An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment
Elena Verdugo and Iker Puerto contributed equally to this work.
Abstract
Glioblastoma multiforme (GBM) is the most aggressive and common malignant primary brain tumor. Patients with GBM often have poor prognoses, with a median survival of ∼15 months. Enhanced understanding of the molecular biology of central nervous system tumors has led to modifications in their classifications, the most recent of which classified these tumors into new categories and made some changes in their nomenclature and grading system. This review aims to give a panoramic view of the last 3 years’ findings in glioblastoma characterization, its heterogeneity, and current advances in its treatment. Several molecular parameters have been used to achieve an accurate and personalized characterization of glioblastoma in patients, including epigenetic, genetic, transcriptomic and metabolic features, as well as age- and sex-related patterns and the involvement of several noncoding RNAs in glioblastoma progression. Astrocyte-like neural stem cells and outer radial glial-like cells from the subventricular zone have been proposed as agents involved in GBM of IDH-wildtype origin, but this remains controversial. Glioblastoma metabolism is characterized by upregulation of the PI3K/Akt/mTOR signaling pathway, promotion of the glycolytic flux, maintenance of lipid storage, and other features. This metabolism also contributes to glioblastoma's resistance to conventional therapies. Tumor heterogeneity, a hallmark of GBM, has been shown to affect the genetic expression, modulation of metabolic pathways, and immune system evasion. GBM's aggressive invasion potential is modulated by cell-to-cell crosstalk within the tumor microenvironment and altered expressions of specific genes, such as ANXA2, GBP2, FN1, PHIP, and GLUT3. Nevertheless, the rising number of active clinical trials illustrates the efforts to identify new targets and drugs to treat this malignancy. Immunotherapy is still relevant for research purposes, given the amount of ongoing clinical trials based on this strategy to treat GBM, and neoantigen and nucleic acid-based vaccines are gaining importance due to their antitumoral activity by inducing the immune response. Furthermore, there are clinical trials focused on the PI3K/Akt/mTOR axis, angiogenesis, and tumor heterogeneity for developing molecular-targeted therapies against GBM. Other strategies, such as nanodelivery and computational models, may improve the drug pharmacokinetics and the prognosis of patients with GBM.
Abbreviations
-
- 2-HG
-
- 2-Hydroxyglutarate
-
- 5hmC
-
- 5-Hydroxymethylcytosine
-
- ACC
-
- Acetyl-CoA carboxylase
-
- ACSS2
-
- Acyl-CoA synthetase short chain family member 2
-
- AHNAK2
-
- AHNAK nucleoprotein 2
-
- AKT
-
- Alpha serine/threonine-protein kinase
-
- ANKRD10
-
- Ankyrin repeat domain 10
-
- ANXA2
-
- Annexin A2
-
- ANXA7
-
- Annexin A7
-
- APC
-
- APC regulator of Wnt signaling pathway
-
- ASCL1
-
- Achaete-scute family BHLH transcription factor 1
-
- ATRX
-
- ATP-dependent helicase ATRX
-
- AXIN
-
- Axis inhibitor
-
- B4GALT3
-
- Beta-1,4-galactosyltransferase 3
-
- BBB
-
- Blood-brain barrier
-
- BCL6
-
- BCL6 transcription repressor
-
- BCORL1
-
- BCL6 corepressor like 1
-
- BIN
-
- Bridging integrator
-
- BMP2
-
- Bone morphogenetic protein 2
-
- BRAF
-
- Serine/threonine-protein kinase B-Raf
-
- BTK
-
- Bruton's tyrosine kinase
-
- CAD
-
- Carbamoyl-phosphate synthetase 2
-
- CCL2
-
- C-C motif chemokine ligand 2
-
- CCNB1
-
- Cyclin B1
-
- CCND1
-
- Cyclin D1
-
- CD24
-
- Small cell lung carcinoma cluster 4 antigen
-
- CD27
-
- Tumor necrosis factor receptor superfamily member 7
-
- CD3+
-
- Cluster of differentiation 3
-
- CD31
-
- Platelet endothelial cell adhesion molecule
-
- CD41
-
- Integrin subunit alpha 2b
-
- CD44
-
- Homing cell adhesion molecule
-
- CD8+
-
- Cluster of differentiation 8
-
- CD99
-
- Single-chain type-1 glycoprotein
-
- CDC6
-
- Cell division cycle 6
-
- CDK4/6
-
- Cyclin-dependent kinases 4 and 6
-
- CDKN2A/B
-
- Cyclin-dependent kinase inhibitors 2A/2B
-
- CELF2
-
- CUGBP Elav-like family member 2
-
- C-GBMs
-
- Cerebellar glioblastomas
-
- CGGA
-
- Chinese Glioma Genome Atlas
-
- c-KIT
-
- Tyrosine-protein kinase KIT
-
- CMV
-
- Cytomegalovirus
-
- c-Myc
-
- Master Regulator of Cell Cycle Entry and Proliferative Metabolism C
-
- CNS
-
- Central nervous system
-
- CSF
-
- Cerebrospinal fluid
-
- CTLA-3
-
- Cytotoxic T-lymphocyte-associated protein 3
-
- CTLA-4
-
- Cytotoxic T-lymphocyte-associated protein 4
-
- Cx43
-
- Connexin 43
-
- CXCL12
-
- C-X-C motif chemokine ligand 12
-
- CXCR4
-
- C-X-C chemokine receptor type 4
-
- DECR1
-
- 2,4- dienoyl-CoA reductase 1
-
- DEGs
-
- Differentially expressed genes
-
- DHODH
-
- Dihydroorotate dehydrogenase
-
- DNA
-
- Deoxyribonucleic acid
-
- DSC-MRI
-
- Dynamic susceptibility contrast magnetic resonance imaging
-
- DTI
-
- Diffusion tensor imaging
-
- E2F7
-
- E2F transcription factor 7
-
- EGFR
-
- Epidermal growth factor receptor
-
- EGFRvIII
-
- Epidermal growth factor receptor variant III
-
- ErbB2
-
- ErbB2 Receptor tyrosine kinase 2
-
- EVs
-
- Extracellular vesicles
-
- FABP3/7
-
- Fatty-acid binding proteins 3/7
-
- FASN
-
- Fatty-acid synthase
-
- FDA
-
- Food and Drug Administration
-
- FN1
-
- Fibronectin 1
-
- FRP
-
- Frizzled-related protein
-
- GABRA1
-
- Gamma-aminobutyric acid type A receptor subunit alpha1
-
- GBM
-
- Glioblastoma multiforme
-
- GBP2
-
- Guanylate binding protein 2
-
- GBSCs
-
- Glioblastoma stem cells
-
- GFAP
-
- Glial fibrillary acidic protein
-
- GICs
-
- Glioma-initiating cells
-
- GLUT1
-
- Glucose transporter 1
-
- GLUT3
-
- Glucose transporter 3
-
- G-MCI
-
- Gene-mediated cytotoxic immunotherapy
-
- GM-CSF
-
- Granulocyte-macrophage colony-stimulating factor
-
- GNB2
-
- G-protein subunit beta 2
-
- GNB3
-
- G-protein subunit beta 3
-
- GNB4
-
- G-protein subunit beta 4
-
- GNB5
-
- G-protein subunit beta 5
-
- GO
-
- Gene Ontology
-
- GPR17
-
- G-protein-coupled receptor 17
-
- GSCs
-
- Glioma stem cells
-
- GSH
-
- Glutathione
-
- GSK-3β
-
- Glycogen synthase kinase-3-beta
-
- H3F3A
-
- H3.3 histone A
-
- HDAC1
-
- Histone deacetylase 1
-
- HDACIs
-
- Histone deacetylase inhibitors
-
- HER-2
-
- Human epidermal growth factor receptor 2
-
- HGGs
-
- High-grade gliomas
-
- Hh
-
- Hedgehog
-
- HIF
-
- Hypoxia inducible factor
-
- HIF-1α
-
- Hypoxia inducible factor 1 subunit alpha
-
- HIF-2α
-
- Hypoxia inducible factor 2 subunit alpha
-
- HK2
-
- Hexokinase 2
-
- HPSE
-
- Heparanase
-
- IDH
-
- Isocitrate dehydrogenase
-
- IDH1
-
- Isocitrate dehydrogenase 1
-
- IDH2
-
- Isocitrate dehydrogenase 2
-
- IDO
-
- Indoleamine 2,3-dioxygenase
-
- IGFBP2
-
- Insulin like growth factor binding protein 2
-
- IgG1
-
- Immunoglobulin G1
-
- IL-10
-
- Interleukin 10
-
- IL-12
-
- Interleukin 12
-
- JAK
-
- Janus activated kinase
-
- KEGG
-
- Kyoto encyclopedia of genes and genomes
-
- KIF20A
-
- Kinesin family member 20A
-
- KIF23
-
- Kinesin family member 23
-
- KMT2C
-
- Lysine methyltransferase 2C
-
- KMT2D
-
- Lysine methyltransferase 2D
-
- LAG-3
-
- Lymphocyte-activation gene 3
-
- LDH-A
-
- Lactate dehydrogenase A
-
- LGGs
-
- Low-grade gliomas
-
- lncRNA
-
- Long noncoding RNA
-
- LOXL1
-
- Lysyl oxidase like 1
-
- MAPK
-
- Mitogen-activated protein kinase
-
- MAPK1
-
- Mitogen-activated protein kinase 1
-
- MARK4
-
- Microtubule affinity regulating kinase 4
-
- MAX
-
- MYC associated factor X
-
- MDM2
-
- Murine double minute 2
-
- MEK1/2
-
- Mitogen-activated protein kinase kinases 1 and 2
-
- MET
-
- MET proto-oncogene receptor tyrosine kinase
-
- MGMT
-
- O-6-Methylguanine-DNA methyltransferase
-
- MHC
-
- Major histocompatibility complex
-
- MHC-I
-
- Major histocompatibility complex class I
-
- MIR4435-2HG
-
- MIR4435 host gene 2
-
- miRNA
-
- Micro RNA
-
- MKI67
-
- Marker of proliferation Ki-67
-
- MMP-2/9
-
- Matrix metalloproteinases 2 and 9
-
- MPC1
-
- Mitochondrial pyruvate carrier 1
-
- MRI
-
- Magnetic resonance imaging
-
- mRNA
-
- Messenger ribonucleic acid
-
- mt-DNA
-
- Mitochondrial DNA
-
- mTOR
-
- Mammalian target of rapamycin
-
- NAD+
-
- Nicotinamide adenine dinucleotide
-
- NADPH
-
- Nicotinamide adenine dinucleotide phosphate hydrogen
-
- ncRNA
-
- Noncoding RNA
-
- NEFL
-
- Neurofilament light-chain gene
-
- NES
-
- Nestin
-
- NF1
-
- Neurofibromin 1
-
- NFATC3
-
- Nuclear factor of activated T cells 3
-
- NF-κB
-
- Nuclear factor kappa-light-chain-enhancer of activated B cells
-
- NF-κB1/2
-
- Nuclear factor kappa B subunits 1 and 2
-
- NG2
-
- Neuron-glial antigen 2
-
- NOTCH1
-
- Neurogenic locus notch homolog protein 1
-
- NPs
-
- Nanoparticles
-
- NR4A1
-
- Nuclear receptor subfamily 4 group A member 1
-
- NSCs
-
- Neural stem cells
-
- OLIG2
-
- Oligodendrocyte lineage transcription factor 2
-
- OPC
-
- Oligodendrocyte progenitor cell
-
- OS
-
- Overall Survival
-
- OSMR
-
- Oncostatin M receptor beta
-
- OX-40
-
- TNF receptor superfamily member 4
-
- PARP-1
-
- Poly(ADP-ribose) polymerase 1
-
- PD-1
-
- Programmed cell death protein 1
-
- PDGF
-
- Platelet-derived growth factor
-
- PDGFR
-
- Platelet-derived growth factor receptor
-
- PDGFRA
-
- Platelet-derived growth factor receptor alpha
-
- PDIA3
-
- Protein disulfide isomerase family A member 3
-
- PDK1/2
-
- 3-Phosphoinositide-dependent kinases 1 and 2
-
- PD-L1
-
- Programmed death ligand-1
-
- PDPN
-
- Podoplanin
-
- PFKP
-
- Phosphofructokinase platelet
-
- PFS
-
- Progression-free survival
-
- PHIP
-
- Pleckstrin homology domain interacting protein
-
- PI3K
-
- Phosphatidylinositol 3-kinase
-
- PIK3CA
-
- Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
-
- PLCG1
-
- Phospholipase C gamma 1
-
- POLR2F
-
- RNA polymerase II, I and III subunit F
-
- poly-ICLC
-
- polyinosinic-polycytidylic acid
-
- PSMA
-
- Prostate-specific membrane antigen
-
- PTEN
-
- Phosphatase and tensin homolog
-
- PTPN11
-
- Protein tyrosine phosphatase non-receptor type 11
-
- PUFA
-
- Polyunsaturated fatty acids
-
- RAF
-
- Rapidly accelerated fibrosarcoma kinase
-
- RAS
-
- Rat sarcoma virus GTPase
-
- Rb
-
- Retinoblastoma protein
-
- RNA
-
- Ribonucleic acid
-
- RNA-LP
-
- RNA-lipid particle
-
- RON
-
- Ron receptor tyrosine kinase
-
- RPL39L
-
- Ribosomal protein L39 like
-
- RTK
-
- Receptor tyrosine kinase
-
- RTK I
-
- Receptor tyrosine kinase I
-
- RTK II
-
- Receptor tyrosine kinase II
-
- RT-qPCR
-
- Real-time polymerase chain reaction
-
- RYR2
-
- Ryanodine receptor 2
-
- SDF-1
-
- Stromal cell-derived factor 1
-
- siRNA
-
- Small interfering RNA
-
- SLC12A5
-
- Solute carrier family 12 member 5
-
- SLC7A11
-
- Solute carrier family 7 member 11
-
- SNHG12
-
- Small nucleolar RNA host gene 12
-
- SNRPB
-
- Small nuclear ribonucleoprotein polypeptides B and B1
-
- SNVs
-
- Single nucleotide variants
-
- SOX1
-
- SRY-box transcription factor 1
-
- SOX10
-
- SRY-box transcription factor 10
-
- SOX40
-
- SRY-box transcription factor 40
-
- SREBP-1
-
- Sterol regulatory element-binding protein 1
-
- STAT
-
- Signal transducer and activator of transcription
-
- STAT3
-
- Signal transducer and activator of transcription 3
-
- SUSD2
-
- Sushi domain containing 2
-
- SVZ
-
- Subventricular zone
-
- SYT1
-
- Synaptotagmin-1
-
- TAMs
-
- Tumor-associated macrophages
-
- TCGA
-
- The Cancer Genome Atlas
-
- TEM7
-
- Tumor endothelial marker
-
- TERT
-
- Telomerase reverse transcriptase
-
- TGFBR2
-
- Transforming growth factor-beta receptor II
-
- TGF-β
-
- Transforming growth factor beta
-
- tGLI1
-
- Glioma-associated oncogene homolog 1 truncated variant
-
- TICs
-
- Tumor-initiating cells
-
- TIM-3
-
- T cell immunoglobulin and mucin-domain containing-3
-
- TME
-
- Tumor microenvironment
-
- TMEM52
-
- Transmembrane protein 52
-
- TMZ
-
- Temozolomide
-
- TNF-α
-
- Tumor necrosis factor alpha
-
- TNTs
-
- Tunneling nanotubes
-
- TP53
-
- Tumor protein p53
-
- TRADD
-
- TNFR1-associated death domain protein
-
- Treg
-
- Regulatory T cells
-
- USP5
-
- Ubiquitin specific peptidase 5
-
- VCL
-
- Vinculin
-
- VEGF
-
- Vascular endothelial growth factor
-
- VEGF-A
-
- Vascular endothelial growth factor A
-
- VEGF-C
-
- Vascular endothelial growth factor C
-
- VEGFR1
-
- Vascular endothelial growth factor receptor 1
-
- VILL
-
- Villin-like protein
-
- VIM
-
- Vimentin
-
- VPA
-
- Valproic acid
-
- WGCNA
-
- Weighted gene correlation network analysis
-
- WHO
-
- World Health Organization
-
- Wnt
-
- Wingless/Integrated
-
- WT1
-
- Wilms tumor gene-1
-
- WWOX
-
- WW domain containing oxidoreductase
-
- YKL40
-
- Chitinase-3-like protein 1
-
- α-KG
-
- Alpha ketoglutarate
1 BACKGROUND
Glioblastoma multiforme (GBM) is the most aggressive and common type of malignant primary brain tumor. The incidence of GBM increases with age and is slightly higher in men than in women [1]. GBM's incidence oscillates between 0.59 and 5 cases per 100,000 people and is rising in many countries owing to the aging population and improvements in diagnosis, among other factors [2].
Despite the considerable increase in knowledge about the molecular pathogenesis and biology of this tumor, patients with GBM continue to suffer from poor prognoses. They have a median survival of ∼15 months [3] and a 5-year relative survival rate of only 6.8%, although this could depend on the patient's sex and age at diagnosis [4]. Since 2005, the treatment regimen for newly diagnosed patients comprises surgery followed by concurrent radiotherapy with temozolomide (TMZ) and further adjuvant TMZ [5]. In recent years, clinical trials testing new drugs and strategies have been rising, particularly those on immunotherapy and targeted therapies [1].
Although our group published a review on the literature related to the molecular biology of glioblastoma in 2019 [6], given the remarkable amount of research related to GBM and its classification, characterization, and treatment conducted within the last 3 years, an update on the topic was advisable. The present review aimed to gather information on the latest advances in understanding the molecular biology of glioblastoma, their clinical implications, and the latest therapeutic advancements.
2 FINDINGS ON GLIOBLASTOMA ORIGINS
The origin of IDH-wildtype GBMs has been described as a neuronal network that starts in the subventricular zone (SVZ) and spreads toward the frontotemporal cortex and lobe, thus creating a “firework” pattern [7]. Tumoral progression is possible because of the presence of astrocyte-like neural stem cells at the astrocytic ribbon, whose mutations gradually accumulate as they reach the cortex. This origin of primary glioblastomas has been confirmed [8]. On the contrary, a second origin was proposed, highlighting the genesis of outer radial glial-like cells from astrocytes showing a high expression of ErbB2, a tyrosine kinase receptor implicated in cell proliferation and motility [9].
A series of studies hypothesized the “double origin” of GBM from a mixed population of ventricular and outer radial glial cells in SVZ. The epidermal growth factor receptor variant III (EGFRvIII) is responsible for reprogramming during proliferation, regardless of whether GBM originates from neural stem cells or GFAP-positive progenitors [8]. Subsequently, glioblastoma consists of a heterogeneous cell population derived from glioma stem cells (GSCs) located within a vascularized tumor niche. GSCs are astrocyte-like neural stem cells that are prevalent in the SVZ. These cells take advantage of a weakened immune system and proliferate because of the overproduction of growth factors in the perivascular region, stimulated by the release of cytokines. Accompanied by this tumor niche, neural stem cells (NSCs) from the germinal vascular zones establish crosstalk with GSCs and also exhibit cell differentiation capabilities [10].
Lombard et al. [11] summarized the similarities between adult NSCs and GSCs and their effect on the prognosis of a patient with glioblastoma due to recurrence and drug resistance. These two stem cell types are associated with vasculature; niche companions, such as pericytes and endothelial cells; migration and proliferation regulation; and nestin expression. Furthermore, GSCs expose the mutated genes expressed in NSCs: TERT, TP53, PTEN, EGFR, and PDGF [12].
CXCL12 and pleiotrophin might play a role in the migration of GSCs from the tumor niche to the SVZ, heading to the exclusive transformation of NSCs in this brain region. NSCs in other neurogenic niches, such as the hippocampus, are not involved in gliomagenesis [10]. In fact, the SVZ is in contact with the cerebrospinal fluid (CSF), which might interfere with healthy cell growth and is partially responsible for the malignancy of GBM in the proximity of the SVZ. Proliferating GSCs near the SVZ might receive altered genetic information via the CSF. In addition, NSCs in the SVZ can undergo somatic mutations, leading to uncontrolled proliferation and genetic alterations similar to the progenitor cells in IDH-wt and IDH-mutant mouse xenografts. Further, Lozano-Ureña et al. [13] demonstrated that adult NSCs could not be recognized from GSCs based on their genetic expressions.
Tumor recurrence might be mediated by glioma-initiating cells (GICs), reactivated by their presence in the parenchyma, where they stayed during the dormant period [8, 14]. Yoon et al. [7] hypothesized an alternative for tumor recurrence, which comes from the remigration of these dormant cells to the tumor niche from the SVZ.
NSCs can be selected as the cell lineage for the origin of gliomas based on their location (SVZ), differentiation properties, and matching variations with glioblastoma. IDH-wildtype patients show changes in the gene expression of TERT, TP53, PDGFR, and EGFR in these cells. Nevertheless, mature astrocytes can dedifferentiate and reprogram into tumor cells, and oligodendrocyte progenitor cell-like cells (OPC-like cells) can redirect their transcriptome to accelerate the uncontrolled proliferation of malignant glioblastomas.
3 ADVANCES IN THE MOLECULAR CLASSIFICATION OF GLIOMAS
Historically, glioma classification was based on histological and immunohistochemical criteria. The classical diagnostic methods for gliomas are based on imaging or screening tests, such as functional magnetic resonance imaging (MRI), positron emission tomography, computed tomography, and the performance of a liquid biopsy, which is a non-invasive technique used to confirm the diagnosis and augment treatment prospects [15, 16]. Additional molecular diagnoses can be performed to provide a more personalized prognosis and enhance the chances of therapeutic efficacy.
Despite the low impact of molecular classification in medical diagnosis, during the past few years, there have been remarkable advances in this field, especially for the central nervous system (CNS) tumor classification, which was included in the fifth edition of the World Health Organization (WHO) Classification of Tumors of the Central Nervous System, published in 2021 [17]. This new edition integrates molecular changes with clinicopathological utility essential for accurately classifying CNS tumors. This edition also introduces changes to the former taxonomy and nomenclature, including the term “type” instead of “entity” and “subtype” instead of “variant.” Traditional names that refer to histological features, such as anaplastic, malignant, or giant cells, can still be used for medical recognition but are likely to disappear in future classifications [18]. Arabic numerals are now used to grade neoplasms within types, whereas the past edition used Roman numerals to grade neoplasms across different tumor types [19].
Fourteen newly identified neoplasms have been incorporated into the categories of gliomas, glioneuronal tumors, and neuronal tumors. The WHO divided this category of CNS tumors into the following six families: 1) Adult-type diffuse gliomas, 2) Pediatric-type diffuse low-grade gliomas, 3) Pediatric-type diffuse high-grade gliomas, 4) Circumscribed astrocytic gliomas, 5) Glioneuronal and neuronal tumors, and 6) Ependymomas (Table 1). As this classification suggests, diffuse gliomas that primarily occur in adults and those that mainly in children have been separated prognostically and biologically into different groups. Moreover, pediatric gliomas are segregated into low-grade gliomas, those that exhibit diffused growth in the brain but have less-specific histological features, and high-grade gliomas. Integrating molecular and histopathological information is essential for precisely diagnosing these tumors. It should also be noted that the term “glioblastoma” has been discarded to identify pediatric-type gliomas, which are referred to as those that affect the 0-14 years old age group [17, 20].
| WHO Classification of Tumors of the Central Nervous System | Most representative criteria for GBM classification | Categories | Nomenclature |
|---|---|---|---|
| Fourth edition (2016) |
Histology IDH mutational status 1p/9q codeletion |
Diffuse astrocytic and oligodendroglial tumors Other astrocytic tumors Other gliomas Ependymal tumors |
Roman numerals (I, II, III, IV) Grade is based on histological criteria |
| Fifth edition (2021) |
TERT promoter mutational status EGFR amplification Gain chromosome 7 and loss chromosome 10 |
Gliomas, glioneuronal and neuronal tumors Adult-type diffuse glioma Pediatric-type diffuse low-grade gliomas Pediatric-type diffuse high-grade gliomas Circumscribed astrocytic gliomas Ependymomas |
Arabic numerals (1, 2, 3, …) Grade is based on the natural history of the tumor |
- Abbreviations: CNS, Central Nervous System; EGFR, Epidermal growth factor receptor; GBM, Glioblastoma multiforme; IDH, Isocitrate dehydrogenase; TERT, Telomerase reverse transcriptase; WHO, World Health Organization.
Glioblastoma, IDH-wildtype, was integrated into the gliomas, glioneuronal tumors, and neuronal tumors category within the adult-type diffuse gliomas. Previously, IDH-mutant diffuse astrocytic tumors were assigned to three different types: 1) diffuse astrocytoma, 2) anaplastic astrocytoma, or 3) glioblastoma. Singer et al. [21] proposed a new classification for IDH-mutant astrocytoma because of its lower aggressiveness compared with diffuse midline gliomas and IDH-wildtype glioblastomas. The current classification identifies all IDH-mutant diffuse astrocytic tumors as astrocytoma, IDH-mutant, which can be graded as CNS WHO Grade 2, 3, or 4 [22].
Microvascular proliferation and necrosis rates are proposed as determinants for oligodendrogliomas, now defined by IDH1/2 mutations, 1p/19q codeletion, TERT promoter mutations, and NOTCH1. On the contrary, histone H3.3 G34-mutant gliomas are characterized by OLIG2 and ATRX mutations, and cerebellar glioblastomas (C-GBMs) are described as “high-grade astrocytoma with piloid features” with IDH, ATRX, and CDKN2A/B mutations [21]. Exclusive alterations in ATRX and PDGFRA can define C-GBMs, with most of these tumors exhibiting IDH1/TP53 mutations and the upregulation of NG2 and NR4A1 [23].
4 MOLECULAR PARAMETERS FOR GLIOBLASTOMA CLASSIFICATION AND CHARACTERIZATION
4.1 Genetic and transcriptomic criteria for the diagnosis and subclassification of glioblastoma
The number of published articles on glioblastoma and its genetics has increased exponentially during the last decade [24]. The 2021 WHO Classification of CNS Tumors defines three genetic parameters for diagnosing glioblastoma, IDH-wildtype: TERT promoter mutation, epidermal growth factor receptor (EGFR) amplification, and the combined gain of entire chromosome 7 and loss of entire chromosome 10 [17]. However, this neoplasm can be further classified into molecular subtypes, which can impact disease progression and clinical practice.
In 2016, Verhaak et al. [25]classified glioblastomas into four subtypes based on their molecular features: neuron, astrocyte, oligodendrocyte, and cultured astrocytic gliomas. Detecting these subtypes relies on the different therapeutic approaches required for each patient and their impacts on tumor progression [26]. Recently, Neftel et al. [27] identified four heterogeneous cellular states using single-cell RNA-sequencing and validated the intratumoral heterogeneity present in GBM and the relevance of this subtyping. They classified the development of neural signatures into neural-progenitor-like, oligodendrocyte-progenitor-like, astrocyte-like, and mesenchymal-like states [26].
The neural subtype is derived from astrocytes and oligodendrocytes and expresses neuron-related genes, whereas the proneural subtype exhibits the characteristics of oligodendroglial cells and develops in young patients [25]. The classical subtype possesses astrocytic features and expresses neuron precursor and stem cell markers, whereas the mesenchymal subtype shows characteristics of cultured astrocytic gliomas [28].
Verhaak's latest update for reclassifying gliomas removed the neural subtype because it is problematic for identifying primary and recurrent gliomas owing to its ongoing genomic signature changes [29]. Further, a study dedicated to the evolution of the tumor determined that the vast majority of The Cancer Genome Atlas (TCGA) subtypes were from a proneural-like precursor and switched to a mesenchymal-like state in a differentiation process regulated by TNF-α/NF-κB signaling or ASCL1. To sum up, Neftel et al.’s studies [27]and the recent update of the WHO CNS tumor classification reflect the fluidity of GBM's transcriptional states and the influence of the tumor microenvironment (TME) on the development and transition from one subtype to another [22].
Jankowska et al. [30] classified glioblastoma subtypes based on immunochemical expression and concluded that the classical subtype is represented by TP53 mutation, which makes this subtype highly sensitive to classical radiotherapy plus chemotherapy with adjuvant TMZ. The mesenchymal subtype shows NF1, PTEN, AKT, MET, and TRADD mutations. Further, PDGFRA, IDH1, TP53, HIF, and OLIG2 mutations are characteristic of the proneural subtype. Another study revealed the neuronal markers for identifying and profiling neural glioblastomas, such as NEFL, GABRA1, SLC12A5, and SYT1 (Table 2) [31].
| Subtype | Cell-like features | Targeted biomarkers |
|---|---|---|
| Proneural | Oligodendroglial cell or neural stem cell | PDGFRA, TP53, HIF, OLIG2, MKI67, B4GALT3 |
| Neural | Astrocyte and oligodendrocyte | NEFL, GABRA1, SLC12A5, SYT1 |
| Mesenchymal | Astrocyte | NF1, PTEN, AKT, MET, TRADD, MGMT, YKL40, GBP2, STAT3 |
| Classical | Cultured astrocytic gliomas | TP53, EGFR, NES, VIM |
- Abbreviations: AKT, Alpha serine/threonine-protein kinase; B4GALT3, Beta-1,4-galactosyltransferase 3; EGFR, Epidermal growth factor receptor; GABRA1, Gamma-aminobutyric acid type A receptor subunit alpha1; GBM, Glioblastoma multiforme; GBP2, Guanylate binding protein 2; HIF, Hypoxia-inducible factor; MET, MET proto-oncogene receptor tyrosine kinase; MGMT, O-6-Methylguanine-DNA methyltransferase; MKI67, Marker of proliferation Ki-67; NEFL, Neurofilament light-chain gene; NES, Nestin; NF1, Neurofibromin 1; OLIG2, Oligodendrocyte lineage transcription factor 2; PDGFRA, Platelet-derived growth factor receptor alpha; PTEN, Phosphatase and tensin homolog; SLC12A5, Solute carrier family 12 member 5; STAT3, Signal transducer and activator of transcription 3; SYT1, Synaptotagmin-1; TP53, Tumor protein p53; TRADD, TNFR1-associated death domain protein; VIM, Vimentin; YKL40, Chitinase-3-like protein 1.
In parallel to this classification, Herrera-Oropeza et al. [32] performed a multi-omics analysis of driver genes. They concluded that mesenchymal subtype development was related to the upregulation of the MGMT promoter and the downregulation of ATRX, H3F3A, TP53, and EGFR. Complementary information was provided for the proneural subtype, characterized by the overexpression of MKI67 and OLIG2, and the classical subtype by the overexpression of EGFR, NES, VIM, and TP53. The characterization of differential molecular characteristics of histologically similar tumors is relevant to improve the diagnosis of GBM in patients. Besides, the determination of expression profiles is useful for creating progression models and enhancing the prognosis of each tumor subtype.
There is a rising tendency to classify gliomas based on their mRNA sequencing and the clustering of samples with computational programs. Using the “Consensus Cluster Plus” package for R v4.0.3, Cai et al. [33] investigated the reclassification of glioma based on the expression levels of Gβ/γ genes from TCGA and the Chinese Glioma Genome Atlas (CGGA) datasets. The result was a differential distribution map correlated with the samples analyzed from these two chosen databases. The Gβ/γ heterodimer can activate the Erk1/2 pathway by inducing the overexpression of guanine nucleotide-binding protein beta 4 (GNB4), which results in the transformation of epithelial and mesenchymal cells into glioma cells. After clustering, they obtained three subgroups: GNB2, GNB3, and GNB5. GNB2 appeared to be the best indicator of malignant tumors, especially in patients with IDH-mutated, non-codeleted 1p/19q low-grade gliomas (LGGs). This subgroup is characterized by high M0/M2 cell infiltration levels and is highly associated with the immunosuppressive phenotype, thus demonstrating enhanced PI3K-Akt/JAK-STAT pathways and high levels of tumor-associated macrophages (TAMs) and M2 macrophages. Therefore, the GNB2 subgroup would represent the immunosuppressive phenotype in gliomas. Each subgroup has a unique tumor-related pathway that can answer the selection of a chemotherapeutic drug and enhance the glioma prognosis by choosing the right target [33].
An analysis of the differentially expressed genes (DEGs) revealed 110 upregulated genes and 75 downregulated genes in the GBM samples. This observation identified a four-protein prognostic signature (SLC12A5, CCL2, IGFBP2, and PDPN) for the segregation of patients into high- and low-risk groups and for the estimation of survival time. These were observed via a weighted gene correlation network analysis (WGCNA) algorithm, a strategy that has also been used to determine disease-related genes in other oncologic diseases [34]. Another DEG screening found 662 DEGs in patients with GBM and concluded that DECR1, POLR2F, HDAC1, and PDIA3 could be the critical genes related to the overall survival (OS) time of patients with GBM [35].
A WGCNA study comparing the transcriptome and proteome of glioblastoma, IDH-wildtype tumors found six proteomic modules correlated with survival, but none of the identified RNA modules did [36]. After performing a Kaplan–Meier analysis, 11 proteins were revealed to have a significant association with survival, despite not being significant at the RNA level. Owing to the apparent lack of correlation between RNA and survival, previously established single-cell-based signatures were used to define the dominant cell subpopulation of each tumor analyzed. This study revealed that mesenchymal and neural progenitor cell-like subpopulation signature genes correlated with shorter survival, whereas oligodendrocytic precursor cell-like and astrocytic subpopulation signature genes correlated with more prolonged survival. Gene Ontology (GO) enrichment analysis from the proteomic and single-cell-based signature data revealed that lysosomal activity and amino and nucleotide sugar metabolism were enriched in a cluster of genes and proteins correlated with short survival [36].
Transcriptomic and proteogenomic profiling techniques have been further developed for the classification of gliomas; these could lead to a robust and objective method for the stratification of patients and improve survival prediction, as commented in the previous survival [36].
After a regular medical diagnosis of glioblastoma is achieved via MRI, computed tomography, or biopsy followed by blood analysis, patients await a more personalized and guided treatment [38]. The repertoire of molecular biomarkers characterized by transcriptomics is quite robust. However, only a few are critical for a detailed diagnosis. Further research is needed to classify glioblastomas into subtypes and grades and to estimate survival rates. The most relevant biomarkers described in this section are the mutation of TP53 as the molecular feature of classical glioblastoma, the PDGFRA mutation for the proneural subtype, the presence of GNB2 as an indicator of aggressive tumors, and the cell subpopulation signature as a measure for survival.
4.2 Age- and sex-related patterns in molecular classification
Molecular classification can also reveal an age-related pattern of biomarkers for glioma. This evidence is reflected in a previous computational clustering study that revealed H3F3A, AHNAK2, SOX1, SUSD2, and KMT2C were the most mutated genes in young-age patients, PIK3CA and TERT were the most mutated genes in middle-age patients, and RYR2 was the most mutated gene in old-age patients. Furthermore, two mutations were relevant for young- and middle-age groups: BCORL1 (as an indicator for HGGs) and KMT2D, whereas three mutational events on TERT, PTEN, and NF1 were more frequent in old-age patients [39].
The characterization of these new biomarkers could provide a more refined molecular classification of HGG/LGGs between age groups when added to the IDH1 mutational status and TERT methylating status. However, these data are based on the IV WHO classification of CNS tumors, and thus they should be updated [18].
RNA-sequencing and real-time polymerase chain reaction (RT-qPCR) quantification followed by Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO) analyses revealed a sex-related pattern that emerged from the differential expression of hub genes in the cell cycle, DNA replication, and the Fanconi anemia pathway. These DEGs were mainly enriched in women because the involved pathways mediated progesterone release, which leads to oocyte maturation. Four strongly correlated genes (CCNB1, CDC6, KIF23, and KIF20A) were upregulated in the glioma samples and mediated cell cycle, ATP-binding, and DNA replication. The CCNB1 protein accelerates mitosis and promotes tumoral invasion, thus suggesting a recurrent role in GBM [40]. CDC6 encodes an enzyme that mediates mitosis via E2F regulation [41]. KIF23 is highly expressed in malignant tumors, and KIF20A promotes reverse transport from the Golgi complex to the endoplasmic reticulum and the presentation of the major histocompatibility complex class I (MHC-I), thereby disguising the tumor from immune response and maintaining its proliferation [42, 43]. These last two hub genes might be potential biomarkers for GBM diagnosis, especially in women [44].
These molecular approaches aim to specify a pattern of DEGs between age or sex groups. On the one hand, distinctive DEG clusters between age groups have been observed that could be useful for classifying the tumors of the CNS following the most recent criteria, especially to distinguish the pediatric-type diffuse LGGs and HGGs. On the other hand, an RNA-sequencing study led to the characterization of four hub genes highly related to glioma samples, two of them being possible new biomarkers for GBM diagnosis in women: KIF23 and KIF20A.
4.3 Epigenetic-based characterization of glioblastoma and prognostic value
Survival prediction can be discussed from an epigenetic perspective. The most frequently observed molecular feature is the status of the MGMT promoter, whose methylation level correlates with the tumor's prognosis and is considered a universal marker to evaluate TMZ sensitivity in glioma chemotherapy. In fact, the MGMT promoter methylation level is more significant than grade or 5-hydroxymethylcytosine (5hmC) for age-related prognosis [14, 45, 46].
Conventional chemotherapy with TMZ as adjuvant treatment is an inductor of DNA damage and leads to genetic alterations in the glioma cells, which adapt to the drug dose and develop resistance when the MGMT promoter is hypermethylated [45].
Methylation profiling is another interesting strategy to stratify GBM tumors. DNA methylation-based GBM subtypes seem related to local T-cell infiltration [47]. In fact, these immunological characteristics lead to the classification into four methylation subgroups: IDH, RTK I, RTK II, and mesenchymal tumors.
Interestingly, IDH methylation groups have the lowest CD3+ T-cell infiltration and a low PD-1 expression. Mesenchymal subtype tumors have the highest CD3+/ CD8+ T-cell infiltration. An increased PD-1 expression along with higher levels of CD8+ infiltration results from radiochemotherapy, suggesting that CD8+ T-cells might evolve to an anergic phenotype and activate the immunosuppressive response. Consequently, the mesenchymal subtype might become more aggressive against immune response after conventional therapy. Thus, this information could help identify patients suitable for specific immunotherapy trials [47].
DNA methylation-based diagnosis could support the histological diagnosis of GBM by combining the transcriptomic and methylation patterns of tumor samples and measuring the methylation degree of the CpG islands [45]. Using the MethylMix algorithm, Wang et al. [48] revealed six highly methylated genes (ANKRD10, BMP2, LOXL1, RPL39L, TMEM52, and VILL) that could be used for the molecular subclassification of GBM. The methylation signature is an independent factor that might predict high- and low-risk glioblastomas and overall survival.
Another epigenetic modification contributing to cancer proliferation is the aberrant methylation of histones, a process regulated by histone methyltransferases. An active research field in GBM therapy relies on applying histone deacetylase inhibitors (HDACIs) to improve the patient's OS. An excellent example of the application of HDACIs to treat GBM is a phase II/III trial designed with HDACIs + gene-mediated cytotoxic immunotherapy (G-MCI) or gene-editing treatment mediated by zinc-finger, CAS enzyme, and new-generation sequencing findings [45]. A phase II study tested valproic acid (VPA), an HDAC inhibitor, in newly diagnosed patients showing improved overall survival outcomes and lower toxicity. VPA sensitized glioblastoma cells to radiation in 81% of the patients, thus increasing the effectiveness of standard radiotherapy [49].
It has been observed that receptor tyrosine kinases I (RTK-I) subtype GBM show global hypomethylation. The SOX10 gene, linked to chromatin remodeling and therapy resistance in melanoma, is hypomethylated and overexpressed in RTK-I subtype GBM. Repression of this gene in an in vivo syngeneic graft GBM mouse model resulted in epigenetic alterations, a phenotypic switch to a mesenchymal subtype, and increased tumor cell invasion. In this case, the RTK-I subtype is related to better overall survival than the mesenchymal subtype [50].
Some super-enhancers involved in the regulation of cell identity genes show subtype-specific enrichment. Consequently, the status of the enhancer landscape plays an essential role in determining tumor subtype identity in GBM, and their enrichment could serve as a biomarker for the molecular diagnosis of specific subtypes [50].
Alternative-splicing profiling represents a novel technique for glioblastoma classification. ANXA7, MARK4, MAX, USP5, WWOX, BIN, RON, and CCND1 have been suggested as altered biomarkers serving as functional targets for personalized treatment depending on the heterogeneity of the phenotype and genotype of each patient [51-53]. Moreover, SNRPB, a vital element of the spliceosome complex SmB/B′ implicated in DNA repair and chromatin remodeling, might be a potential target for novel therapies [54]. Additionally, CELF2, a regulator of splicing events, could be a valuable predictor of the prognosis, along with the IDH status and the zinc-finger motif deletions (3′ ZNF domain alterations) [55].
In summary, the MGMT promoter methylation level is an indicator of age-related gliomas, prognosis, and immunoresistance. However, this is not the only biomarker identified by epigenetic changes. Hypermethylation, hypomethylation, and alternative splicing play an essential role in tumor heterogeneity. The latest clinical trials focused on the methylation of histones and the evaluation of CELF2 as a potential predictor of prognosis. Epigenetic changes are a hallmark of cancer, and alternative splicing is one of their most frequent manifestations. These subtle changes lead to a wide heterogeneity of phenotypes, making epigenomic profiling and characterization two essentials for personalized prognosis.
4.4 Noncoding RNA's role in the progression of glioblastoma
Noncoding RNAs (ncRNAs), such as microRNAs (miRNAs) and long ncRNAs (lncRNAs), have interesting regulatory effects on GBM. These nucleic acids can potentially modify the expression levels of proteins involved in the proliferation and migration of tumor cells, like metalloproteinases, cytokines, and growth factors [56]. GBM cells exchange miRNA molecules with oligodendrocytes and endothelial cells within the TME. These molecules can promote angiogenesis and cell differentiation, but some work as tumor suppressors [57].
Identifying miRNA with highly altered expression in glioma provides another method of analyzing patient samples via microarray. The diagnosis can be made by detecting only three miRNAs: miR-4763-3p, miR-1915-3p, and miR-3679-5p. The first and second miRNAs appear oncogenic and are higher in patients with diffuse glioma, while the last might be a tumor suppressor because of its lower levels in patients. Although this could be a promising technique, the current results seem inefficient in discriminating diffuse gliomas from healthy tissue. Nevertheless, these three serum miRNAs represent a powerful tool for GBM diagnosis in combination with histological and molecular characterization [58].
The lncRNA MIR4435-2 Host Gene (MIR4435-2HG) is upregulated in GBM tissues. Besides, higher expression of this lncRNA correlated with shorter OS. MIR4435-2HG targets miR-1224-5p, which inhibits TGFBR2. The inhibition of this receptor results in a diminished cell invasive potential compared to MIR4435-2HG overexpressing U87 cells. This result agrees that MIR4435-2HG knockdown resulted in the inhibition of cell proliferation and increased cell apoptotic rates in U87 and U251 cell lines. Furthermore, this lncRNA can be found in other tumors (e.g., gastric and colorectal cancer), in which its upregulation is also linked to poor [59].
The small nucleolar RNA host gene 12 (SNHG12) is overexpressed in TMZ-resistant GBM samples after TMZ treatment. Hypomethylation of the promoter region of this lncRNA induces transcriptional activation of SNHG12 by the SP1 transcription factor. SNHG12 acts as a molecular sponge for miR-129-5p, increasing the expression of MAPK1 and E2F7, which regulate TMZ-induced cell apoptosis and cell proliferation. Even though tumor heterogeneity implies that each patient presents distinct differentially expressed lncRNAs, ncRNAs are promising biomarkers that could have relevant clinical significance [60].
In brief, noncoding RNAs seem to play a role in tumoral proliferation. lncRNAs exhibit a poor prognosis linked to higher apoptosis rates, whereas miRNAs can be targeted for tumor suppression. MIR4435-2HG and SNHG12 are highly expressed in glioblastomas, increasing the tumoral genotypes and heterogeneity. Targeting these molecules could prevent aggressive or high-grade glioblastomas.
4.5 Computational methods for glioblastoma diagnosis
Deep convolutional radiomics features of diffusion tensor imaging (DTI) [61] and machine-learning assisted dynamic susceptibility contrast-magnetic resonance imaging (DSC-MRI) are novel constructional methods that are worth mentioning. These methodologies provide a better molecular classification of gliomas [62, 63] and subtypes of GBM based on analyzing pathological images and their computational modeling via a deep learning method integrating different biomarkers [37, 64]. Deep learning machine analysis is based on computational artificial intelligence that learns from data samples and builds up neural network models that elucidate the diagnosis, decision-making, and clinical predictions related to GBM therapy [65].
An example of computational modeling is described in a study by Randles et al. [66], where the authors investigated the dynamics of glioblastoma stem cells within the perivascular niche as they designed a computational model on the Vulcan supercomputer, which let them examine different treatments and their outcomes. Each simulation analyzed the spatial distribution and interactions between cells, giving a fitness value to each cell. Following the motion and the spatial landspace of these cells, the supercomputer could determine tumor growth through time. Thus, they concluded that giving chemotherapy with TMZ right before radiotherapy improved survival because of the timing glioblastoma stem cells spread in space. In this manner, the immunoresistant response to TMZ was more effectively blocked.
Computational models can interpret both proteogenomic and metabolomic characterizations of GBM. Wang et al. [67], using computational analysis, revealed how PTPN11 and PLCG1 are signaling hub genes in RTK-altered tumors, how immune cells characterize GBM subtypes, and how histone H2B acetylation is a biomarker for classical glioblastoma. The processing of data collection and interpretation was facilitated by a non-negative matrix factorization for multi-omics subtyping, the use of iPROfun for covariates, and a deep learning histopathology image analysis.
An enormous variety of studies can be approached by computational modeling, a potential tool for long-period analysis and multiple condition evaluations. The computational model is designed in terms of the population census and the experimental conditions. Nevertheless, deep-learning methods make a difference. Advances in processing and refining data compilations might help researchers head in a direction when giving a diagnosis and prognosis to GBM patients.
The list of possible molecular biomarkers for diagnosing and classifying gliomas is endless. The following table (Table 3) collects those molecular targets for diagnosis, prognosis, and the personalized treatment mentioned in the previous sections:
| Application | Function | Molecular biomarkers |
|---|---|---|
| Diagnosis | Confirmatory after previous MRI, CT or biopsy | TERT, EGFR, IDH, P53 |
| Additional to histology | ANKRD10, BMP2, LOXL1, RPL39L, TMEM52, VILL, HDAC | |
| Age-related | ||
| Young group (0-17 years old) | H3F3A, AHNAK2, SOX1, SUSD2, KMT2C | |
| Middle age group (17-64 years old) | PIK3CA, TERT, BCORL1, KMT2D | |
| Old age group (>64 years old) | RYR2, TERT, PTEN, NF1 | |
| Sex-related | ||
| Female patients | KIF23, KIF20A, CCNB1, CDC6 | |
| Prognosis prediction | High risk or malignancy | SNHG12, MIR4435-2HG, SOX10, MGMT, KIF23, GNB2 |
| Shortened OS | CELF2, MIR4435-2HG, cell signature subpopulation | |
| Personalized treatment | ANXA7, MARK4, MAX, USP5, WWOX, BIN, RON, CCND1 |
- Abbreviations: ANKRD10, Ankyrin repeat domain 10; ANXA7, Annexin A7; BCORL1, BCL6 corepressor like 1; BIN, Bridging integrator; BMP2, Bone morphogenetic protein 2; CCNB1, Cyclin B1; CCND1, Cyclin D1; CDC6, Cell division cycle 6; CELF2, CUGBP Elav-like family member 2; CT, Computed tomography; EGFR, Epidermal growth factor receptor; GBM, Glioblastoma multiforme; GNB2, G-protein subunit beta 2; HDAC, Histone deacetylase; IDH, Isocitrate dehydrogenase; KIF20A, Kinesin family member 20A; KIF23, Kinesin family member 23; KMT2C, Lysine methyltransferase 2C; KMT2D, Lysine methyltransferase 2D; LOXL1, Lysyl oxidase like 1; MARK4, Microtubule affinity regulating kinase 4; MAX, MYC associated factor X; MGMT, O-6-Methylguanine-DNA methyltransferase; MIR4435-2HG, MIR4435 host gene 2; MRI, Magnetic resonance imaging; NF1, Neurofibromin 1; OS, Overall survival; P53, Tumor protein p53; PIK3CA, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PTEN, Phosphatase and tensin homolog; RON, Ron receptor tyrosine kinase; RPL39L, Ribosomal protein L39 like; RYR2, Ryanodine receptor 2; SNHG12, Small nucleolar RNA host gene 12; SOX1, SRY-box transcription factor 1; SOX10, SRY-box transcription factor 10; SUSD2, Sushi domain containing 2; TERT, Telomerase reverse transcriptase; TMEM52, Transmembrane protein 52; USP5, Ubiquitin specific peptidase 5; VILL, Villin like; WWOX, WW domain containing oxidoreductase.
5 KEY GBM-RELATED PATHWAYS AND THEIR ROLE IN TUMOR HETEROGENEITY
5.1 GBM-related molecular pathways
Tumor heterogeneity deviates from the “cell niche” regulation and develops from several signaling and immunosuppressive pathways interceptions. Control of the deactivation of cell proliferation, self-renewal, and differentiation of glioma-initiating cells is mediated by the Wnt, Notch, and TGF-β signaling pathways. The mesenchymal subtype is characterized by an overexpression of TGF-β and vascular endothelial growth factor (VEGF) pathways and attenuation to both Wnt and Notch signaling as well as the expression of YKL40, a specific biomarker for this subtype. On the contrary, Notch and Wnt signaling pathways were prominently activated in the proneural subtype [68]. There is differential activation of GICs specific to each GBM subtype. The concurrency of TGF-β signaling and lower activation of both Notch and Wnt signaling pathways suggests that targeting GIC subtypes might improve clinical outcomes.
Apart from these pathways, p53 signaling remains essential for immortality by amplifying murine double minute 2 (MDM2), which binds the TP53 gene and inhibits its regulatory role in mutations. The retinoblastoma protein (Rb) pathway is also crucial for regulating the cell cycle and proliferation. Rb protein inhibits the E2F transcription factor, which stimulates the transcription of genes involved in the progress from the G1 to S phase during mitosis [69]. These two latest pathways control the cell cycle and their targeted interception might mitigate the invasiveness and migration of glioblastoma cells.
There is an opposing interplay between the IDH1 mutation and the Wnt/β-catenin pathway [70]. The Wnt signaling pathway is crucial in cell proliferation, migration, and apoptosis. However, this pathway inhibits glycogen synthase kinase-3β (GSK-3β), an inflammation and cell membrane signaling regulator. IDH1 mutation is related to a better response to cytotoxic therapy and longer survival in GBM patients.
The PI3K/Akt pathway is involved in the phosphorylation of GSK-3β, which leads to the nuclear transport of β-catenin. This transport promotes the activation of STAT3, an oncogenic transcriptional factor involved in GBM growth, stimulation of cyclin D1 and c-Myc (related to angiogenesis and proliferation), and overexpression of MMP-2/9, which induces cell invasion [71, 72]. A clinical trial based on the combined treatment using sulindac + LY294002 [73] aims to inhibit PI3K for the blockade of GBM invasion. Another biological drug inhibiting invasion is celecoxib, a PI3K inhibitor that can also diminish Akt signaling [71]. The concomitant reduction in tumor proliferation is accompanied by increased cell death [72].
The most remarkable epigenetic silencing of the Wnt pathway occurs because of the hypermethylation of soluble frizzled-related protein (FRP) genes. FRPs create a receptor complex that binds to Wnt ligands and consequently activates the AXIN/APC/GSK-3β complex via phosphorylation. This last step promotes the accumulation of β-catenin in the cytosol and leads to the activation of RTKs, therefore, the stimulation of the HIF-1α via the PI3K/Akt pathway. HIF-1α is a hypoxia factor that enhances the Warburg effect by overproducing glycolytic enzymes, such as LDH-A. The final result of FRP silencing is the inhibition of glucose metabolism in the glioma cells [74, 75].
In recent studies, IDH1-R132H mutation was found to be correlated with better prognosis owing to the decreased expression of the Wnt/β-catenin pathway. This result could be explained by the lower intracellular glutathione (GSH) levels due to the reduced availability of NADPH, an essential cofactor in the oxidative carboxylation of α -ketoglutarate, and higher levels of reactive oxygen species, which induce apoptosis and reduce cell proliferation. IDH1 is an independent predictor of improvement in the clinical outcomes of TMZ therapy. As mentioned above, IDH1-mutated tumors correlate with a better prognosis for low-/high-grade gliomas. Consequently, this type of mutation in patients with glioma reduces proliferation and induces apoptosis [76].
The Notch signaling pathway regulates cell migration, differentiation, apoptosis, self-renewal, and homeostasis. This pathway consists of four cytoplasmic receptors (Notch 1-4) and their ligands, Jagged-1, Jagged-2, and DII 1-4. The expression level of Notch 1, predominantly expressed in neurons, astrocytes, precursor/ependymal, and endothelial cells could be related to the GBM survival period. Notch signaling activity might be useful to predict the overall survival and tumor resistance. Results with a novel therapeutic antibody, functionally validated with a computational-guided approach, suggest that Notch signaling via Hes1/Hey1 targeted genes could be a druggable and clinically relevant target in GBM. Brontictuzumab (BRON) is the first humanized anti-Notch 1 blocking antibody directed against cell surfaces to diminish tumor cell invasion [77].
The Hedgehog (Hh) signaling pathway plays a crucial role in embryogenesis and tumorigenesis; furthermore, this pathway plays a pivotal role in tissue repair and regeneration. The terminal effectors of the Hh pathway in glioma are glioma-associated oncogene homolog 1 (GLI1) zinc-finger transcription factors. An alternative-splicing, truncated variant, tGLI1, is expressed in most GBM samples, but it is undetectable in normal brain cells. This tGLI1 is a gain-of-function variant able to activate several genes not regulated by GLI1. The targeted genes upregulated by tGLI1 include VEGFR1, VEGF-A, VEGF-C, TEM7, HPSE, CD24, and CD44, thus promoting glioblastoma cell proliferation, migration, invasion, and angiogenesis [78]. The aberrant role of the Hh pathway leads to the need to understand the impact of GLI variants, potentiating the development of novel therapies that stop metastasis.
In summary, one of the main reasons that glioblastomas are so heterogeneous is that they can modulate the core regulatory signaling pathways involved in immune response, apoptosis, cell growth, proliferation, and migration. The evolution of glioma depends on the upregulation or downregulation of three main pathways: TGF-β, Wnt, and Notch.
5.2 The role of the RTK/PI3K/Akt/mTOR axis in glioblastoma
A relevant part of the research devoted to molecular-targeted therapy of GBM focuses on identifying intrinsic biomarkers in the RTK/PI3K/Akt/mTOR, JAK-STAT3, and RAS/RAF/MEK pathways as well as the p53 and cell cycle regulation pathways. The RTK/PI3K/Akt/mTOR pathway regulates cell growth, metabolism, and survival in gliomas (Figure 1) [46]. mTOR kinase functions in two complexes: as the nutrient sensor of the cell regulating cell growth (mTOR complex 1 + protein RAPTOR) and coordinates the cytoskeleton's organization and Akt activation via phosphorylation (mTOR complex 2). A remarkable distinction between normal and glioma cells is the loss of function of phosphatase and tensin homolog (PTEN). Consequently, the deactivation of PTEN results in increased Akt activity that triggers mTOR activity that enhances cell proliferation [79].
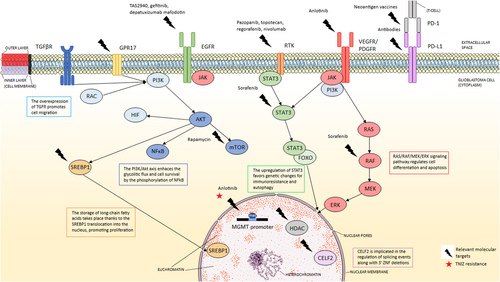
Relevant molecular targets in GBM (indicated with a thunderbolt) and their respective treatments (above the thunderbolt) if applied. The star highlights the resistance to temozolomide therapy present in MGMT+ glioblastoma cells. This figure describes a compendium of the GBM-related signaling pathways (PI3K/AKT/mTOR, JAK-STAT3, and RAS/RAF/MEK), altered gene expression, and characteristic cell surface receptors (EGFR, VEGFR, PDGFR, RTKs, GPR17, TGFβR). The selection of these pathways and the molecular biomarkers compiles the diversity of targeted therapies. This figure was created using Microsoft PowerPoint and Servier Medical Art (https://smart.servier.com) under a Creative Commons Attribution 3.0 Unported License (http://creativecommons.org/licenses/by/3.0/). Author: Elena Verdugo. Abbreviations: AKT, Alpha serine/threonine-protein kinase; CELF2, CUGBP Elav-like family member 2; EGFR, Epidermal growth factor receptor; ERK, Extracellular regulated kinase; ERK1/2, Extracellular signal-regulated kinases 1/2; FOXO, Class O of forkhead box transcription factor; GBM, Glioblastoma multiforme; GPR17, G-protein-coupled receptor 17; HDAC, Histone deacetylase; HIF, Hypoxia inducible factor; JAK, Janus activated kinase; MEK, Mitogen-activated protein kinase kinase; MGMT, O-6-Methylguanine-DNA methyltransferase; mTOR, Mammalian target of rapamycin; mTORC1, Mammalian target of rapamycin complex 1; NF-κB, Nuclear factor kappa-light-chain-enhancer of activated B cells; PD-1, Programmed cell death protein 1; PDGFR, Platelet-derived growth factor receptor; PD-L1, Programmed death ligand-1; PI3K, Phosphatidylinositol 3-kinase; RAC, Subfamily of GTPases; RAF, Rapidly accelerated fibrosarcoma kinase; RAS, Rat sarcoma virus GTPase; RTK, Receptor tyrosine kinase; SREBP-1, Sterol regulatory element-binding protein 1; STAT3; PHIP, Pleckstrin homology domain interacting protein; TAS2940, HER2 and EGFR inhibitor; TGFβR, Transforming growth factor-beta receptor; TMZ, Temozolomide; VEGFR, Vascular endothelial growth factor receptor
On the contrary, RTKs activate PI3K and lead to the activation of Akt depending on the phosphorylation of protein kinases 1 and 2 (PDK1/2). Thus, the hyperactivation of Akt is pertinent in understanding why glioma cells are permanently proliferating, changing their metabolism, and promoting a cancer phenotype. Resistance to TMZ treatment stems from the role of autophagy in glioma cells induced by the inhibition of this last pathway [79].
The relevance of the hyperactivation of this axis relies on the control of glioblastoma cell survival. This survival is characterized by changes in metabolic, cell cycle, and cell growth pathways and is translated into radiotherapy + TMZ chemotherapy resistance.
6 MECHANISMS INVOLVED IN TUMOR HETEROGENEITY
This next section introduces the models available to explain tumor heterogeneity, a hallmark of GBM, which is influenced by epigenetics and metabolism. There are two proposed mechanisms for intratumor heterogeneity in GBMs. First is “the clonal evolution model,” wherein cumulative epigenetic changes in normal cells lead to the genesis of cancer cells, which proliferate and acquire their tumorigenic potential. The second is “the cancer stem cell model,” which suggests that only a portion of cancer cells possess infinite self-renewal potential and can start and maintain tumor development [80].
Even when there is no clear definition for the origin of the tumor, tumor-initiating cells (TICs), a subset of highly tumorigenic glioblastoma stem cells (GBSCs), are highly resistant to conventional therapy because TAMs (30%–40%) and tumor-infiltrating lymphocytes, contributing to the intratumoral vascular density by connecting the neoplastic cells and provide endothelial markers for immunity resistance, such as CD31, CD41, and CD99 [80-82].
The complex structure of the tumor cell niche can be studied via the connections between tunneling nanotubes (TNTs) established for proliferation and long-distance communication. TNTs are long cytoplasmic F-actin extensions of astrocytes and oligodendroglioma cells that may be open-ended or connected by connexins 43 (Cx43). These extensions invade normal tissue cells and mediate the repopulation of the tumor after radiotherapy through the transfer of cellular material from GBSCs to the targeted cells. The exchange of the altered mitochondrial DNA (mt-DNA) is particularly relevant since it affects and modifies metabolism and restores tumor adaptation and resistance, providing the tumorigenic phenotype to sensitive-to-treatment tumor cells [14].
Interestingly, intratumor spatial heterogeneity can be measured by targeting Bruton's tyrosine kinase (BTK), attributed to GBM core cells. BTK has a pivotal role in the maturing process of B cells. BTK profiling is based on RNA-sequencing of four transcriptional factors in its pathway: NFATC3, NF-κB2, BCL6, and NF-κB1, and distinguishes edge from core-cell populations. BTK silencing might improve chemotherapy results by promoting core-cell apoptosis (Figure 2) [83].
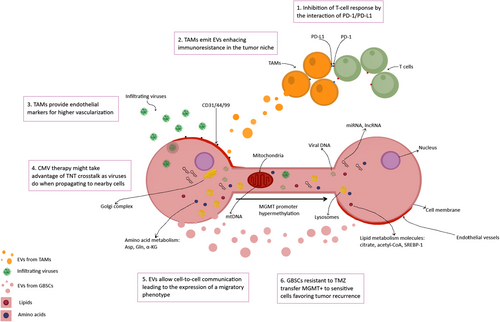
Glioblastoma stem cells (GBSCs) transfer TMZ resistance via tunneling nanotubes (TNTs). The temozolomide (TMZ)-exposed tumoral tissue shows a specific phenotype: MGMT+, CD31/41/99+, and altered miRNA/lncRNA expression. MGMT+ cells transfer this feature to sensitive-to-TMZ GBSCs, which might explain tumor recurrence after conventional therapy. They also provide metabolic precursors, modified mitochondrial DNA, and differentially expressed genes by cell-to-cell communication and extracellular vesicles. This figure was created using Krita (https://krita.org/). Author: Elena Verdugo. Abbreviations: Asp, Aspartate; CD31, Platelet endothelial cell adhesion molecule; CD44, Homing cell adhesion molecule; CD99, Single-chain type-1 glycoprotein; CMV, Cytomegalovirus; DNA, Deoxyribonucleic acid; EVs, Extracellular vesicles; Gln, Glutamine; GSBCs, Glioblastoma stem cells; lncRNA, Long noncoding RNA; MGMT, O-6-Methylguanine-DNA methyltransferase; miRNA, Micro RNA; mtDNA, Mitochondrial DNA; PD-1, Programmed cell death protein 1; PD-L1, Programmed death ligand-1; RNA, Ribonucleic acid; SREBP-1, Sterol regulatory element-binding protein 1; TAMs, Tumor-associated macrophages; TMZ, Temozolomide; TNTs, Tunneling nanotubes; α-KG, Alpha-ketoglutarate
One of the challenges of treating glioblastoma is its heterogeneity, affecting genetic expression, modulation of metabolic pathways, and immune system evasion. Cell-to-cell communication, extracellular vesicles (EVs), and TNTs mediate the transfer of molecular information between radiotherapy-resistant and -sensitive tumor cells, propagating the tumorigenic phenotype from the tissue's core cells to the marginal zones. Therefore, these changes expand throughout healthy tissues around the tumor.
6.1 A closer look at metabolic changes in glioma cell
The conventional classification of LGGs, based on the IDH1/2 mutational status, leads to the metabolic characterization of differentiated astrocytes, one of the cell types that could possibly originate from GICs, as previously described. The traditionally defined IDH-mutated astrocytoma represents the best example of altered metabolism within the tumoral heterogeneity of gliomas [84].
The brain is highly dependent on glucose intake to function correctly. Glioma cells adapt their metabolism according to glucose availability, which gives them extra resistance to hypoxia or altered redox situations. Selective pressure on tumors makes them overexpress glucose transporters, such as GLUT1/3, on their plasmatic membranes, regulated by the hypoxia factor HIF-2 α. Even when glucose levels are low, HIF-1 α guarantees the upregulation of hexokinase 2 (HK2), increasing the glycolytic pathway. Furthermore, many gliomas are characterized by the loss of PTEN function, which causes the constitutive activation of Akt1 and the stabilization of PFKP [85, 86].
MYC is a proto-oncogene that promotes a bidirectional flow in the mitochondrial transport of lactate-pyruvate. Deletion of MPC1 and the accumulation of LDH-A leads to the transformation of pyruvate into lactate, which enhances the Warburg effect [87].
The next modifying step comes with the modulation of the Krebs cycle by extracting C atoms from it (cataplerosis) or introducing C atoms (anaplerosis) to it. These reversible reactions play a crucial role in the de novo biosynthesis of fatty acids, amino acids, and nucleotides. Intermediate metabolites, such as citrate and α-KG, escape oxidation and serve as precursors for fatty-acid biosynthesis and aspartate/glutamate synthesis. Aspartate initiates nucleotide biosynthesis, while glutamate provides the C-skeleton of non-essential amino acids. Hence, α-KG DNA repair and demethylating activities become highly inhibited by the overproduction of 2-HG (sensitizing IDH-mutant cells to PARP-1 inhibition and NAD+ deficiency), which also affects the transamination of this compound into keto acids and glutamate [88]. The marginal extensions of astrocyte-like glioma cells contain high levels of cytosolic citrate, especially in the pseudopodia, where limited access to glucose leads to the uptake of acetate and oxidation, mediated by the ACSS2 enzyme [89].
Lipid metabolism is also altered in GBM. The marked metabolic heterogeneity of GBMs allows the use of this altered lipid metabolism to mark GBM stem and non-stem cells in separate tumor niches [88]. GBMs can use ketone bodies and fatty acids to maintain growth, thus allowing their progression during ketogenic diet therapy [90]. Two essential enzymes mediate the biosynthesis of fatty acids, acetyl-CoA carboxylase (ACC) and fatty-acid synthase (FASN). FASN can be used as a biomarker since it is enriched in GBM-derived EVs [91]. ACC and FASN are regulated by SREBP-1, which responds to the EGFR-PI3K-Akt1 signaling pathway. By increasing the EGFR signaling, SREBP-1 favors the tumor evolution of GBSCs into a proliferative status by synthesizing long-chain ω3/6 polyunsaturated fatty acids. Meanwhile, the marginal and hypoxic regions store fatty acids via FABP3/7, which binds to polyunsaturated fatty acids (PUFAs) in a particular structure, defined as pseudopalisades, a hallmark of GBM [92, 93]. A super-enhancer in GBM and GSCs promotes PUFA synthesis. These PUFA maintain EGFR signaling and membrane organization in GSCs. This observation suggests that dual targeting of EGFR and PUFA metabolism could be a novel potential therapeutic approach for glioblastoma management [94].
Regarding nitrogen metabolism, glioblastoma cells show both altered expression and activity of the amino acid transporter SLC7A11 [95], key enzymes involved in glutamine metabolism (glutamine synthetase and glutaminase) [96, 97] and cysteine metabolism [98]. The resulting balance of nitrogen metabolism gives rise to cataplerosis (a decreased availability of carbon atoms to enter the Krebs cycle oxidative pathway) [84].
Glutamine dependence exhibited by some tumor cells has motivated the development of therapeutic approaches based on the metabolism of this amino acid, and GBM is no exception. A phase I clinical trial combines a glutaminase inhibitor, Telaglenastat (CB-839), with radiotherapy and TMZ chemotherapy in IDH1-mutant astrocytomas. Telaglenastat may stop tumor growth by blocking the enzymes needed for this process (NCT03528642).
Altered metabolism is a consequence of tumor heterogeneity and is favored by the complexity of the tumor niche, which is highly vascularized, with infiltrating M2 lymphocytes and TAMs, and a heterogeneous cell population. This characterization of gliomas is exemplified by the study of the mutation in IDH1/2. Knowledge relative to the altered metabolism shown by glioma cells is important to determine how these metabolic changes can affect the development of the tumor and to find new therapeutic targets. Several key intermediates and enzymes from the four main metabolic pathways previously described are discussed and suggested for glioblastoma targeting and treatment. Targeting PTEN could reduce the glucose intake in glioblastoma cells, while targeting PUFA and EGFR might affect the storage of lipids. Finally, the design of inhibitors in the synthesis mechanism of 2-HG might recover the DNA-repairing system.
7 MOLECULAR FEATURES OF GLIOBLASTOMA INVASION
Aggressive invasion potential is a hallmark feature of all subtypes of GBM and entails a struggle for its treatment. GBM invasion mechanisms are well understood in vitro, but this knowledge has yet to be transferred to new treatments in healthcare [99].
Tumor cell-to-cell crosstalk within the TME via EVs is involved in migratory phenotypes. EVs generated by mesenchymal subtype cells can affect their environment and contribute to the tumor invasion potential [100].
Annexin A2 (ANXA2) is one of the most abundant proteins in glioma EVs [101]. It is an important mediator in the plasminogen activator system, which mediates the conversion of plasminogen to plasmin and is essential for activating metalloproteinases involved in extracellular matrix degradation. However, the role of the transport of ANXA2 through EVs remains unknown [102].
ANXA2 regulates the molecular phenotype and aggressiveness of GBM via the ANXA2-STAT3-OSMR axis, which promotes mesenchymal transition. Consequently, ANXA2 and the ANXA2-STAT-OSMR axis could be attractive targets to manage GBM cells’ aggressiveness and migration [103].
Several genes are involved in GBM cell proliferation and invasion. B4GALT3 expression increases in GBM samples, especially in the proneural subtype, and this high expression predicts poor survival for patients with glioma. B4GALT3 depletion reduces cell viability and invasion of U251 glioblastoma cells, presumably due to the reduced expression of β -catenin, vimentin, and matrix metalloproteinase-2, along with an increased expression of E-cadherin [92].
GBP2 expression is also elevated in GBM samples, particularly in mesenchymal GBM, and this overexpression promotes cell migration and invasion in vitro. Fibronectin (FN1) expression and other genes are induced by GBP2 overexpression in U87 and U251 glioblastoma cell lines. FN1 is an extracellular glycoprotein involved in cell migration, and its depletion avoids GBP2-induced invasiveness in the studied cell lines. STAT3, which contributes to the maintenance of GBM's mesenchymal subtype, is also involved in GBP2-promoted FN1 expression [104].
PHIP is another gene involved in GBM motility through its regulatory activity on the focal adhesion complex. Besides, it also promotes cell invasion in melanoma, which shares its neuroectodermal origin with GBM. PHIP physically interacts with VCL, which is located at the force transductor domain of focal adhesions. PHIP downregulation significantly suppresses the migratory potential of U251 cells, an expected effect considering the role of focal adhesions in cell migration. This gene expression has also been suggested to be a biomarker of glioma progression [105].
Another target implicated in regulating GBM invasion and proliferation is ephrinB2, which tends to have a lower methylation status and, consequently, a higher expression in GBM compared with other gliomas. Paradoxically, this gene can act as an oncogene and a tumor-suppressor gene. EphrinB2 overexpression increases the activation of Eph4 and reduces tumor growth but enhances invasion, while its knockdown has an anti-invasive but proliferative effect. EphrinB2 knockdown followed by administration of ephrinB2-Fc fusion protein results in tumor growth suppression along with an anti-invasive response in U87 ephrinB2 tumor-bearing mice [106].
Glioblastoma cells are forced to overexpress the neuronal glucose transporters GLUT1/3, as described in section 6.1. Libby et al. [107] observed that the overexpression of GLUT3 promotes GBM invasion in vitro. This invasive phenotype is independent of glycolytic metabolism, as the overexpression of GLUT3 did not have notable effects on glycolytic metabolic flux, which could be associated with the invasive phenotype. Interestingly, the substitution of GLUT3 C-terminus with GLUT1 eliminated the pro-invasive phenotype of GBM cells, while on the inverse, the substitution of GLUT1 C-terminus with that of GLUT3 increased invasive potential. Thus, the GLUT3 C-terminus could be a valuable target for inhibiting the invasion potential of GBM and other overexpressing GLUT3 cancers, such as breast, lung, liver, colon, head, and neck cancers.
8 PROGRESS IN GBM TREATMENT
Conventional GBM treatment comprises surgical intervention, which considers the age and medical condition of the patient, followed by radiotherapy and chemotherapy plus adjuvant TMZ. After surgery, the most important postoperative predictor associated with OS is the extent of resection. Recent findings propose hypofractionated radiation for older patients, administered daily in case of focal radiotherapy, and preventing adverse effects as much as possible [108, 109].
Traditional TMZ therapy has two significant issues: high concentrations are toxic to hematopoietic cells, and its administration engenders drug resistance in patients with newly diagnosed or recurrent GBM. Bevacizumab, a humanized anti-VEGF monoclonal antibody and the first antiangiogenic-approved therapy for colon cancer, is another drug whose application for GBM treatment remains uncertain based on the OS outcome [108, 109]. However, bevacizumab therapy for recurrent GBM has been approved, and it attempts to improve prognosis and survival [69, 110]. In fact, Meng et al. [111] observed that bevacizumab targets VEGF-A (a promoter of angiogenesis and vascular permeability) and prevents edemas but does not affect the survival rate of glioblastoma patients [112].
Immunotherapy is gaining relevance, and future clinical trials may orient toward a more personalized diagnosis and treatment. This rising tendency honors the privileged immunoresistance present in CNS cancers. Early studies focused on the design of inhibitors (anti-PD-1, anti-PD-L1, and anti-CTLA-4), among which the combination of anti-CTLA-4 and anti-PD-1 was the only promising and effective therapy, showing a long-term cure rate of 75% in GBM. Unfortunately, there are several immune-resistant mechanisms in GBM, and the effects propagate systematically. The fast expansion and associated infiltrating immune cells quickly eradicate antibody monotherapies by disrupting the antigen flow and immune cell traffic toward the tumor niche. The latest immunotherapy strategies utilize vaccines to enhance T-cell response [112, 113].
An ongoing clinical trial is evaluating immunotherapy in regard to the OS and progression-free survival (PFS) of patients with GBM following intervention with durvalumab, an approved IgG1 for treating metastatic urothelial carcinoma (NCT02336165). The following sections describe the last approaches for GBM treatment [114].
8.1 Immunotherapy methods
Microglia are the principal antigen-presenting cells in the CNS. GBSCs escape the immune system by increasing STAT3 expression, which translates into the upregulation of Wnt and TGF- β pathways (NCT01904123), resulting in the secretion of immunosuppressive factors, such as TGF β-2 and interleukins IL-10. This cell-to-cell mediated release is regulated by activating the cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) in regulatory T cells (Tregs). Additional help comes from TAMs that show high levels of programmed cell death-ligand 1 (PD-L1). PD-L1 binds to receptors on the surface of T cells, restricting their activity [69, 113].
Based on this previously exposed mechanism, the most cited approach in immunotherapy involves blocking TAMs by creating suitable antibodies against CTLA-4 and PD-L1 [35]. The double blockade of CTLA-4 and PD-L1 is another suitable checkpoint for immunotherapy trials (NCT03233152, NCT04145115, NCT04003649). PD-L1 was targeted by a phase I clinical trial that evaluated the effects of nivolumab plus ipilimumab for recurrent GBM. Its purpose was to compare this new treatment's adverse effects and efficacy with bevacizumab monotherapy (NCT02017717) [69]. Another interesting approach is combined therapy comprising PD-1/PD-L1 inhibitors plus radiotherapy or antibodies targeting CTLA-3, TIM-3, LAG-3, IDO, or OX-40 (NCT02658981, NCT04003649) [108, 113]. Antibodies that block the PD-1/PD-L1 interaction or the T-cell response are only effective in some patients. A series of ongoing trials targeting PD-1 aim to restore immunogenicity, especially CD8 and CD4 T-cell responses (NCT02287428, NCT04201873, NCT03422094, NCT03899857).
The most recent interventions are the development of highly personalized cytomegalovirus (CMV) therapy and neoantigen vaccines. CMV protein pp65 causes a robust CD8+ T-cell response that benefits survival, especially for CMV transferred into dendritic cells (NCT03688178). This last intervention serves as an adjuvant for vaccination, the same way CpG oligonucleotides and granulocyte-macrophage colony-stimulating factor (GM-CSF) do. Neoantigen vaccines show potent antitumoral activity by inducing both CD8+ and CD4+ T cell responses. However, before these strategies reach clinical practice, further research and optimization are required [112].
To sum up, most immunotherapy therapies in development to treat GBM aim to avoid the inhibition of T cells, mediated by Tregs and TAMs, using antibodies against CTLA-4 and PD-L1. However, new strategies that could provide personalized treatment with a robust immune response, such as CMV therapies and neoantigen vaccines, constitute a promising research area.
8.1.1 The future of immunotherapy for glioblastoma: neoantigen and nucleic acids vaccines
As previously described, the design of tumor-specific antigens is a promising application for glioblastoma immunotherapy since they are exclusively expressed on tumor cells. The leading two platforms selected for the design of vaccines are neoantigens or tumor-specific antigens and plasmid DNA, while the vehicles used are dendritic cells and heat shock proteins.
Neoantigens are proteins that originate from mutations within tumor cells and differ from cell to cell. This feature makes them excellent targets to address tumor cells selectively. The development of these vaccines requires sequencing data from the whole exome and the RNA of both healthy and tumor cells from the patient. The sequencing aims to find targetable proteins that produce a T-cell response in the organism by the specific recognition and binding to the MHC [115].
There is an ongoing clinical trial (NCT02287428) testing the safety of a neoantigen vaccine against glioblastoma in combination with radiation therapy and pembrolizumab or TMZ. This vaccine works well for newly diagnosed patients with unmethylated MGMT promoters. The patients might be receiving immunoadjuvant poly-ICLC and radiotherapy. These personalized peptide vaccines may be effective for treatment with limited toxicity, modulating the clinical outcomes of glioblastoma patients [116].
The development of nucleic acid-based vaccines is a promising strategy being tested for treating GBM. This strategy involves developing DNA plasmids encoding tumor-specific antigens and cytokines, which promote recognition and the CD8+ T-cell response [115]. DNA-based vaccines are most beneficial among nucleic acid-based vaccines since they enter the nucleus and allow the presentation of antigens to the MHC.
There is an active phase I/II clinical trial (NCT03491683) evaluating the safety, immunogenicity, and preliminary efficacy of two DNA-based vaccines: INO-5401, which is a combination of three DNA plasmids targeting Wilms tumor gene-1 (WT1) antigen, prostate-specific membrane antigen (PSMA) and human telomerase reverse transcriptase (hTERT) genes; and INO-9012, a DNA plasmid for the expression of human interleukin-12 (IL-12). Both treatments are administered in combination with cemiplimab, radiation therapy, and TMZ. This is the first study in human GBM to combine DNA plasmids with PD-1 blockade. The primary outcomes of this trial are related to measuring the percentage of participants with adverse events, which resulted in the usual spectrum for PD-1 inhibitory agents and the vaccine itself not having significant adverse events either. Secondarily, the preliminary 12-month overall survival was 84.4%, and the activation of INO-5401-specific CD8+ T-cells was successful. The current outcomes are promising even though there are no conclusions drawn yet [117].
Another phase I clinical trial (NCT04573140) is recruiting patients for the evaluation of the manufacturing feasibility, safety, and maximum-tolerated dose of an RNA-lipid particle (RNA-LP) vaccine in adult glioblastoma patients [115].
The design of personalized peptide or DNA-based vaccines is currently being tested in clinical trials. Their objective is to improve the outcomes by answering in a more precise mechanism within the patient. This is possible thanks to the immune response behind the treatment. In most cases, vaccines might be a safer and more effective way to increase overall survival with low adverse events or cytotoxicity.
Table 4 summarizes the ongoing and finished clinical trials under the label of “immunotherapies.”
| Clinical Trials ID or NCT number | Condition of glioblastoma | Experimental treatment | Control or comparator treatment | Study phase | Patients, n | Primary outcomes measures | Targets |
|---|---|---|---|---|---|---|---|
| CMV therapy and CAR T cells | |||||||
| NCT03688178 | Multiforme | Human CMV pp65-LAMP mRNA-pulsed autologous DCs + TMZ + Varlilumab | - | Phase II | 112 | OS, Safety | LAMP, CD27 |
| NCT05063682 | Malignant, multiforme | EGFRvIII-CAR T cells | - | Phase I | 10 | AEs, OS | CD3ζ-stimulatory/41BB co-stimulatory T-cell antigen receptor |
| NCT04003649 | Recurrent, refractory | IL13Rα2-CAR T cells + Ipilimumab + Nivolumab | - | Phase I | 60 | AE, DLT, feasibility, OS | CTLA-4, PD-1 or PD-L1 |
| Neoantigen and DNA-plasmid vaccines | |||||||
| NCT03491683 | Multiforme | INO-5401 + INO-9012 + Cemiplimab + RT + TMZ | - | Phase I/II | 52 | AEs | WT1, PSMA, hTERT genes +IL-12 |
| NCT04573140 | Adult, MGMT | mRNA and pp65 fl LAMP/DOTAP liposome vaccine | - | Phase I | 28 | Feasibility, safety, MTD, DLT | LAMP |
| NCT02287428 | Multiforme | RT + Personalized NeoAntigen Vaccine + Pembrolizumab + TMZ | - | Phase I | 56 | AEs, feasibility | PD-1 |
| NCT04201873 | Recurrent | Pembrolizumab + ATL-DC vaccine | Placebo | Phase I | 40 | Cell cycle-related signature, expansion of TCR clones, AEs | PD-1 |
| NCT03422094 | Newly diagnosed, unmethylated | NeoVax + Nivolumab + Ipilimumab | - | Phase I | 3 | Safety, tolerability, feasibility | PD-1 |
| NCT02149225 | Multiforme | APVAC1 vaccine + Poly-ICLC + GM-CSF | - | Phase I | 16 | SAEs, AEs, frequency of CD8 T cells | CD8 T cell response |
| NCT04280848 | Multiforme | UCPVax | - | Phase I/II | 28 | Immunogenicity | TH1 CD4 T-cell response, TERT |
| NCT04015700 | Newly diagnosed, Unmethylated | Vaccine (GNOS-PV01 + INO-9012) | - | Phase I | 12 | Safety, tolerability (DLTs), feasibility | IL-2, CD8 T cell response |
| NCT03899857 | Newly Diagnosed | Pembrolizumab + ATL-DC + poly ICLC | - | Phase II | 56 | OS | PD-1 or PD-L1/2 |
| Antibody therapy | |||||||
| NCT04547777 | Recurrent, Malignant | D2C7-IT + 2121-V11 | - | Phase I | 30 | DLT | CD40 |
| NCT02017717 | Recurrent | Nivolumab + Ipilimumab | Bevacizumab | Phase III | 530 | AEs, OS | CTLA-4, angiogenesis |
| NCT02658981 | Multiforme | Anti-LAG-3 monoclonal antibody BMS 986016 | - | Phase I | 63 | MTD, DLT | PD-1, LAG-3, CD137 |
| NCT03707457 | Multiforme | Anti-GITR Monoclonal Antibody MK-4166 + IDO1 inhibitor INCB024360 + Nivolumab + Ipilimumab | - | Phase I | 3 | DLT | IDO1, GITR |
| NCT02336165†* | Multiforme | Durvalumab alone or Durvalumab + SR or Bevacizumab | - | Phase II | 159 | OS, PFS6 | PD-L1 |
| NCT03233152 | Recurrent | Ipilimumab + Nivolumab | - | Phase I | 6 | PFS, OS | CTLA-4 + PD-1 |
| NCT04145115 | Recurrent, Secondary | Ipilimumab + Nivolumab | - | Phase II | 37 | OS | CTLA-4 + PD-1 |
| NCT04440943 | Multiforme | CDX-527 | - | Phase I | 40 | Safety and tolerability | PD-L1xCD27 |
- Abbreviations: AEs, Adverse events; ATL-DC, Autologous tumor lysate-dendritic cell; CAR, Chimeric antigen receptor; CD4, cluster of differentiation 4; CD8, cluster of differentiation 8; CD27, Tumor necrosis factor receptor superfamily member 7; CD40, Tumor necrosis factor receptor superfamily member 5; CD137, Tumor necrosis factor receptor superfamily member 9; CMV, cytomegalovirus; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; DCs, Dendritic cells; DLT, Dose-limiting toxicity; DNA, Deoxyribonucleic acid; DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane; EGFRvIII, epidermal growth factor receptor variant III; GITR, glucocorticoid-induced TNFR-related protein; GM-CSF, Granulocyte macrophage colony-stimulating factor; hTERT, Human telomerase reverse transcriptase; IDO1, Indoleamine 2,3-dioxygenase 1; IL-2, Interleukin 2; IL-12, Interleukin 12; IL13Rα2, Interleukin 13 receptor alpha 2 chain; LAG-3: Lymphocyte-activation gene 3; LAMP, Lysosomal-associated membrane protein; MTD, Maximum tolerated dose; OS, Overall survival; PD-1, Programmed cell death protein 1; PD-L1/2, Programmed death-ligand 1/2; PFS, Progression-free survival; PFSn, Progression-free survival at “n” months; pp65 fl LAMP, pp65 full length lysosomal associated membrane protein; pp65-LAMP, pp65-lysosomal-associated membrane protein; PSMA: prostate-specific membrane antigen; RT, Radiation therapy; SAEs, Severe adverse events; SR, Standard radiotherapy; TCR, T cell receptor; TERT, Telomerase reverse transcriptase; Th1, T helper 1; TMZ, Temozolomide; WT1, Wilms tumor gene-1.
8.2 Molecular-targeted therapy
8.2.1 Targeting the PI3K/Akt/mTOR axis
An approximation to molecular-targeted therapy comes from the novel strategies for identifying biomarkers in the RTK/PI3K/Akt/mTOR signaling pathway. One way to forestall the recurrence of GBM is through developing RTK-targeting drugs, such as imatinib, approved by the Food and Drug Administration (FDA) for chronic myeloid leukemia. Imatinib is directly related to PDGFR inhibitors. The latest phase II studies evaluating the effectiveness of combined imatinib and hydroxyurea showed little improvement in patients with recurrent GBMs. Nevertheless, hydroxyurea is being administered because it sensitizes GBM to TMZ treatment [108]. Recent trials on dasatinib monotherapy and dasatinib plus lomustine therapy showed no considerable effectiveness for recurrent GBM [79]. Besides, enasidenib is used to treat acute myeloid leukemia and is being evaluated in IDH2-mutated tumors, such as gliomas (NCT02273739). Other metabolic-related clinical trials are being conducted, designing IDH- and PARP-specific inhibitors (NCT03224104, NCT04740190, NCT03914742).
The next possible molecule that intercepts the pathway is EGFR. Gefitinib and erlotinib are efficient first-generation EGFR reversible inhibitors that impede the binding of ATP to the EGFR tyrosine kinase domain receptors. In fact, a clinical trial has assessed the administration of gefitinib and radiotherapy and its effectiveness in inhibiting cell growth (NCT00052208). Afatinib, a second-generation EGFR inhibitor, binds irreversibly to Cys 773 EGFR residues and Cys 805 HER2 residues. However, these EGFR inhibitors have only shown some efficacy in in vitro assays since they cannot efficiently cross the blood-brain barrier (BBB) [79].
A phase II clinical trial demonstrated that direct treatment with rindopepimut was ineffective against EGFRvIII expressing GBM [69]. In addition, sorafenib therapy against tumor growth and proliferation is being tested for GBM. This small molecule can bind to multiple tyrosine kinases (RAF, VEGFR, PDGFR, and c-KIT) and inhibit them. Unfortunately, a phase III clinical trial on sorafenib also failed [110]. The expectancy of these two trials improved the overall survival and prognosis for patients with severe or metastatic tumors. One ongoing trial is testing TAS2940 safety in candidates who are not approved for currently available therapies, targeting HER-2 and VEGFR in solid tumor cancers (NCT04982926). This trial aims to predict lower adverse events and improved overall survival.
Another important drug is the main mTORC1 inhibitor, rapamycin, which affects kinase conformation. Its monotherapy remains insufficient for recurrent GBM but shows more significant activity in combination with other analogs [79].
Clinical approaches to the PI3K/Akt/mTOR axis illustrate how targeted molecular therapy can improve prognosis in patients with aggressive tumors, such as glioblastoma. Multiple tyrosine kinases can be targeted and inhibited by a biological drug, for example, RTK, EGFR, RAF, or PDGFR. Despite the positive outcomes for imatinib, enasidenib, and sorafenib in other tumors, they remain ineffective for glioblastoma treatment. This recalls for an update on the drugs administered and their respective targets.
8.2.2 Targeting angiogenesis and cell growth factors: antibodies take action
There are many possible ways of intercepting or impairing tumor growth in GBM. A promising study assessed the efficacy of G-protein-coupled receptor 17 (GPR17) agonist GA-T0, which crosses the BBB and promotes GBM cell death in murine models through modulation of the MAPK/ERK, STAT, PI3K/Akt, and NF-κB pathways. This work showed that GPR17 expression in GBM was related to improved overall survival and could be used as a predictive biomarker [118].
Biological drugs or antibody therapies are potent treatments for many types of tumors. However, they have provided little efficacy in GBM so far. Cetuximab (anti-EGFR), panitumumab, and nimotuzumab are examples of three antibodies designed to target EGFRvIII. The first one, cetuximab, has a five-month limited effect (intravenous treatment, phase II study) in reducing EGFR mutations, tumor survival, and proliferation. Some small-molecule inhibitors are non-specific compounds that target different biomarkers of tumor-related signaling pathways, for example, lenvatinib, dovitinib, and brivanib [79]. Tyrosine kinases are other antiangiogenic targets in the VEGF signaling pathway for patients with newly diagnosed GBM, characterized by the higher levels of HIF-1α and SDF-1, which are responsible for microvascularization. The SDF-1 pathway induces the recruitment of endothelial progenitor cells from the bone marrow to the tumor niche, where SDF-1 interacts with CXCR4 receptors on the surface of endothelial matured cells. A way to prevent vasculogenesis would be the design of a new biomolecule that targets CXCR4 or SDF-1. For example, CXCR4 antagonist AMD3100 treatment could prevent tumor vasculogenesis and growth [110].
Anlotinib is a multi-target tyrosine kinase inhibitor that blocks the migration and proliferation of endothelial cells and can inhibit tumor angiogenesis and cell growth by targeting specific growth factor receptors, such as VEGFR and PDGFR. Currently, there are clinical trials testing the viability of this treatment against GBM (NCT04725214, NCT05033587, NCT04547855, NCT04959500).
Depatuxizumab mafodotin is an antibody-drug conjugate against activated EGFR, which has not shown positive results in a recent trial (NCT02343406). However, this treatment did show interesting results that suggest that a subgroup of patients could benefit from this therapy. Specific molecular predictors of treatment efficacy, like EGFR SNVs, could help determine which patients would benefit from this conjugate.
Pazopanib is an FDA-approved oral drug for metastatic-advanced kidney cancer and angiosarcoma. Pazopanib has proven to be an effective therapy when combined with prior exposure to bevacizumab (NCT01931098). Another trial (NCT04704154) is investigating the safety and tumoral response of nivolumab plus regorafenib combo for solid tumors treatment, GBM among them.
Both monoclonal antibodies and small-molecule inhibitors are under research to develop molecular-targeted therapies focused on angiogenesis to treat glioblastoma. EGFRvIII, CXCR4, SDF-1 and VEGFR are among the analyzed targets involved in angiogenesis. Nonetheless, even though ongoing clinical trials are testing these strategies, there have not been conclusive results yet.
8.2.3 Nanoparticles, staying one step ahead of antibody therapy
Alternatively, small interfering RNA (siRNA) nanoparticles designed for gene silencing might be more effective than antibodies. A recent study focused on the comparison of antibody therapy vs. siRNA treatment. The effectiveness of siRNA treatment against cancer invasion and progression was approximately 40%–65%, while cetuximab or trastuzumab (anti-HER2) therapies showed no reduction in invasiveness. The main reason behind siRNA treatment's efficacy relies on the nature of the materials used for the nanoparticles’ design, such as liposomal carriers, which can transfer siRNA across the BBB [110]. Nanodelivery eliminates the main barrier against pharmacokinetics, the crossing of the BBB, being a suitable mechanism for effective and individualized therapy. This new field is further explored in section 9.4.
8.2.4 Targeting tumor heterogeneity
GICs appear to be resistant to conventional therapy and responsible for the reappearance of the tumor after surgery resection [109]. Consequently, GBM resistance depends on tumoral heterogeneity and the phenotype of GBSCs. The disruption of key enzymes in the pyrimidine synthesis, such as CAD or DHODH, intercepts GBM resistance. Leflunomide and teriflunomide are two effective inhibitors of the enzyme DHODH, which has a crucial role in stem-like phenotype maintenance in GBSCs. Despite the significant prognosis of these treatments, GBSCs can still reprogram their pyrimidine metabolism and develop resistance [120].
8.2.5 Pediatric healthcare: ongoing therapies for pediatric gliomas
A phase IV clinical trial (NCT03975829) is assessing the long-term effects in pediatric patients diagnosed with astrocytoma, oligodendroglioma, neurofibromatosis type 1, and other gliomas. The trial consists of the administration of dabrafenib plus trametinib. The goal of these two inhibitors is to block MEK1/2 (trametinib) and BRAF kinase (dabrafenib), two proteins related to the activation of the RAS/RAF/MEK/ERK signaling pathway. This pathway mediates cell growth and is usually upregulated in tumors, meaning that its inhibition could prevent tumor expansion. This combo has improved the overall survival of metastatic melanoma [121]. The same primary outcomes are expected for glioblastoma treatment. Children diagnosed with high-grade gliomas may be treated with depatuxizumab mafodotin, designed to be combined with TMZ or lomustine (NCT02343406), or alternatively, repotrectinib (NCT04094610). The primary outcomes of both studies are low toxicity and enhanced overall survival [122]. Moreover, a tentative FDA approval calls for palbociclib isethionate (NCT02255461), which has been tested for young patients with recurrent, progressive, or refractory CNS tumors. This oral drug is a CDK4/6 inhibitor, thereby promoting cell cycle arrest in the G1 phase and decreasing tumor proliferation. Its safety has been tested in pediatric patients with low-/high-grade glioma, showing positive results. This trial aims to study the side effects of palbociclib isethionate to determine the maximum tolerated dose and the plasma pharmacokinetics. There is a favorable background for this clinical intervention regarding the safety of palbociclib administered to children and adolescents for other brain tumors [123].
To sum up, these therapeutic trials being tested on children and adolescents aim to design a safer treatment with positive outcomes and less toxicity for the administered drugs.
Table 5 depicts information on the ongoing and finished clinical trials under the label of “molecular-targeted therapies”:
| Clinical Trials ID or NCT number | Condition of glioblastoma | Experimental treatment | Control or comparator treatment | Study phase | Patients, n | Primary outcomes measures | Targets |
|---|---|---|---|---|---|---|---|
| Targeting metabolism | |||||||
| NCT02273739†* | Glioma | Enasidenib (AG-221) | - | Phase II | 21 | AEs, MTD, RP2D | IDH2 |
| NCT03224104 | Multiforme | TG02 + RT + TMZ | - | Phase I | 71 | MTD, PFS6 | IDH1-R132H |
| NCT04740190 | Recurrent | Talazoparib + Carboplatin | - | Phase II | 33 | PFS6 | PARP1/2 |
| NCT03914742 | IDH1/2, Recurrent | BGB-290 + TMZ | - | Phase I/II | 100 | MTD, AE, TRR | PARP1/2 |
| Targeting angiogenesis and cell growth | |||||||
| NCT01931098†* | Recurrent | Pazopanib + Topotecan | Bevacizumab | Phase II | 35 | PFS6, PFS3 | DNA topoisomerase type I, RTKs |
| NCT04704154 | Multiforme | Regorafenib + Nivolumab | - | Phase II | 200 | ORR | RTKs |
| NCT04982926 | Multiforme | TAS2940 | - | Phase I | 42 | MTD, ORR | ERBB proteins (HER2 and EGFR) |
| NCT04547855 | Multiforme | Anlotinib + dose-dense TMZ | - | Phase II | 54 | PFS6, ORR6 | Cell growth factors and VEGFR |
| NCT04959500 | Newly diagnosed | Anlotinib Hydrochloride + RT + TMZ | Placebo + RT + TMZ | Phase II | 150 | PFS | Cell growth factors and VEGFR |
| NCT04725214 | MGMT-Unmethylated | Anlotinib | - | Phase II | 33 | OS | Cell growth factors and VEGFR |
| NCT05033587 | MGMT-Unmethylated | Anlotinib + AK105 + RT | - | Phase II | 28 | PFS | Cell growth factors, PD-1 |
| NCT00052208† | Adult giant cell, multiforme | Gefitinib + RT | - | Phase I/II | 158 | MTD, toxicity, OS | EGFR-TK |
| NCT04424966 | Multiforme | Infigratinib | - | Early Phase I | 20 | [Infigratinib] in tumor tissue, plasma and CSF | FGFR |
| NCT01904123 | Recurrent | STAT3 inhibitor WP1066 | - | Phase I | 8 | MTD, DLTs, AEs | JAK2 |
| Pediatric gliomas | |||||||
| NCT04094610 | Pediatric and young adult, multiforme | Repotrectinib | - | Phase I/II | 75 | DLT, RP2D, ORR | ALK, ROS1 or NTRK1-3 |
| NCT02343406†* | Recurrent, Pediatric | Depatuxizumab mafodotin | TMZ or Lomustine | Phase II | 266 | Cmax, PFS, OS, AUC and others | EGFR |
| NCT02255461†* | Recurrent, Childhood | Palbociclib isethionate | - | Phase I | 35 | MTD, AEs | Cell cycle (CDK4/6, cyclin D1-3 and Ink4a-ARF) |
| NCT03975829 | Multiforme, Pediatric | Dabrafenib + Trametinib | - | Phase IV | 250 | SAEs | MEK, MAPK/ERK and BRAF kinase |
- Abbreviations: AEs, Adverse Events; ALK, Anaplastic lymphoma kinase; AUC, Area Under the Curve; BRAF, B-Raf Proto-Oncogene; CDK4/6, Cyclin-dependent kinase 4/6; Cmax, Maximum concentration; CSF, Cerebrospinal fluid; DLT, Dose-Limiting Toxicity; EGFR, Epidermal growth factor receptor; EGFR-TK, Epidermal growth factor receptor tyrosine kinase; FGFR, Fibroblast growth factor receptor; HER2, Human epidermal growth factor receptor 2; IDH-1/2, Isocitrate dehydrogenase 1/2; Ink4a-ARF, Cyclin-Dependent Kinase 4 Inhibitor A-alternate reading frame tumor suppressor; JAK2, Janus kinase 2; MAPK/ERK, Mitogen-Activated Protein Kinase 1/Extracellular Signal-Regulated Kinase; MEK, Mitogen-Activated Protein Kinase Kinase 1; mRNA, Messenger ribonucleic acid; MTD, Maximum Tolerated Dose; NTRK1-3, Neurotrophic Receptor Tyrosine Kinase 1-3; ORR, Objective Response Rate; ORRn, Objective Response Rate at “n” months; OS, Overall Survival; PARP1/2, Poly ADP ribose polymerase 1/2; PD-1, Programmed cell death protein 1; PFS, Progression-Free Survival; PFSn, Progression-Free Survival at “n” months; ROS1, ROS Proto-Oncogene 1; RP2D, Recommended Phase 2 Dose; RT, Radiation therapy; RTK, Receptor tyrosine kinase; TRR, Tumor Radiographic Response; SAEs, Severe Adverse Events; STAT3, Signal transducer and activator of transcription 3; TMZ, temozolomide; VEGFR, Vascular endothelial growth factor receptor.
9 CURRENT AND FUTURE TRENDS IN GLIOBLASTOMA DIAGNOSIS AND THERAPY
Despite the recent findings on the molecular biology of gliomas and the testing of several biomarkers, their predictive value and efficacy need to be validated and approved. Figure 3 summarizes the main expected trends in glioblastoma therapy. The conventional treatment for glioblastoma remains insufficient to cover the whole spectrum of cases. Due to GBM heterogeneity, patients should be treated on a case-by-case basis. This way, patients can obtain a more personalized diagnosis, guiding neurologists into the best ways to diminish tumoral progression and provide the proper treatment.
Relevant future trends in glioblastoma diagnosis and therapy and key concepts related to them. This figure was created using Dia Diagram Editor (http://dia-installer.de/). Author: Iker Puerto San Román. Abbreviations: CTCs, Circulating tumor cells; CTLA-4; Cytotoxic T-lymphocyte-associated protein 4; GBM, Glioblastoma multiforme; miRNA, Micro RNA; PD-L1, Programmed death ligand-1; siRNA, Small interfering RNA
The following section describes the present trends being explored in the latest ongoing trials and could open the door to promising therapies in the future.
9.1 Immunotherapy and molecular-targeted therapies
Immunotherapy and molecular-targeted therapies are two major trends in the treatment of glioblastoma. Immunotherapy has not provided positive results thus far due to the immunoresistance of CNS tumors. Even so, the development of new approaches, such as tumor-specific neoantigen-based and nucleic acid-based vaccines, could be a step toward personalized GBM treatment. In fact, an increase in the number of clinical trials based on nucleic acid-based vaccines may be expected thanks to the remarkable advances in this strategy achieved during the last years. Implementing multiple-antigen targeting could improve vaccine therapy by increasing OS and PFS, two benchmarks currently failing in recent trials. Most commonly found molecular biomarkers might be targeted simultaneously: EGFR, IL-10, CD27, SOX40 and WT1, combined with tumor-specific antigen vaccines, thus providing a more individualized treatment [112, 115]. The challenges of a multifaceted approach remain overwhelming, but the potential benefits that may result from them are substantial.
9.2 Circulating biomarkers for GBM diagnosis and prognosis
Circulating tumor cells, EVs, and circulating nucleic acids and proteins are candidate biomarkers for glioma diagnosis, although none of them have been approved for clinical practice thus far [31]. Circulating tumor nucleic acids are a source of comprehensive information relative to glioblastoma cells' genome, while circulating miRNAs can be used to find potential targets related to tumorigenesis and proliferation and to elucidate the grade and response to glioma chemotherapy [124, 125].
As previously mentioned, liquid biopsy can be a helpful tool for diagnosing and developing personalized treatments. Besides being the brain immersed in CSF, this body fluid could be used as a source of tumor metabolites and other biomarkers [126]. This routinely applied clinical intervention could amplify the chances of identifying specific targets with minimal sample invasion. However, further studies with larger cohorts and the standardization of detection approaches are needed to enhance the potential of liquid biopsy applied to GBM [127].
9.3 Mathematical models and IA applications on diagnosis
Mathematical models may provide useful information about GBM cell invasion dynamics of prognostic value, which could facilitate the prediction of tumor progression [128]. MRIs contain structural and functional information for diagnosis and can be analyzed by machine-learning algorithms.
Artificial intelligence applied to MRI data could be relevant in the future for diagnosing and prognosticating patients with GBM. However, as gliomas are relatively rare, it is challenging to generate a large amount of clinical data necessary to develop these tools [129]. Nonetheless, certain models and algorithms have already reached diagnostic accuracies higher than 80% [130]. These approaches could also become an excellent non-invasive method for identifying GBM subtypes [131].
9.4 Nanodelivery for GBM treatment
Many pharmacological therapies are inefficient due to the hydrophilic nature of the compound and its low capacity to go through the BBB. Nanodelivery is designed to optimize drug pharmacokinetics across the BBB. The design of nanoparticles (NPs) includes proper factors to assimilate the therapeutic agent and enhance its outcome. Hence, a wide range of nanodelivery systems is being investigated in interventional trials. CMV therapy might be tested for nanodelivery, although there are uncertain values for toxicity and immunogenicity that should be solved [109, 132].
A current trial of siRNA NPs describes an example of liposomal carriers [119]. Another interesting application of nanodelivery comes with autophagy modulators found in natural products, such as resveratrol, curcumin, capsaicin, and others. The limited availability of these phenolic compounds relies on their low solubility. This can be overcome by administering NPs that form complexes and interactions to enhance their solubility and potential efficacy [133].
The promising outcomes from nanodelivery approaches may enhance specific targeting, the reversion of drug resistance, a reduction in the adverse effects, and a more prolonged circulation time [134]. Nevertheless, nanodelivery is still a pilot-stage field that should be further researched and documented before approval. The lack of treatments based on nanodelivery relies on the extended testing they require, the lack of standardization of the nanotoxicological assays, and the manufacturing costs of the techniques [135].
10 CONCLUSION
This review reflects the efforts made to elucidate the molecular biology and genetics underlying the development and complexity of GBM. Although a large body of knowledge related to the molecular basis of this disease has been attained, this knowledge has not resulted in effective remedies for patients who suffer from this unstoppable disease.
To achieve effective therapies against GBM, several of its hallmarks must be overcome, such as metabolic heterogeneity, tumor invasion potential, drug resistance, poor pharmacokinetics, and immunoresistance. As long as these issues remain resolved, efficacious treatment for GBM cannot be guaranteed.
However, the development of different approaches and strategies to address this disease, such as the future trends discussed herein, guarantees promising advances in the understanding and treatment of this CNS tumor.
AUTHOR CONTRIBUTIONS
Conceptualization, EV, IP and MAM; bibliographic investigation, EV, and IP; writing—original draft preparation, EV, and IP; writing—review and editing, MAM; drawing of figures, EV, and IP; supervision, MAM; funding acquisition, MAM. All authors have read and agreed to the published version of the manuscript.
ACKNOWLEDGMENTS
Not applicable.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
The experimental work carried out by our group and this review are supported by grants PID2019-105010RB-I00 (Spanish Ministry of Science, Innovation and Universities), and UMA18-FEDERJA-220 (Andalusian Government and FEDER) and funds from group BIO 267 (Andalusian Government). The “CIBER de Enfermedades Raras” (Spanish Biomedical Research Network Center for Rare Diseases) is an initiative from the ISCIII (Spain). The funders had no role in the study design, data collection and analysis, publication decision, or manuscript preparation.
Open Research
DATA AVAILABILITY STATEMENT
Not applicable.




