanced interferon signaling, Nuclear Factor-κB (NF-κB) activation and TRAF6-mediated interferon regulatory factor 7 (IRF7) activation compared to IR (Figure 2I). Taken together, the transcriptome results demonstrated profound alterations in innate immune signaling mediated by IR + ATRi.
3.4 ATRi potentiates IR-induced cGAS-STING-pTBK1/pIRF3 pathway
Recent studies have reported that cell cycle-specific DNA damage is required for canonical cGAS-STING activation by induction of micronuclei and the STING signaling is essential in innate immune activation against tumors [30, 31]. We hypothesized that IR + ATRi might increase micronuclei formation as ATRi promoted unexpected mitotic reentry before DNA damage was efficiently repaired (Figure 3A). This was confirmed by IF analysis, with increased micronucleated cells and cGAS localization to micronuclei following IR + ATRi treatment (Figure 3B-C, Supplemenatry Figure S4). dsDNA quantification assay also confirmed higher levels of dsDNA after the IR + ATRi treatment in both HCT116 and CT26 cells (Figure 3D). After recognizing cytosolic dsDNAs, cyclin GMP-AMP (cGAMP) is produced and then activates STING [32], which in turn phosphorylates TANK-binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3) [20, 33]. As expected, the most robust activation of pTBK1 and pIRF3 expression was observed after IR + ATRi treatment in multiple CRC cell lines, indicating strong activation of the cGAS-STING-pTBK1-pIRF3 axis (Figure 3E, Supplementary Figure S5). A similar effect was observed in vivo, with CT26 tumors derived from syngeneic mice in the IR + ATRi group showing the highest levels of pTBK1 and pSTING (Figure 3F) and increased fluorescence of pIRF3 foci (Figure 3G).
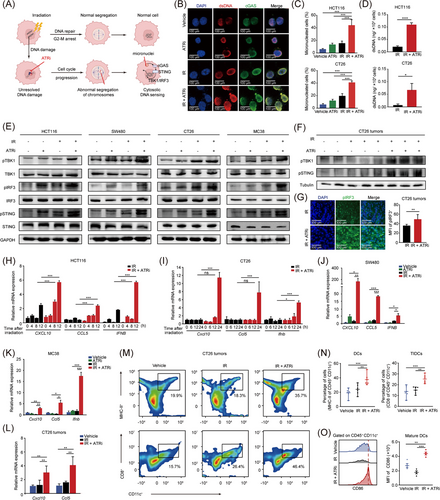
IR +ATRi activates the canonical cGAS-STING signaling and promotes innate immune activation (A) Schematic diagram of micronuclei formation after IR-induced cell cycle checkpoint and DNA repair are disrupted by ATRi. (B) IF images of dsDNA and cGAS staining in HCT116 cells. (C) Quantification of micronucleated cells in HCT116 cells and CT26 cells. (D) dsDNA quantification in HCT116 and CT26 cells after treatment with IR or IR + ATRi. (E) Immunoblot of the key proteins from the canonical cGAS-STING axis in multiple CRC cell lines. (F) Immunoblot of pTBK1 and pSTING from CT26 tumors. (G) Representative images of pIRF3 staining in CT26 tumors and quantification of pIRF3 MFI. (H-I) Relative expression levels of the CXCL10, CCL5 and IFNB genes at different time points in HCT116 (H) and CT26 cells (I). (J-K) Relative expression levels of CXCL10, CCL5 and IFNB in SW480 (J) and MC38 (K) cells after different treatments. (L) Relative expression levels of Cxcl10, Ccl5 in CT26 tumors. (M-N) Representative images (M) and statistical analysis of DCs and tumor infiltrating-DCs (N) from flow cytometry analysis of CT26 tumors. (O) Representative images of CD86 staining and statistical analysis of CD86 MFI in CT26 tumors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Abbreviations: IR, radiation; cGAS, cyclic GMP-AMP synthase; STING, stimulator of interferon genes; TBK1, TANK binding kinase 1; IRF3, interferon regulatory factor 3; dsDNA, double stranded DNA; IF, immunofluorescence; MFI, mean fluorescence intensity; CXCL10, C-X-C motif chemokine ligand 10; CCL5, C-C motif chemokine ligand 5; IFNB, interferon beta 1; DC, dendritic cell; TIDC, tumor-infiltrating dendritic cell.
3.5 IR + ATRi induces activation of innate immune signaling
The cGAS-STING axis promotes the induction of innate immune genes, including the interferon stimulated genes (ISGs) C-X-C motif chemokine ligand 10 (CXCL10) and C-C motif chemokine ligand 5 (CCL5) and the type I IFN gene Interferon Beta 1 (IFNB) [20]. Consistently, compared to IR alone, IR + ATRi caused a sharp increase in the mRNA levels of CXCL10, CCL5, and IFNB in HCT116 and CT26 cells (Figure 3H-I), which was confirmed in other human (SW480) and mouse (MC38) cell lines (Figure 3J-K). in vivo activation of type I IFN signaling also confirmed the effect in CT26 tumors (Figure 3L). STING-related production of type I IFN was demonstrated to promote innate immune cell maturation, such as DCs, to facilitate adaptive immune response against tumors [34, 35]. Consistent with the effect, IR + ATRi improved levels of CD11c+ MHC-II+ DCs and tumor-infiltrating DCs (TIDCs, CD11c+ CD8+ DCs) (Figure 3M-N) and stimulated the maturation marker CD86 within the CT26 tumors (Figure 3O). Taken together, these results demonstrated that IR + ATRi induced robust activation of type I interferon signaling and innate immune signaling.
3.6 IR + ATRi potentiates non-canonical STING activation by inhibiting SHP1-TRAF6/STING interaction
Multiple DNA damage agents, including ATM inhibitors, etoposide, platinum-based therapy, and IR, have been proven to induce the non-canonical STING activation, which is also indispensable in the anti-tumor innate immune activation [21, 36, 37]. To fully understand the ATRi's impact on the STING-mediated innate immune signaling, we next evaluated non-canonical STING axis under different treatment regimens. In multiple CRC cell lines, IR + ATRi improved pP65 activation, a non-canonical STING singling marker required for innate immune gene production, and upregulated pSTAT1 activation (Figure 4A, Supplementary Figure S6). IF analysis also confirmed the effect with a marked increase in p65 translocation to the nucleus following IR + ATRi treatment in HCT116 cells and HT29 cells (Figure 4B, Supplementary Figure S7). These results indicate strong activation of non-canonical STING signaling after treatment with IR + ATRi.
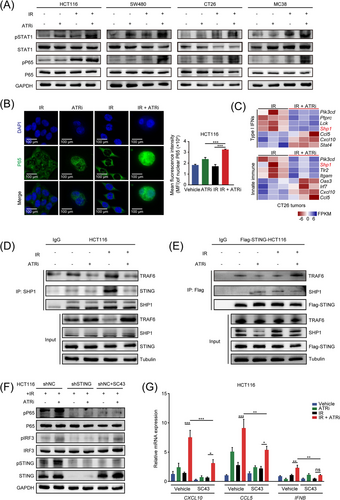
IR +ATRi activates the non-canonical STING-p65 signaling by inhibiting SHP1 (A) Immunoblot analysis of pSTAT1 and pP65 in multiple CRC cell lines after treatments. () Representative IF images of P65 staining in HCT116 cells and quantitative analysis of nuclear P65 MFI in HCT116 cells after treatments. (C) Heatmap of genes related to type I interferon signaling and innate immune signaling from RNA-seq results. (D) Co-IP analysis of SHP1-interacted STING and TRAF6 under different treatment regimens. (E) Co-IP analysis of STING-interacted SHP1 and TRAF6 under different treatment regimens. (F) Immunoblot analysis of pP65, pIRF3 and pSTING after SHP1 agonist SC43 treatment in HCT116 and STING-knockdown HCT116 cells. (G) Relative expression levels of CXCL10, CCL5 and IFNB after treatment with SHP1 agonist SC43 in HCT116 and STING-knockdown HCT116 cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Abbreviations: IR, radiation; STAT1, signal transducer and activator of transcription 1; MFI, mean fluorescence intensity; SHP1, src homology 2 (SH2) domain containing protein tyrosine phosphatase-1; IF, immunofluorescence; RNA-seq, RNA sequencing; IP, immunoprecipitation; co-IP, co-immunoprecipitation; TRAF6, TNF receptor associated factor 6; STING, stimulator of interferon genes; IRF3, interferon regulatory factor 3; CXCL10, C-X-C motif chemokine ligand 10; CCL5, C-C motif chemokine ligand 5; IFNB, interferon beta 1; NC, negative control.
Next, we sought to determine the mechanism of non-canonical activation of STING signaling induced by IR + ATRi. Surprisingly, the analysis of the transcriptome data revealed decreased mRNA levels of Shp1 (also known as protein tyrosine phosphatase non-receptor type 6, Ptpn6), which was involved in both type I IFN signaling and innate immune signaling (Figure 4C). This led us to explore the role of SHP1 in ATRi-related STING axis activation. A recent study reported that SHP1 was specifically activated upon HIV infection and dephosphorylated TRAF6 and STING and inhibited their ability to induce antiviral immunity [38], but how SHP1 affects tumor-specific STING activation has not been described. Thus, we sought to determine whether IR + ATRi affects the non-canonical STING axis by inhibiting SHP1. Surprisingly, co-IP analysis revealed SHP1-STING and SHP1-TRAF6 interactions decreased after IR + ATRi treatment (Figure 4D-E). Evidence existed that the TRAF6-STING-TBK1 complex assembly plays a crucial role in regulating the recruitment of P65 and IRF3 activation [37]. Consistently, we observed higher levels of STING-TRAF6 interaction after IR + ATRi treatment (Figure 4E), indicating strong activation of the TRAF6-STING axis. Next, we used an SHP1 agonist, SC43, and STING-knockdown HCT116 cells to identify the impact of SHP1 on the activation of non-canonical STING induced by IR + ATRi. As expected, STING knockdown fully abrogated pSTING, pIRF3, and pP65 activation. SC43 largely reduced pP65 activation, while the canonical pIRF3 and overall pSTING activation were only partially inhibited (Figure 4F, Supplementary Figure S8). Similarly, the upregulation of CXCL10, CCL5, and IFNB was abrogated by adding SC43 (Figure 4G, Supplementary Figure S9). These results demonstrated that IR + ATRi inhibited the recruitment of SHP1 to the TRAF6/STING complex to facilitate the activation of the non-canonical STING axis.
3.7 IR + ATRi inhibited SHP1 function by promoting SUMOylation of SHP1 at lysine 127
Next, we conducted mass spectrometry to investigate the protein changes binding to SHP1 after IR + ATRi to understand the mechanism by which ATRi inhibited the SHP1-TRAF6/STING interaction. Surprisingly, SHP1-conjugated small ubiquitin-like (SUMO)-related proteins, including E1-ligase SUMO1 activating enzyme subunit 2 (SAE2), E2-ligase ubiquitin conjugating enzyme E2 I (UBC9), E3-ligase zinc finger protein 451 (ZNF451), and SUMO1, were found increased following IR + ATRi treatment (Figure 5A). These results may indicate that IR + ATRi could increase the SUMOylation of SHP1. By transfection of Flag-SHP1 plasmids and subsequent co-IP analysis, we confirmed increased SUMO1-SHP1 interaction after IR + ATRi, compared to IR alone (Figure 5B). For further investigation, we used three online prediction tools, SUMOplotTM Analysis program (Abcepta), GPS-SUMO and JASSA and found lysine 127 (K127) as one consensus SUMOylation site, which was predicted as a common SUMO conjugation site from the results of all three tools (Figure 5C). To confirm the prediction, we mutated lysine 127 to arginine and generated an HCT116-mut cell line (ΔK127R-Mut). IP analysis showed decreased SUMO1 conjugation in SHP1-ΔK127R-mut HCT116 cells compared to SHP1-wildtype (SHP1-WT) cells. Consistently, SHP1-ΔK127R-mut reversed the decreasing SHP1-TRAF6/STING interaction induced by IR + ATRi (Figure 5D). Notably, we also found that the signal transducer and activator of transcription 3 (STAT3), a generally acknowledged substrate of SHP1 [39], was increased in SHP1-ΔK127R-mut cells, indicative of enhanced phosphatase activity when SUMOylation of SHP1 was blocked. ΔK127R-mut also abrogated the protein levels of pP65 and pIRF3 (Figure 5E, Supplementary Figure S10) and the mRNA expression of CXCL10 and CCL5 (Figure 5F), similar to previous results using the SHP1 agonist SC43. These results demonstrate that IR + ATRi promotes SUMOylation of SHP1 at K127 to inhibit its activity, thus contributing to robust STING pathway activation.
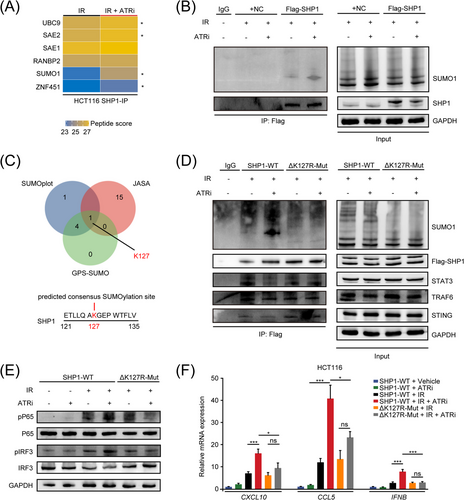
IR + ATRi inhibits SHP1 function via promoting the SUMOylation of SHP1 at serine 127 (A) Heatmap of SHP1-conjugated proteins in HCT116 cells from mass spectrometry data. *, differently altered proteins. (B) SUMOylation analysis of SHP1 in HCT116 cells after treatment with IR + ATRi. (C) Venn diagram of common SUMOylation sites predicted by SUMOplot analysis program, JASA, and GPS-SUMO. (D) Co-IP analysis of SUMO1 conjugation and the SHP1 phosphatase substrates STAT3, TRAF6 and STING in SHP1-WT and SHP1-K127R-Mut HCT116 cells. (E) Immunoblot analysis of key STING signaling proteins in SHP1-WT and SHP1-K127R-Mut HCT116 cells under different treatments. (F) Relative mRNA levels of CXCL10, CCL5 and IFNB in SHP1-WT and SHP1-K127R-Mut HCT116 cells under different treatment different regimens. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Abbreviations: IR, radiation; SHP1, src homology 2 (SH2) domain containing protein tyrosine phosphatase-1; IP, immunoprecipitation; SUMO1, small ubiquitin like modifier 1; STAT3, signal transducer and activator of transcription 3; TRAF6, TNF receptor associated factor 6; STING, stimulator of interferon genes; IRF3, interferon regulatory factor 3; CXCL10, C-X-C motif chemokine ligand 10; CCL5, C-C motif chemokine ligand 5; IFNB, Interferon Beta 1, WT, wild type.
3.8 ATRi enhances the efficacy of combined IR and PD-L1 blockade therapy in CT26 and MC38 tumors
Innate immune system activation induced by STING signaling may cooperate with ICIs to enhance the antitumor effect [35]. Therefore, we next evaluated the antitumor efficacy of the combined ATRi, IR, and anti-PD-L1 therapy (Figure 6A). In both CT26 tumors and MC38 tumors, triple therapy using IR + ATRi + anti-PD-L1 dramatically delayed tumor growth compared to dual therapy and monotherapy regimens (Figure 6B-C). Survival analysis showed that IR + ATRi + anti-PD-L1 largely extended the survival of mice compared to any other combinational treatment (Figure 6D). Notably, complete tumor regression was observed in mice treated with IR + ATRi + anti-PD-L1 (Figure 6E). Bioluminescence imaging of MC38 tumors at the 50-day endpoint demonstrated reduced tumor burden in the triple combination therapy group (Figure 6F-G), indicating optimal tumor control by the triple therapy. For further investigation, MC38 tumors were isolated for Ki67 and TUNEL staining. The triple combination group exhibited lowest population of Ki67-positive cells (Supplementary Figure S11A-B) and highest expression of TUNEL (Supplementary Figure S11C-D) compared to other groups. These results confirmed the optimal anti-tumor effect induced by the triple therapy.
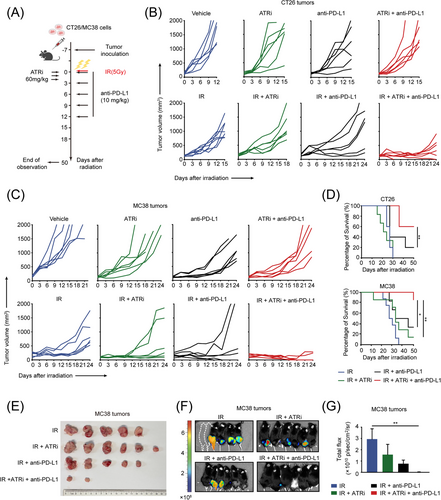
IR +ATRi +anti-PD-L1 exhibits optimal antitumor efficacy and prolonged survival in CT26 and MC38 tumors (A) Treatment schedule for the subcutaneous tumor models. (B) Tumor volume curve of CT26 tumors treated with IR (n = 6), ATRi (n = 6), anti-PD-L1 (n = 6), or combination therapies (n = 6 in each group). (C) Tumor volume curve of MC38 tumors treated with IR (n = 6), ATRi (n = 6), anti-PD-L1 (n = 6), or combination therapies (n = 6 in each group). (D) Survival analysis of mice bearing CT26 (n = 6) or MC38 tumors (n = 6). (E) Views of tumors derived from the flanks of MC38-tumor bearing mice (n = 6). (F-G) Bioluminescence imaging of MC38 tumors (F; n = 6) and statistical analysis (G; n = 6) of total flux of bioluminescence imaging. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Abbreviations: IR, radiation; PD-L1, programmed death-ligand 1.
To assess the requirement of STING in the combinational plan, we next stably knocked down Sting expression in CT26 cells, confirmed by immunoblotting analysis (Supplementary Figure S12A). Consistent with previous results, Sting knockdown remarkably abrogated the upregulation of type I IFNs after IR + ATRi treatment (Supplementary Figure S12B) as well as CD8 cell infiltration (Supplementary Figure S12C). These data suggest that the immunomodulatory effect of IR + ATRi is STING-dependent. For further investigation, shSting and shNC tumors were treated with triple combination therapy. As expected, Sting depletion completely impaired the antitumor efficacy of triple therapy (Supplementary Figure S12D). Notably, CD8 depletion using anti-CD8 antibody also dramatically abrogated the antitumor efficacy (Supplementary Figure S12E-F), confirming the requirement of CD8 cytotoxicity in the combinational plan. From these results, we concluded the indispensable role of STING and cytotoxic CD8+ cells in the antitumor efficacy of IR + ATRi + anti-PD-L1.
4 DISCUSSION
In the present study, we uncovered a predominant role of ATRi in the activation of STING-mediated innate immune signaling. IR + ATRi boosts the canonical and non-canonical activation of the STING axis. Canonically, ATRi promoted DNA damage, abrogated IR-induced G2/M arrest and promoted mitotic re-entry, leading to accumulation of cytosolic DNA and subsequent cGAS-STING-pTBK1/pIRF3 axis activation. Non-canonically, IR + ATRi promoted SUMOylation of SHP1 at lysine 127, inhibiting its recruitment to TRAF6 and STING, leading to higher levels of NF-κB-P65 activation. Both axes contribute to the innate immune activation induced by IR + ATRi, which was then exploited by adding anti-PD-L1 therapy to the combination of ATRi and IR for an optimal antitumor effect (Figure 7). The study raised a new combination strategy to enhance ICI efficacy in CRC.
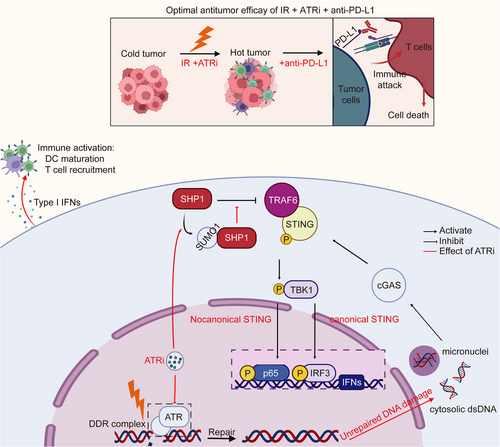
Abstract figure showing the antitumor efficacy of IR +ATRi +anti-PD-L1 and the mechanism
IR + ATRi activates the canonical cGAS-STING-pTBK1-pIRF3 axis by inducing of cytosolic dsDNA, and the non-canonical axis by abrogating SHP1-mediated TRAF6/STING-p65 complex inhibition, through SUMOylation of SHP1. By boosting the STING-type I interferon signaling, IR + ATRi promotes immune activation, turning the cold tumor hot, and in turn facilitate the efficacy of anti-PD-L1 therapy. The red lines indicate the effects of ATRi.
Abbreviations: IR, radiation; PD-L1, programmed death-ligand 1; SHP1, src homology 2 (SH2) domain containing protein tyrosine phosphatase-1; IP, immunoprecipitation; SUMO1, small ubiquitin like modifier 1; TRAF6, TNF Receptor Associated Factor 6; STING, stimulator of interferon genes; TBK1, TANK Binding Kinase 1; IRF3, interferon regulatory factor 3; DDR, DNA damage response; dsDNA, double stranded DNA; DDR, DNA damage response; cGAS, cyclic GMP-AMP synthase; IFN, interferon.
IR plays a potent role in inducing antitumor immunity in addition to producing lethal damage to kill tumors [40, 41]. Hence, many clinical trials are ongoing investigating the efficacy of combining IR with immune-based therapy [42]. A growing body of evidence has shown the pro-inflammatory role of IR in shaping innate immune activation against tumors, primarily by induction of the canonical cGAS-STING-mediated DNA sensing [19, 20, 43]. ATR kinase plays an upper-stream role in response to IR-induced DNA damage, making it a promising target in combination with radiotherapy [44]. A recent report found that IR + ATRi could induce in vivo activation of STING signaling, and combining another ATRi, celarasertib, with IR and anti-PD-L1 successfully induced optimal antitumor in hepatocellular carcinoma, but the mechanism remains to be investigated [45]. While our study tested the similar effect in the context of CRC, we performed in-depth investigations of the molecular basis of the type I interferon signaling induced by IR + ATRi. Our results revealed that inhibition of ATR in the setting of radiotherapy led to robust activation of the STING signaling, and thus higher levels of innate immune activation and TIL infiltration.
While the canonical STING activation has been extensively studied in the context of DNA damage, the non-canonical STING activation, which pivots on the recruitment of NF-κB-p65 by the TRAF6/STING/TBK1 complex to facilitate the expression of p65-related inflammatory genes [46], is less defined. Recent studies have uncovered the role of ATM in non-canonical STING signaling, which provides therapeutic potential in combination with IR and ICIs [21, 37]. Regarding these reports, we explored the role of ATRi in inducing non-canonical STING activation. Our findings suggest that the ability of ATRi to induce STING-p65 activation was largely dependent on its inhibitory effect on SHP1, a dominant inhibitor controlling TRAF6/STING assembly, which was only defined in the context of viral infection [38]. Further analysis revealed that IR + ATRi enhanced the SUMOylation of SHP1 to inhibit its phosphatase activity, which also partly explained previous findings that IR + ATRi promoted RNA-sensing pathway activation because SHP1 is a suppressor of RIG-1 mediated RNA-sensing signaling [47, 48]. However, the mechanism underlying the induction of SHP1 SUMOylation, for example, whether there is an E3-ligase specifically targeting SHP1 for SUMOylation and how ATRi affects the SUMOylation process of SHP1, remains to be investigated. One possible explanation is that ATRi may enhance the SUMOylation machinery because ATRi were reported to induce SUMOylation of a subset of proteins [49], which was also confirmed in the GSEA analysis of our transcriptome data. While we did not directly show the direct effect of SUMOylation at K127 on SHP1, we found increased SHP1-STAT3 interaction after SUMOylation site was mutated. It can be concluded that SUMOylation of SHP1 affects its phosphatase activity as STAT3 is a generally acknowledged substrate of SHP1 phosphatase. Despite its inhibitory effect on tumor-intrinsic STING signaling, SHP1 also functions as a signaling hub in T cells, DCs and macrophages [50]. Therefore, multiple inhibitors targeting SHP1 were developed and tested for synergy with immunotherapy [51, 52]. Therefore, the ability of ATRi to inhibit SHP1 activity could be applied in the context of other immuno-based therapies. Notably, we also found decreased Shp1 mRNA levels following IR + ATRi treatment compared to IR treatment, indicating that ATRi may also control SHP1 expression on the translational level. Further studies require a comprehensive understanding of how the ATRi affects the gene expression, post-modification and function of SHP1.
Due to the success of ICIs in solid tumors, extensive studies have focused on developing prognostic biomarkers for ICI treatment. Targeting the PD-L1/PD-1 axis inhibits the exhaustion of TIL to functionally kill the tumors, making TIL level one of the most important biomarkers predicting ICI therapy efficacy. Less-infiltrated tumors (or “cold” tumors), such as pMMR/MSS CRCs with low immunogenicity generally respond poorly to ICIs. Thus, combinational strategies to turn the cold tumor hot may provide therapeutic potential to improve ICI efficacy. In alignment with this concept, we combined IR and ATR inhibition to enhance TILs and evaluated their efficacy in sensitizing CRC tumors to anti-PD-L1 therapy. While the results revealed successful synergy, the efficacy was dependent on STING-mediated innate immune activation because Sting knockdown nearly completely abrogated the therapeutic efficacy of the combinational plan. This is consistent with recent findings that resistance to ICIs in patients with dMMR CRCs may be due to loss of the cGAS-STING function, and this axis is essential and could act as a prognostic biomarker for ICI treatment [29, 53]. Moreover, CT26 and MC38 are considered as MSS/pMMR (low immunogenicity) and MSI-H/dMMR (high immunogenicity) models, respectively [28, 29, 54]. This indicates that this plan could be used independently of the microsatellite or immunogenicity statuses, which provides informative methods for the clinical use of ICI therapy.
Our work has high clinical relevance. First, clinical data on the combination therapy of IR and ICIs have demonstrated promising therapeutic potential [55-57]. The strategy of adding ATRi to the combination of ICI and radiotherapy has the potential to overcome the resistance in pMMR mCRC or improve the current unsatisfactory efficacy of ICIs in dMMR mCRC. Recently, a review of the interim data of a phase II trial showed a 25% pathological complete response (pCR) rate in patients with locally advanced CRC (LARC) MSS/pMMR receiving neoadjuvant short-course radiotherapy followed by chemotherapy and anti-PD-1 antibody treatment [58]. Here, we provided a rationale for the clinical investigation of adding ATRi as an alternative in this neoadjuvant combination, which may potentially increase the pCR rate and lead to a better quality of life for patients with LARC. Second, in metastatic tumors, emerging results from ongoing clinical trials suggest better synergy with immunotherapy when IR was applied simultaneously to multiple metastatic sites, mainly attributable to STING-mediated innate immune response against tumors [59, 60]. By inducing strong activation of STING signaling, combining ATRi with IR and ICIs may synergistically achieve a systematic control. Third, multiple clinical trials are investigating the efficacy of ATR inhibition monotherapy or ATR inhibition combined with DNA damage agents such as platinum-based therapy in solid tumors and inspiring results were observed in CRCs with several DDR gene defects [61-63]. Notably, a recent clinical data have reported that the ATRi ceralasertib (AZD6738) may overcome anti-PD-1 therapy resistance in patients with advanced melanoma [64]. In this case, we provided a new perspective on exploiting ATRi in cancer immunotherapy, which could be further tested and applied in clinical use.
Despite intriguing findings, there were some limitations in the present study: (1) The immune profile of subcutaneous tumors in syngeneic mice cannot perfectly represent that of human CRCs. Therefore, the response to ICIs may vary in human and needs further investigation in clinical practice. (2) While we focus on the tumor-intrinsic STING activation induced by IR + ATRi, STING signaling in immune cells were not well-defined and needs further investigation. (3) We focused on DCs and T cells in the tumor microenvironment. Future studies require evaluation of an intact immune landscape in CRC models treated with IR + ATRi + anti-PD-L1.
5 CONCLUSIONS
The present study demonstrated how the ATRi berzosertib exploits the cGAS-STING signaling to promote T cell infiltration in colorectal tumors, turning the cold tumor hot and facilitating the efficacy of IR and anti-PD-L1 combination therapy. This interplay of two checkpoint inhibitors on the DNA damage stage resulted in promising tumor control and prolonged survival in murine CRCs with different microsatellite statuses. Our work raised a new strategy for CRC treatment. As clinical trials are ongoing to investigate the efficacy of ATRi, such a strategy could be evaluated and optimized for clinical use.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Xianglin Yuan designed and guided the research. Chaofan Liu performed most of the experiments and analyzed the data. Xi Wang, Jingyao Tu, Chunya Li, and Li Ma performed irradiation and most of the animal study. Weiheng Zhao and Wan Qin contributed methodology support. Bo Liu and Hong Qiu wrote the original drat and performed most of the revised work. All authors have reviewed and approved the manuscript.
ACKNOWLEDGEMENTS
We thank the Medical Subcenter of Huazhong University of Science and Technology Analytical & Testing Center in data acquisition. This work was supported by the Innovative Capacity Building Project of the Hubei Engineering Research Center for Radiotherapy and Radiation Protection of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (grant No.: 2018-420114-35-03-071705), the State Key Program of National Natural Science of China (grant No.: 82130092), the National Natural Science Foundation of China (grant No.: 81372664) and the National Natural Science Foundation of China (grant No.: 81902619).
COMPETING INTERESTS
All authors have no conflicts of interests to declare.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Laboratory Animal Welfare & Ethics Committee of Tongji Hospital Huazhong University of Science and Technology (permit number: TJH-201910007), and performed in accordance with all applicable guidelines and regulations.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
All relevant data to this study are included in this article or in the supplementary information. Raw data are available on reasonable request from the corresponding author.




