Reversing epigenetic repression of transposable elements for improving tumor immunogenicity
List of abbreviations
-
- KDM5B
-
- lysine demethylase 5B
-
- SETDB1
-
- SET domain bifurcated histone lysine methyltransferase 1
-
- TE
-
- transposable element
-
- MHCI
-
- major histocompatibility complex class I
-
- IFN
-
- interferon
-
- dsRNA
-
- double-stranded RNA
-
- dsDNA
-
- double-stranded DNA
-
- ISG
-
- interferon-stimulated gene
-
- MDA5
-
- melanoma differentiation-associated gene 5
-
- cGAS
-
- cyclic GMP- AMP synthase
-
- STING
-
- stimulator of interferon genes
-
- ER
-
- endoplasmic reticulum
In a recent study published in Nature, Zhang et al. [1] revealed a new epigenetic mechanism for restraining tumor intrinsic immunogenicity: Lysine demethylase 5B (KDM5B) recruits SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) to repress the transcription of transposable elements (TEs) by H3K9me3 modification, while TE de-repression by targeting KDM5B consequently activated nucleic acid sensing pathways and antigen presentation to elicit tumor intrinsic immunity which enhanced host anti-tumor immune response.
Immune evasion is an important strategy for tumor survival and progression. Cancer immunotherapies have been developed to block immune escape by manipulating the host immune system to recognize and eventually eliminate tumor cells, which have shown outstanding efficacy in multiple cancers [2]. However, a majority of patients fail to respond to immunotherapies, largely due to tumor evasion through tumor-intrinsic resistance or immune exhaustion [3]. One of the efficient strategies to overcome immunotherapy resistance is to unleash tumor-intrinsic immunity, mainly including: (1) upregulating tumor antigen presentation by major histocompatibility complex class I (MHCI) pathway; (2) inducing secretion of immune-stimulating cytokines such as interferons (IFNs), chemokines and activation of their related signaling pathways in tumor cells. TEs, existing ubiquitously in eukaryotic genomes, have been known as the latent intrinsic agonist to IFN response mediated by viral mimicry mechanism in tumor. After activation, TE transcripts can form double-stranded RNA (dsRNA) and reverse-transcribed double-stranded DNA (dsDNA) to induce IFN production by nucleic acid-sensing pathways in tumor cells and then activate downstream IFN signaling to induce the expression of interferon-stimulated genes (ISGs), which boosts tumor-intrinsic immunogenicity to induce an anti-tumor immune response and synergize with immunotherapy [4].
Epigenetic dysregulation is one representative feature of tumorigenesis. Tumor epigenome generally shows a global DNA hypomethylation with focal DNA methylation at the CpG-rich site and dysregulated post-translational histone modifications. Epigenetic alterations in tumor cells have a genome-wide influence on gene expression, including tumor suppressors and other pro-oncogenic genes [5]. Furthermore, new types of epigenetic repression have been being revealed grandually [9]. Growing interests have focused on the epigenetic repression of TE in tumor cell biology. Many studies reported the important role of epigenetic factors in regulating TE expression in tumors, e.g. removing active epigenetic markers such as H3K4me2 represses transcription of TEs to avoid activation of the dsRNA sensing pathway [6]. Chromatin modifications, such as DNA methylation and H3K9 methylation, in TE regions show strong association with TE activity. We highlight this study for revealing a new epigenetic mechanism in tumor TE repression by KDM5B-SETDB1 interaction.
Zhang et al. [1] found that KDM5B expression was higher in melanoma patients who showed poor response to immune checkpoint blockade therapy, and KDM5B loss could improve anti-tumor adaptive immunity, thereby limiting the growth of melanoma xenografts. KDM5B has been traditionally thought to be transcriptional repressors depending on its function in erasing H3K4 methylation. Chromatin modifiers can mediate epigenetic regulation in a catalytic activity-independent strategy [7]. Similarly, KDM5B-mediated regulation of TE transcription is independent of its demethylase activity: KDM5B depletion upregulates TE expression and elicits type I IFN response without increasing H3K4me3 levels at these loci while robustly decreasing H3K9me3 levels. SETDB1, a well-known H3K9 methyltransferase, associates and colocalizes with KDM5B in TEs and writes H3K9me3 there. Activation of innate immune response downstream TE activation by KDM5B loss is through both the melanoma differentiation-associated gene 5 (MDA5) RNA-sensing and cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) DNA-sensing pathways, inducing type I IFN expression and activating IFN response to improve tumor intrinsic immunogenicity. KDM5B loss-induced TE activation may also lead to the appearance of TE-encoded peptides that can be presented by MHC-I molecules; thus, activating antigen-specific cytotoxic T cells (Figure 1). Furthermore, even targeting the same modifier showed various downstream effects of TE activation. In early 2021, another group reported that SETDB1 loss triggers TE-specific cytotoxic T cell response by de-repressing the presentation of TE-encoded retroviral antigen, which hardly depends on type I IFN response [8]. These differences suggest the diversity in both TE clusters and activation landscapes among a series of tumors.
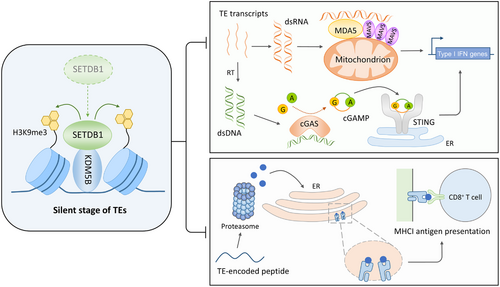
Epigenetic therapeutics focus on inhibiting enzymatic activities of specific regulators. Zhang's work brings a new perspective that epigenetic regulators impact TE expression in a catalytic activity-independent manner, which adds a new layer to the development of epigenetic treatments. The finding of KDM5B-SETDB1 co-regulation model also opens up questions: (1) The sequence-specific binding mechanism of KDM5B is not fully understood; (2) Different chromatin marks may be differently enriched in the same or different TE clusters for regulating their activation in different tumor types. How to fine-tune the cross-talk among different modifications? Detailed understanding of the precise epigenetic regulation in TE loci in a wide variety of tumor types is strongly needed to uncover the deeper mechanisms of tumor intrinsic immune resistance. In addition, how to utilize this mechanistic pathway to pharmaceutically change the “cold” tumor to “hot” tumor for improving the efficacy of cell or antibody-mediated cancer immunotherapy needs further investigations.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported by the grant from the National Natural Science Foundation of China (81922032 and 81788101).
AUTHORS' CONTRIBUTIONS
X.W. and Q.Z. drafted the manuscript and figures. X.C. supervised and revised the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
Not applicable.




